Abstract
Recent studies indicate that steroid receptor-mediated transcriptional initiation is a cyclical process involving multiple rounds of coactivator assembly and disassembly. Steroid receptor coactivator 3 (SRC-3) coactivator phosphorylation has been shown to regulate coactivator complex assembly, but the mechanisms by which coactivator disassembly is triggered are not well understood. In this study, we provide in vitro and in vivo evidence that members of the SRC coactivator family serve as substrates for the enzymatic coactivator coactivator-associated arginine methyltransferase 1 (CARM1). Methylation of SRC-3 was localized to an arginine in its CARM1 binding region and correlated with decreased estrogen receptor alpha-mediated transcription, as seen with both cell-based and in vitro transcription assays. Consistent with this finding, we demonstrated that methylation promotes dissociation of the SRC-3/CARM1 coactivator complex. Methylation of SRC-3 is regulated by estrogen signaling in MCF7 cells and serves as a molecular switch for disassembly of the SRC-3 transcriptional coactivator complex. We propose that CARM1 is a dual-function coactivator, as it not only activates transcription by modifying core histone tails but also terminates hormone signaling by disassembly of the coactivator complex.
The estrogen signaling pathway is pivotal for maintaining female reproductive function and contributes extensively to breast tumorigenesis (10, 18). Estrogen exerts its effect mainly through binding to its cognate receptors, estrogen receptor (ER) alpha and beta, resulting in recruitment of coactivators and transcriptional activation of estrogen-dependent genes (26, 27, 35). Over the past 10 years, many transcriptional coactivators have been identified and studied. Steroid receptor coactivator 1 (SRC-1) was the first bona fide steroid receptor coactivator cloned as an interacting partner for the progesterone receptor (29). The subsequent identification and characterization of SRC-2 (GRIP1, TIF2) (16, 36) and SRC-3 (p/CIP, RAC3, ACTR, AIB1, and TRAM-1) (1, 6, 23, 32, 34) established the SRC/p160 family of coactivators. The critical role of coactivators in estrogen signaling was substantiated by the observed partial hormone resistance in SRC-1 null mice (41). The SRC-3/AIB1 gene and its transcripts was reported to be amplified and overexpressed in 10% and 64% of all primary breast cancers, respectively (1). Subsequent studies verified SRC-3 as an authentic oncogene (19, 35).
Considerable experimental data have established that agonist-bound steroid receptors directly recruit the SRC/p160 family coactivators, which subsequently recruit secondary coactivators, including the E1A binding protein p300 and its homolog, the cyclic AMP-response element binding protein (CREB)-binding protein (CBP), as well as the coactivator-associated arginine methyltransferase 1 (CARM1) (5). Many of these coactivators contain intrinsic enzymatic activities. For instance, p300/CBP contain potent histone acetyltransferase (HAT) activity (28), whereas SRC-1 and SRC-3/ACTR exhibit relatively weak HAT activity (6, 31). CARM1 and PRMT1 (5, 38) have histone methyltransferase activity, and SWI/SNF complexes possess ATP-dependent nucleosome remodeling activity (3, 17). It has been generally accepted that following recruitment to the promoter, these coactivators can modify local chromatin structure and increase DNA accessibility to the basal transcription machinery (26).
Recent studies indicated that coactivator recruitment to estrogen receptor-bound promoters is a cyclical and ordered process involving multiple rounds of coactivator assembly and disassembly (27, 30). SRC-3 coactivator phosphorylation has been shown to affect coactivator complex assembly (40). However, the mechanisms by which coactivator complex disassembly is regulated have not been well established. In this study, we provide in vitro and in vivo evidence that SRC-3/AIB1 is methylated by CARM1 during estrogen signaling and that this modification promotes dissociation of the SRC-3/CARM1 coactivator complex, thereby attenuating the transcriptional response and completing a dynamic equilibrium of receptor-mediated coactivator assembly and disassembly at the promoter.
MATERIALS AND METHODS
Plasmids.
The construction of glutathione S-transferase (GST)-SRC-3 fragments with five distinctive functional domains has been described previously (40). All other GST-fused SRC-3 deletion and point mutants, including GST-H, H1, H2, H3, H (R1171A), H (R1177A), H (R1188A), H (R1207A), H (R1225A), H (R1227A), H (R1171/1177A), and H (R1235/1238A) were generated by a double-PCR strategy. Amplified fragments were inserted into the XbaI and XhoI sites of pGEX-KG vector. pSG5 vector with an N-terminal Flag tag was used to construct mammalian expression vectors for wild-type and mutated SRC-3. The SRC-3 cDNA was transferred from pCMV-TAG2-SRC-3 to pSG5-Flag vector through NotI and BglII sites. Flag-tagged SRC-3 methylation mutants (R1171A, R1177A, R1171/1177A, R1225A, and ΔMe [Δ1171-1238]) were created by use of the double-PCR strategy. Sequencing confirmed all mutations. pSG5-HA-CARM1 was kindly provided by Michael R. Stallcup (University of Southern California). The ERE-Luc reporter has been previously described (44).
Recombinant proteins and siRNA.
Recombinant ERα and PR-A proteins were generously provided by Dean Edwards (Baylor College of Medicine). Recombinant SRC-1 protein was a gift from Steven Nordeen (University of Colorado). SRC-2 and SRC-3 recombinant proteins were generated by the Bac-to-Bac baculoviral system (Invitrogen). SRC-2 and SRC-3 were doubly tagged, with an N-terminal His tag and a C-terminal Flag tag, to ensure production of full-length SRC proteins. After 3 rounds of amplification, the baculoviruses were used to infect log-phase Sf9 cells maintained in Grace's medium containing 10% fetal calf serum (FCS) for 48 h. SRC-2 and SRC-3 proteins were purified with a Ni-nitrilotriacetic acid column (QIAGEN) followed by purification using anti-Flag-M2 affinity gel (Sigma). Wild-type CARM1 protein was purchased from Upstate, and CARM1 (E267Q) protein with an N-terminal Flag tag was expressed by using the Bac-to-Bac system and purified as described above. All GST fusion proteins were expressed in BL21 cells (Stratagene) and purified using glutathione Sepharose according to the manufacturer's protocol (Amersham Bioscience). p300 baculovirus was obtained from W. Lee Kraus (Cornell University) and purified with a Ni-nitrilotriacetic acid column (QIAGEN). HeLa core histones were prepared as previously described (38). Small interfering RNA (siRNA) of CARM1 used in the knockdown experiments was SMART pool siRNA from Dharmacon. CARM1 siRNA was used at 20 nM in each sample.
Cell culture, transfection, and luciferase assay.
CARM1 wild-type and knockout mouse embryonic fibroblasts (MEFs) were kindly provided by Mark Bedford (M.D. Anderson Cancer Center) (43). MCF7, CV-1, SRC-3−/− MEFs, and HEK293T cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% FCS. For estradiol-induced experiments, cells were maintained in phenol red-free medium containing 5% charcoal-dextran-stripped FCS until hormone addition. Fugene 6 transfection reagent (Roche) was used for all of the transient transfections. Transfected cells were treated with 10 nM estradiol 24 h after transfection and harvested after another 24-h incubation. Luciferase activity was determined with the Promega luciferase assay kit according to the manufacturer's protocols. Three independent experiments were done, and the most representative results were shown.
Methylation assays.
Methylation assays were performed as previously described (11). In brief, 0.5 μg recombinant SRC family proteins or other proteins (see Fig. 1) were incubated with recombinant CARM1 (0.2 μg for each reaction) in a reaction mixture containing 20 mM Tris-HCl (pH 8.0), 4 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, and 1 μl [3H]AdoMet (13.3 Ci/mM; Perkin Elmer) for 1 h at 30°C. Reactions were stopped by the addition of 6× sodium dodecyl sulfate (SDS) loading buffer, and proteins were separated in a 4 to 15% SDS-polyacrylamide gel electrophoresis (PAGE) gel. Following staining with Coomassie blue, gels were treated with autoradiography Amplify reagent (Amersham Biosciences) for 20 min, dried, and exposed to X-ray films. For peptide competition assay, different concentrations of unmodified or methylated peptide were added to reactions containing Flag-tagged SRC-3 (see Fig. 5).
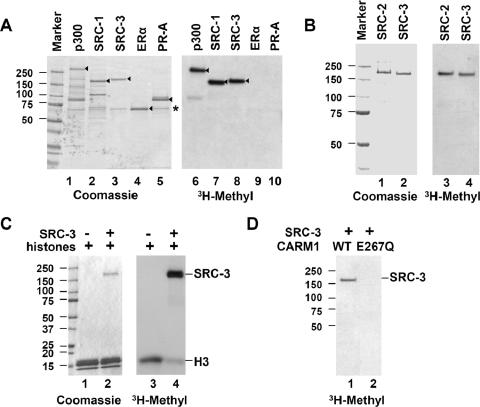
FIG. 1.
SRC family proteins are methylated by CARM1 in vitro. (A) Recombinant p300, SRC-1, SRC-3, ERα, and PR-A were incubated with recombinant CARM1 in the presence of [3H]AdoMet for 1 h at 30°C. Products were analyzed by Coomassie blue staining and fluorography. Arrowheads indicate the positions of full-length proteins. (B) SRC-2 is methylated by CARM1 in vitro. Recombinant SRC-2 and SRC-3 (0.5 μg) were methylated by recombinant CARM1. (C) SRC-3 is a preferred substrate for CARM1 in vitro. Four micrograms of HeLa core histone protein was incubated in the absence (−) or presence (+) of 0.2 μg of SRC-3 protein in the methylation assay. (D) CARM1 is responsible for methylation of SRC-3 in vitro. SRC-3 protein (0.5 μg) was incubated with either wild-type CARM1 or an inactive mutant (E267Q). The positions of protein size standards (in kilodaltons) are indicated beside each panel.
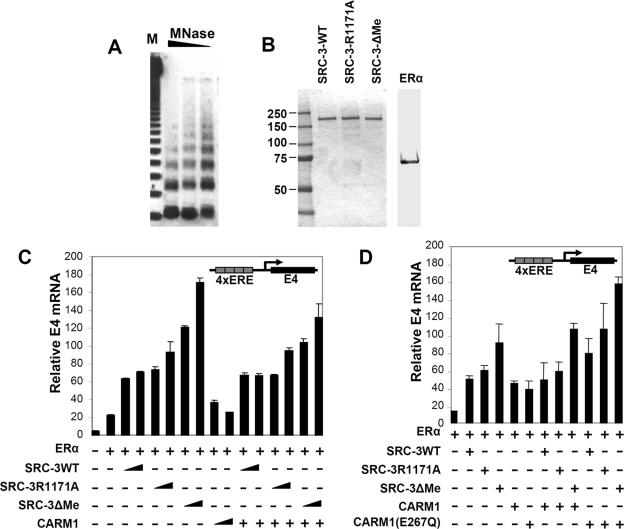
FIG. 5.
Mutation of SRC-3 methylation sites increases its coactivator function in cell-free transcription assays. (A) Analysis of in vitro-assembled chromatin by partial digestion with MNase. Twenty-five microliters of assembled chromatin was digested with 3 U, 1 U, and 0.3 U of MNase (lanes from left to right, respectively). Ladders of nucleosomal bands were visualized by ethidium bromide staining in a 1% agarose gel. (B) Recombinant proteins used in the in vitro transcription assay. Purity was determined by Coomassie blue staining. (C) The schematic in the upper right corner shows the plasmid pERE-E4 that was assembled into chromatin and transcribed. Triangles indicate 20 or 60 ng of SRC-3 protein and 50 or 150 ng of CARM1 protein employed in the different transcription reactions for which results are shown in the bottom panel. The relative amount of E4 transcript was determined by real-time RT-PCR. Error bars shown throughout Fig. 5 represent standard deviations. (D) A similar experiment was performed as for panel C, except that 20 ng of SRC-3 protein and 50 ng of wild-type or mutated CARM1 was added to the mixture individually or sequentially. +, present; −, absent.
Immunoprecipitation and immunoblotting.
Forty-eight hours after transfection, cells were harvested and washed with ice-cold phosphate-buffered saline before being disrupted with lysis buffer (50 mM Tris, 100 mM NaCl, 0.5% NP-40, 50 mM NaF, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 0.5 μg of leupeptin/ml, and 0.7 μg of pepstatin/ml). After incubation for 30 min at 4°C, the cell debris was removed by centrifugation. About 0.8 mg of protein extracts was incubated with 5 μl of anti-Flag-M2 affinity gel (Sigma) at 4°C for 4 h. For detection of methylated SRC-3 protein in cells, MEFs or MCF7 cell extracts were immunoprecipitated with either an antibody against mono- or dimethylated arginine (Novus) or a polyclonal antibody against SRC-3 (39) for 4 h, followed by the addition of 10 μl of protein A/G slurry (Santa Cruz Biotechnology) for 1 h. After three washes with lysis buffer, the immunoprecipitated proteins were analyzed by Western blotting. Other antibodies used in Western blotting included anti-CARM1 (Bethyl Labs), anti-p300 (Bethyl Labs), anti-Flag (Affinity BioReagents), anti-β-actin (Sigma), and anti-ERα (Santa Cruz Biotechnology).
Chromatin assembly and analysis.
Chromatin was assembled using the pERE-E4 plasmid DNA template (a gift from W. Lee Kraus, Cornell University), HeLa core histones, an ATP-regenerating system, and S190 extract derived form postblastoderm Drosophila melanogaster embryos (0 to 6 h), as previously described (25). Untreated rabbit reticulocyte lysate (Promega) was added into the assembly reaction mixture to potentiate the transcription activity of ERα (33). Assembly reaction mixtures (100 μl) were incubated at 27°C for 4 h before further experiments, such as chromatin structure analysis or in vitro transcription, were performed. The quality of assembled chromatin was analyzed by partial digestion with micrococcal nuclease (MNase). After MNase digestion for 10 min at room temperature, assembled chromatin was treated with RNase A and proteinase K, followed by phenol-chloroform extraction. Precipitated DNA was separated by agarose gel electrophoresis, and ladders of nucleosomal bands were visualized by ethidium bromide staining.
In vitro transcription.
Each transcription reaction mixture consisted of 10 μl of assembled chromatin, 3 ng of ERα, 100 nM of estradiol, 20 to 60 ng of SRC-3 proteins, and 50 to 150 ng of CARM1, if applicable. After incubation with coactivators for 5 min at room temperature, HeLa nuclear extract (50 μg per reaction) was added, followed by a 20-min incubation to form transcriptional initiation complexes. Transcription was initiated by the addition of recombinant nucleoside triphosphates (0.625 mM final) in a 40-μl final reaction mixture. Templates were transcribed at 30°C for 45 min, and the synthesized RNA was extracted using Tri-reagent (Molecular Research Center) according to the manufacturer's protocol. DNA-free reagent (Ambion) was used to remove any residual DNA in the RNA preparation before further quantitative analysis of E4 gene transcripts by real-time reverse transcription (RT)-PCR. The sense primer of the E4 gene for quantitative PCR was 5′-CGCTGTGGAAGCGCTGTAT-3′, and the antisense primer was 5′-AAAAACCCTCCTGCCTAGGC-3′. The sequence of the probe was 5′-6-carboxyfluorescein-TTGTTCTGGAGCGGGAGGGTGCT-6-carboxy-tetramethyl rhodamine-3′. Quantitative PCR was performed using the TaqMan one-step RT-PCR master mix and processed on an ABI 7500 real-time PCR system (Applied Biosystems). Each sample was duplicated, and three independent experiments were done to show the most representative results. As a control, reactions lacking RT failed to give signal above the background (data not shown).
RESULTS
SRC/p160 family proteins are substrates for CARM1 in vitro.
It is well known that the SRC family of coactivators recruits secondary coactivators, such as CARM1 and p300/CBP, to target gene promoters in response to estrogen signaling (26). CARM1 contains intrinsic methyltransferase activity (5), and p300/CBP has intrinsic histone acetyltransferase activity (28). Since CARM1, p300/CBP, and SRC-3 are physically associated with each other (5, 34) and, moreover, p300/CBP has been reported to be methylated by CARM1 (8, 21, 42, 43), we asked whether SRC-3 could serve as a substrate for CARM1. Purified recombinant proteins p300, SRC-1, SRC-3, ERα, and PR-A were incubated with recombinant CARM1 protein in the presence of [3H]AdoMet (S-adenosyl-l-[methyl-3H]methionine), and the products were separated by SDS-PAGE and analyzed by autofluorography. In agreement with previous reports, p300 was methylated by CARM1 (Fig. 1A). Both SRC-1 and SRC-3 also were methylated by CARM1, and the methylation efficiency appeared comparable to that of p300 (Fig. 1A, compare lanes 6, 7, and 8). The other SRC family member, SRC-2 could also be methylated by CARM1 in vitro (Fig. 1B). Neither ERα nor PR-A could be methylated by CARM1 in this assay, indicating that the SRC family proteins and p300 are relatively specific substrates for CARM1.
Since histone H3 is a well-characterized CARM1 substrate and methylation of histone H3 contributes to CARM1's coactivator function (5), we compared CARM1's substrate preference for core histones versus SRC-3. The addition of equal amounts of purified SRC-3 recombinant protein dramatically decreased histone H3 methylation by CARM1 in vitro (Fig. 1C), indicating that SRC-3 is a preferred substrate for CARM1 in comparison to histone H3.
To confirm that CARM1, and not some other copurifying methylase, is solely responsible for methylation of the SRC proteins in our in vitro assays, we generated a methylation-defective CARM1 protein by mutating its substrate binding site (E267Q) and produced and purified it from a baculovirus expression system. This mutation has been reported previously to completely abolish CARM1 methylase activity (22). As expected, the mutant could not methylate SRC-3 (Fig. 1D), indicating that CARM1 is a bona fide methyltransferase for SRC family proteins in vitro.
SRC-3 is a substrate for CARM1 in vivo.
Having demonstrated that SRC-3 could be methylated by CARM1 in vitro, we next asked whether this modification occurred in vivo. MEFs derived from wild-type and CARM1 knockout mice (43) were used for detection of endogenous SRC-3 methylation. An antibody that specifically recognizes mono- or dimethylated arginine was used to immunoprecipitate methylated proteins from cellular extracts derived from the wild-type and CARM1−/− MEFs, and the presence of SRC-3 or p300 was detected by Western blotting. Methylated SRC-3 and methylated p300 were observed following immunoprecipitation from wild-type cells but not from the CARM1 knockout cells (Fig. 2A). SRC-3 methylation also was detected in a reciprocal immunoprecipitation experiment in which an SRC-3 antibody was used for immunoprecipitation (Fig. 2B). Taken together, these data demonstrate that SRC-3 is a substrate for CARM1 in vivo. Interestingly, methylated SRC-3 was not detected in the MCF7 breast cancer cell line, despite the fact that SRC-3 protein level is much higher in these cells (Fig. 2B, compare lanes 1 and 2), suggesting that the methylation of SRC-3 by CARM1 might be under tight regulation in MCF7 cells.
FIG. 2.
SRC-3 is methylated by CARM1 in vivo. (A) Cell extracts of CARM1 wild-type and knockout MEFs were immunoprecipitated with an antibody against mono- or dimethylated arginine (α-mArg), followed by Western blotting with specific antibodies against p300 or SRC-3 (upper panel). Input lanes (lower panel) represent 5% of the total amount of SRC-3 used for immunoprecipitation (IP). (B) Cell extracts of MCF7 or CARM1 wild-type and knockout MEFs were immunoprecipitated with antibody against SRC-3 (α-SRC-3), followed by Western blotting with an antibody against mono- or dimethylated arginine. The amount of immunoprecipitated SRC-3 is also shown (upper panel). The lower panel shows the amount of CARM1 protein in 5% of the immunoprecipitation input from the three different cell line extracts. (C) SRC-3 methylation is induced by estrogen signaling. MCF7 cells were cultured in medium containing 5% charcoal-dextran-stripped FCS for 3 days before the addition of estradiol (10 nM), followed by different incubation times prior to harvesting. Cell lysates were immunoprecipitated with an antibody against methylated arginine, followed by Western blotting (IB) with antibody against SRC-3. The amount of ERα, CARM1, and SRC-3 in different samples was determined by Western blotting. In all panels, β-actin serves as a loading control. (D) CARM1 is responsible for SRC-3 methylation in response to estrogen signaling. The same experiment was done as for panel C except that the MCF7 cells were transfected with 20 nM scramble siRNA or siRNA against CARM1 for 3 days prior to hormone induction.
MCF7 is an ER-positive breast cancer cell line, and estrogen signaling is important for its growth. Since the level of SRC-3 methylation appeared to be very low in MCF7 cells, we asked whether this modification could be regulated by the estrogen signaling pathway. To this end, MCF7 cells were grown in charcoal-stripped media and treated with 10 mM estradiol (E2) for various times prior to harvesting. The methylation of SRC-3 was examined by immunoprecipitation-Western blotting, as above. Without hormone stimulation, the SRC-3 methylation level was low (Fig. 2C, lane 1), but it increased after incubation with E2 for 45 min. Methylation levels peaked at 60 min and then decreased (Fig. 2C, compare lanes 4, 5, and 6). This result strongly suggested that methylation of SRC-3 is regulated by estrogen signaling and that it is cyclic.
We next wanted to see if CARM1 is responsible for E2-induced SRC-3 methylation in vivo. We used siRNA to knockdown CARM1 in MCF7 cells and examined E2-induced SRC-3 methylation by immunoprecipitation-Western analysis. siRNA against CARM1 efficiently reduced the CARM1 protein level in MCF7 cells (Fig. 2D). Consequently, SRC-3 methylation in response to E2 treatment was significantly reduced (Fig. 2D). The results confirmed that CARM1 is responsible for E2-induced SRC-3 methylation in MCF7 cells and that it likely acts as the only arginine methyltransferase for SRC-3 in vivo.
Identification of a key SRC-3 methylation site in its Q-rich region.
Since there is no consensus CARM1 substrate sequence, we initially identified the region of SRC-3 that was methylated by deletion mapping. GST fusion proteins containing five different functional domains of SRC-3 were purified and subjected to methylation by CARM1 in the in vitro methylation assay (Fig. 3A). Strong methylation was observed on the C-terminal domain of SRC-3, comprising residues 1081 to 1417 (Fig. 3A, lane 5) and indicating that the majority of CARM1 methylation occurs in this region. After additional deletion mapping (Fig. 3B), a small fragment containing 74 amino acids in the Q-rich domain of SRC-3 appeared to be the major target for CARM1 (Fig. 3B, lane 3).
FIG. 3.
Identification of SRC-3 methylation sites. (A) Mapping regions on SRC-3 that are methylated by CARM1. Five fragments of SRC-3 representing different functional domains were generated as GST fusion proteins. The amount of each protein (1 μg) used in the methylation assay was determined by Coomassie blue staining (middle panel). Arrowheads indicate the positions of full-length proteins. The incorporation of [3H]methyl into the proteins is shown on the bottom panel. (B) Deletion mapping further defines the region of SRC-3 that is methylated. The C-terminal region of SRC-3 from 1041 to 1417 was further divided into three fragments and fused with GST. These protein fragments contain 5, 8, and 3 arginine residues, respectively (upper panel). Full-length proteins are indicated by arrowheads, and the incorporation of [3H]methyl into the proteins is shown on the bottom panel. (C) Primary sequence alignment of the methylation region of SRC-3 compared to the other SRC family proteins. Highly conserved regions are shadowed. (D) Site-directed point mutagenesis (arginine-to-alanine) of the SRC-3 methylation region further refines which arginine residues serve as methylation acceptor sites. Eight methylation mutants were generated, and their activities were compared with the wild-type protein in the CARM1 methylation assay. Arrowheads indicate the positions of full-length proteins. (E) Confirmation of methylation sites in the content of full-length SRC-3 protein. Wild-type and methylation mutants of SRC-3 were expressed and purified from a baculoviral system and tested in the CARM1 methylation assay. The amount of protein (0.3 μg) used in each assay was determined by Coomassie blue staining.
SRC family proteins share 36% identity over the 74 residues identified as the CARM1 methylation region of SRC-3 (Fig. 3C). Two regions containing the major amino acid identities harbor four of the eight conserved arginine residues (R1171, R1177, R1225, and R1227) found in the SRC protein family. Since all three SRC family proteins can be methylated by CARM1 in vitro, we reasoned that, within these two small conserved regions, these arginines are likely to be the targets of methylation. We mutated every one of the potential eight arginine acceptors to an alanine and added these mutants to our in vitro methylation assay (Fig. 3D). From this analysis, it appeared that R1171, R1177, and R1225 were potential methylation sites (Fig. 3D, lanes 2, 5, and 7). The double mutation R1171/1177A did not show a further decrease in methyl incorporation (Fig. 3D, lane 9), suggesting that one of these two sites could function as a regulatory residue rather than a substrate methylation site.
To confirm the identity of the methylation sites in the context of the full-length protein, we expressed full-length SRC-3 proteins bearing these corresponding mutations in a baculoviral system and purified them to homogeneity (Fig. 3E). The in vitro methylation assay revealed that all proteins containing the R1171A mutation exhibited decreased methyl incorporation, further confirming that R1171 was a major substrate site for CARM1 modification (Fig. 3E, lanes 2, 4, and 5). Similar to the GST fusion mutants, the R1177 mutant showed compromised methylation compared with the wild-type protein, but methyl incorporation in the R1171/1177A double mutant was not further decreased compared to the R1171A single mutation (Fig. 3E, compare lanes 2 and 4), providing additional evidence that R1177 may play only a regulatory role. Although the R1225A mutant displayed reduced methylation in the context of the truncated SRC-3 GST fusion, the R1225/1227A double mutant of the full-length protein incorporated the same level of methylation as the wild-type protein (Fig. 3E, compare lanes 1 and 6). The data suggest that R1225 is not a preferred methylation site for CARM1. Similarly, a 68-amino-acid deletion mutant (ΔMe/Δ residues 1171 to 1238) eliminating all 8 arginines was generated and tested in the methylation assay. No detectable methylation was observed for the deletion mutant (Fig. 3E, lane 7), indicating that this region either contains all major methylation sites or, less likely, is required for methylation on another region of in SRC-3. In summary, we mapped the methylation of SRC-3 by CARM1 to a 68-amino-acid Q-rich region of SRC-3, and within this region, we identified R1171 as the major methylation site.
SRC-3 proteins defective for methylation have increased coactivator activity in vivo.
Having demonstrated that SRC-3 can be methylated by CARM1 in vitro and in vivo, we next asked whether this modification has any effect on SRC-3 coactivator function. We compared the SRC-3 methylation mutants R1171A and ΔMe with wild-type SRC-3 for their ability to facilitate transcriptional activation by ERα in a transient-transfection assay in CV-1 cells. Although SRC-3 methylation mutants showed coactivator activity comparable to that of wild-type SRC-3 in the absence of CARM1, they exhibited 1.5-fold-higher activity when CARM1 was coexpressed (Fig. 4A). We reasoned that the relatively mild effect seen with the methylation mutants could be due to endogenous wild-type SRC-3 masking, at least to some extent, the effect of the SRC-3 methylation mutants. Therefore, we performed the same experiment in SRC3−/− MEFs instead of CV-1 cells. As shown in Fig. 4B, the SRC-3 methylation mutants, in the presence of CARM1, exhibited about threefold-higher activity than wild-type SRC-3 in these cells. Our results suggest that methylation of SRC-3 by CARM1 antagonizes SRC-3 coactivator activity in ER-mediated transcriptional activation.
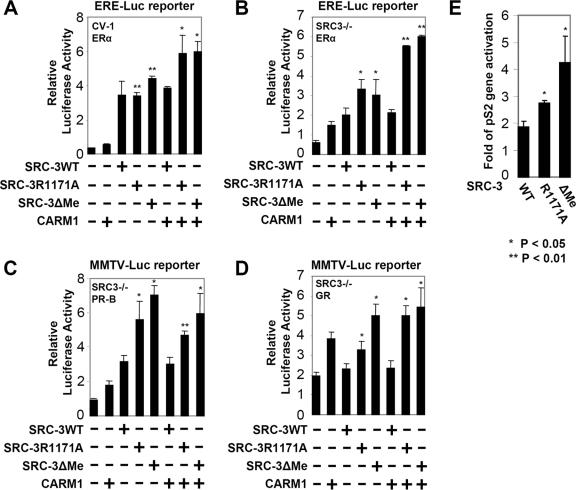
FIG. 4.
Mutation of SRC-3 methylation sites increases its coactivator function in vivo. (A) SRC-3 methylation site mutants, when coexpressed with CARM1, display higher coactivation activity for ERα-mediated transcription. CV-1 cells were transiently transfected with 200 ng of ERE-Luc reporter, 6 ng of pCR3.1-ERα, 100 ng of pSG5-HA-CARM1, and 100 ng of pSG5-Flag-SRC3 wild type (WT) or indicated mutant in each well of a 12-well plate. Ten nanomolar estradiol (E2) was added 24 h after transfection, and luciferase activity was measured at 48 h posttransfection. Luciferase activity was normalized for protein content. t test was performed, and the statistical significance compared with wild-type SRC-3 (n = 4) was shown as follows: *, P value of <0.05; **, P value of <0.01. (B) A similar experiment was performed as for panel A, except that SRC-3−/− MEFs were used. (C and D) The same experiments were carried out as for panel B, except that a mouse mammary tumor virus-luciferase reporter was used, and PR-B or GR was cotransfected instead of ERα. (E) SRC-3 methylation mutants display higher coactivation activity on an endogenous promoter. HEK293T cells were transiently transfected with 100 ng of pCR3.1-ERα and 400 ng of pSG5-Flag-SRC-3 wild type or indicated mutants. Forty-eight hours after transfection, cells were treated with 10 nM E2 overnight to induce endogenous gene transcription. The amount of pS2 gene expression was determined by real-time RT-PCR, and the relative induction is shown. The expression of cyclophilin was used for normalization. +, present; −, absent.
We next investigated if the negative effect of methylation on SRC-3 coactivator activity is specific to the estrogen receptor. To address this, we tested other nuclear receptors, including PR-B and GR, in similar transient-transfection experiments. In both cases, methylation mutants of SRC-3 exhibited higher coactivation activity than wild-type SRC-3 (Fig. 4C and D), suggesting that the repressive effect of SRC-3 methylation by CARM1 is likely to be a more general mechanism to nuclear hormone receptors.
We next asked whether the SRC-3 methylation mutants exhibit enhanced coactivation of an estrogen-induced endogenous gene. Induction of the pS2 gene was examined by real-time RT-PCR in the presence of exogenously expressed wild-type or methylation site-mutated SRC-3 in HEK293T cells. Both the R1171A and ΔMe mutants showed increased ability to activate the endogenous pS2 gene (Fig. 4E), further confirming that the loss of methylation of SRC-3 increased its coactivator function.
SRC-3 proteins defective for methylation have increased coactivator activity in vitro.
To further assess the coactivation activity of the methylation-defective SRC-3 mutants, we took advantage of an ERα-mediated in vitro transcription assay system. In this assay, a reporter plasmid containing the adenoviral E4 gene with 4 estrogen-responsive elements (ERE) was assembled into chromatin by using a Drosophila embryo S190 extract, followed by sequential addition of purified ERα and coactivators. The transcriptional activity of the coactivator was measured by quantifying transcription from the reporter gene by real-time RT-PCR. To confirm the high quality of the assembled chromatin, partial digestion with MNase was performed (Fig. 5A). In agreement with the results obtained from the ERE-Luc reporter assay conducted in cultured cells, the SRC-3 methylation mutants (Fig. 5C) showed higher coactivation activity than wild-type SRC-3 in the chromatin-based in vitro transcription assay; this effect appeared to be dosage dependent (Fig. 5C). Thus, assaying different transcription readout systems, we substantiated that methylation site mutations enhanced the ability of SRC-3 to function as a steroid receptor coactivator.
Next, a CARM1 E267Q mutant, in which the mutation eliminates the methyltransferase activity, was tested in the same assay. As shown in Fig. 5D, the CARM1 mutant showed consistently higher activity than the wild type in the presence of SRC-3. This can be explained by our model that, as a dual-function coactivator, CARM1 exerts both positive and negative effects on ER-mediated transcription. The luciferase readout is the combination of the dual effects. Alternatively, since CARM1 has been shown to contain a methylation activity-independent coactivator function, the E267Q mutation may retain the secondary coactivator function.
SRC-3 methylation modulates the interaction between SRC-3 and CARM1.
We next performed experiments to understand the mechanisms by which SRC-3 methylation mutants exhibit stronger coactivator activity toward ERα. The methylation region of SRC-3 locates within the interaction region between the SRC family proteins and CARM1 (4). This finding prompted us to test whether methylation would affect the association between SRC-3 and CARM1. We transfected Flag-tagged SRC-3 and methylation mutants into 293T cells and analyzed their interaction with CARM1 by coimmunoprecipitation. Following immunoprecipitation of SRC-3 with anti-Flag antibody, endogenous CARM1 associated with the wild type or methylation site SRC-3 mutants was compared. SRC-3 methylation mutants displayed a decreased capacity to bind to CARM1, indicating that R1171 plays a critical role in mediating the association between SRC-3 and CARM1 (Fig. 6A). These results suggest that methylation of SRC-3 on R1171 could modulate the association between SRC-3 and CARM1. To test this hypothesis, we synthesized two peptides containing residues R1171 and R1177. One peptide (P2) contains asymmetrically dimethylated R1171 to mimic the methylated form of SRC-3, whereas the other one (P1) is unmodified (Fig. 6B, upper panel). We then investigated which peptide could effectively inhibit the methylation of SRC-3 by CARM1 in a competitive methylation assay. Interestingly, P1 peptide strongly inhibited the methylation of SRC-3, while the P2 peptide showed little inhibitory activity (Fig. 6B, bottom panel), suggesting that only the unmodified peptide can bind to and sequester functional CARM1 from SRC-3.
FIG. 6.
SRC-3 methylation dissociates CARM1 from the coactivator complex. (A) HEK293T cells were transfected with the indicated Flag-SRC-3 constructs. Following immunoprecipitation (IP) with anti-Flag-M2 (α-Flag) affinity gel, the protein samples were separated on a 4 to 15% SDS-PAGE gel. Endogenous CARM1 associated with Flag-SRC-3 was examined by Western blotting (upper panel). The same membrane was also used to probe for SRC-3 (lower panel). (B) R1171 is a bona fide methylation site on SRC-3. Two peptides, one with an asymmetric dimethylated arginine at R1171 and the other one without any methyl group, were incubated with recombinant SRC-3 protein in the CARM1-mediated methylation assay. Triangles indicate three different peptide concentrations used (1.5 μM, 15 μM, and 150 μM). (C) Peptide with unmodified R1171 competes with SRC-3 for binding CARM1. Peptide P1 or P2 (150 μM) was incubated together with recombinant Flag-SRC-3 and CARM1 for 1 h at 30°C. SRC-3 protein was immunoprecipitated with anti-Flag-M2 affinity gel, and the presence of associated CARM1 was determined by Western blotting. (D) The methylation region of SRC-3 overlaps with the CARM1 docking region. Wild-type (WT) or methylation site-mutated SRC-3 proteins were incubated with GST or GST-CARM1 for 2 h at 4°C. Glutathione beads were used to pull down GST control and GST-CARM1 proteins. Associated SRC-3 proteins were determined by Western blot analysis. (E) A similar experiment was performed as for panel A, except that ERα was also transfected. The levels of p300 and ERα were determined by Western blotting as well.
Since the P1 peptide sequence localizes in the CARM1 binding region of SRC-3, we further investigated the important role of this small peptide sequence in mediating the CARM1-SRC-3 interaction. We tested whether the P1 peptide could interfere with CARM1 and SRC-3 interaction in an in vitro pull-down assay. As shown in Fig. 6C, unmodified P1 peptide efficiently disrupted the physical interaction between SRC-3 and CARM1 in vitro. In contrast, the P2 peptide with the methylated residue at R1171 could not do so (Fig. 6C). Combined with the inhibition assay result (Fig. 6B), our data clearly indicate that a small peptide sequence (residues 1168 to 1179) of SRC-3 is not only necessary but also sufficient to mediate the interaction between SRC-3 and CARM1. Importantly, methylation of a single arginine (R1171) within this region diminishes the interaction between SRC-3 and CARM1 coactivators.
Our results indicated that the methylation region of SRC-3 also serves as a docking site for CARM1. To confirm this deduction, a GST pull-down experiment was performed. Figure 6D showed that GST-CARM1 efficiently pulled down wild-type SRC-3 protein but not the methylation mutants of SRC-3. This result substantiated that the methylation region of SRC-3 overlaps with the protein-protein interacting region between SRC-3 and CARM1 and that the methylation event regulates their interaction.
Methylation of SRC-3 affects its interaction with p300.
As SRC-3 methylation mutants do not have increased interaction with the CARM1 coactivator, we wished to further investigate why these mutants exhibit higher coactivator activity. To this end, we analyzed their interaction with p300 by coimmunoprecipitation using anti-Flag captured SRC-3. Interestingly, more p300 was observed to interact with the SRC-3 methylation mutants, whereas no obvious change was detected on associated ERα (Fig. 6E, lanes 3, 4, and 6). Thus, the increased coactivator activity of SRC-3 methylation site mutants may be due, in part, to enhanced association with p300.
DISCUSSION
Dual functions of transcriptional coactivators.
Current models suggest that agonist-bound estrogen receptors not only recruit their primary coactivators, the SRC/p160 family proteins, but also lead to the subsequent conformational recruitment of secondary coactivators, such as p300/CBP and CARM1 (5, 6). p300/CBP are potent histone acetyltransferases that can acetylate all four core histones in nucleosomes (28), while CARM1 is a histone methyltransferase that methylates histone H3 at several arginines (R2, R17, and R26) in the N-terminal tail (5, 20). These posttranslational modifications are thought to positively mark the nucleosomes for transcriptional activation.
In addition to modifying the core histones in promoters, p300/CBP and CARM1 also can directly regulate the function of nonhistone transcriptional activators and coactivators by covalent modifications. p300/CBP can acetylate p53, leading to increased DNA binding and coactivator recruitment (2, 14). Androgen receptor and estrogen receptor alpha are also acetylated by p300/CBP (13, 37). In terms of transcriptional coactivator modifications, p300/CBP acetylates lysines preceding the LXXLL motif of SRC-3/ACTR, resulting in dissociation of SRC-3/ACTR from estrogen receptor (7). CARM1 has been reported to methylate various arginines in p300/CBP where methylation of arginines in the KIX domain perturbs CREB binding and affects the interaction between p300/CBP, CREB, and steroid receptors (42). Arginines close to the KIX domain of CBP are also modified and regulate its coactivator function (8). Finally, when the GRIP1 binding region of p300 is methylated, this leads to an attenuated interaction between GRIP1 and p300 (21).
In this study, we provided in vitro and in vivo evidence that SRC-3/AIB1 is a natural substrate for CARM1 and that SRC-3 methylation is induced by estrogen signaling. We identified R1171 of SRC-3 as the major methylation site. This arginine resides in a 12-amino-acid region of SRC-3 that is essential for CARM1 binding and is conserved within the SRC-3/p160 coactivator family. Our in vitro assays demonstrated that SRC-1 and SRC-2 also can be methylated by CARM1, likely through this arginine. CARM1 methylation thus may regulate the coactivator function of all SRC/p160 family proteins. Our SRC-3 data further suggest, along with that published for p300/CBP, that CARM1 regulates hormone-dependent transcription not only by modifying histones to enhance initiation of transcription but also by modifying coactivators to subsequently attenuate transcription. Thus, CARM1, and likely p300/CBP, are dual-function coactivators.
Coactivator assembly and disassembly.
Coactivator assembly on promoters is an early step in hormone-induced transcriptional activation that is mediated by numerous protein-protein interactions. For instance, the coactivator LXXLL motif is the most common interacting motif bridging the interactions between steroid/nuclear hormone receptor ligand binding domains and a large number of different coactivators (15). Precisely ordered and synergistic folding has been proposed to be the molecular mechanism for functional interactions between the SRC/p160 coactivators and p300/CBP (9, 24). Coactivator assembly also is regulated by signaling pathways. For example, previous work from our laboratory showed that phosphorylation is a major regulator of SRC-3 activation by promoting coactivator assembly (40, 45). Different signaling pathways induce SRC-3 phosphorylation at distinct sites. This differential phosphorylation of SRC-3 in turn leads to differential recruitment of specific transcriptional coactivators at specific transcription factors.
In contrast, the process of disassembling coactivator complexes is not well understood. Prior data suggested that the p23 and Hsp90 molecular chaperones may promote the disassembly of transcriptional regulatory complexes (12). Two other studies, as noted above, suggest that coactivator modification may play a role in this process, especially in the context of attenuating hormone signaling (7, 21, 42). Our current study provides additional mechanistic detail for this process. We show that CARM1 methylates SRC-3 and that this modification, in turn, induces dissociation between CARM1 and SRC-3. Only an unmethylated SRC-3 peptide, but not the methylated peptide, could inhibit CARM1 methylation of SRC-3 by disrupting the SRC-3-CARM1 association. We propose that methylation of the SRC family of coactivators by CARM1 is a molecular switch for further triggering coactivator complex disassembly.
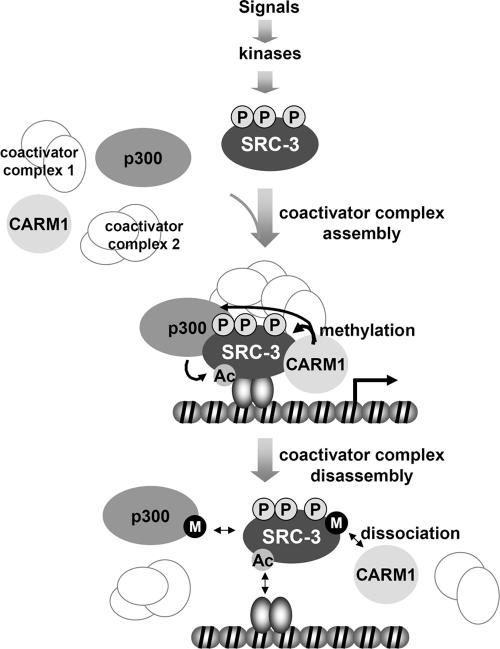
Based on our work and that of others, we propose a model to illustrate the molecular events involved in the assembly and disassembly of SRC-3-containing complexes acting at hormone-responsive genes (Fig. 7). First, SRC-3 activity is modulated by selective phosphorylation of different sites in response to distinct mitogenic signals. The hormone-bound estrogen receptor specifically interacts with promoter/enhancer sequences and recruits the appropriately phosphorylated SRC-3 coactivator to a target gene. SRC-3, in turn, recruits secondary coactivators, such as p300/CBP and CARM1. As a result, CARM1 methylates the core histone H3, while p300/CBP acetylates all core histones. The positive marks resulting from these histone modifications designate transcriptional activation of the target gene. At the same time, as shown by our current study, CARM1 methylates SRC-3 and dismantles the SRC-3-CARM1 coactivator complex. This model is consistent with the kinetics observed when these coactivators are bound and rapidly dissociate from target promoters in estrogen-treated MCF-7 cells (27, 30). In cells, we observe SRC-3 methylation peaking after 1 h of estrogen treatment; disappearance of methylation allows the possibility of another cycle of transcription. It is unclear whether the dissociated p300/CBP and SRC-3 would be rerecruited to the promoter for another round of transcription initiation because it is currently unknown if an arginine demethylase exists for recycling of previously methylated SRC-3. It also is possible that methylated SRC-3 is targeted for proteasome-dependent degradation in a transcription-linked mechanism. In vivo, this cycle is dynamic and leads to cyclic transcription driven by posttranslational coactivator modifications.
FIG. 7.
Model for coactivator assembly and disassembly on estrogen-responsive promoters. SRC-3 is phosphorylated at distinct sites by different kinases induced by various signaling pathways. Dependent upon the differential phosphorylation patterns, SRC-3 may assemble different coactivator complexes at regulated target gene promoters. Estrogen signaling results in estrogen-bound receptors interacting with both promoter DNA and SRC-3 coactivator complexes. Enzymatic activities enriched in the complex modify core histones and thereby facilitate transcription initiation. After one (or limited) round(s) of transcription initiation and to terminate signaling, p300/CBP and CARM1 modify components within the coactivator complex, leading to disassembly of the complex and probably also dissociation of receptor from the promoter DNA. We propose that CARM1, and likely also p300/CBP, is a dual-function coactivator: it not only activates transcriptional initiation by modifying core histone tails but also terminates hormone signaling by disassembling the coactivator complex. It remains unknown as to how many of the transcriptional coactivators in the complex are reutilized for another round of transcription initiation or instead subjected to transcription-linked protein degradation.
SRC-3 methylation and breast cancer.
The putative oncogene SRC-3/AIB1 is highly expressed in the breast cancer cell line MCF-7 in comparison to other cell lines or normal cells such as MEFs (1). Surprisingly, unlike MEFs, methylated SRC-3 was undetectable in MCF-7 cells in the absence of estrogen treatment. Upon estrogen addition, SRC-3 was transiently methylated in these cells. It may be of interest to determine if a lower level of SRC-3 methylation in breast tumors correlates with the more transcriptionally active state. In comparison to methylated SRC-3, unmethylated SRC-3 is a more potent coactivator, as seen by the fact that methylation site mutants exhibit higher transcriptional activity (Fig. 4). This higher activity may be due, in part, to a stronger interaction with p300 (Fig. 6). Enhanced association with p300 could be due to at least two possible mechanisms. SRC-3 methylation may directly regulate the interaction between SRC-3 and p300, since the CBP/p300 binding domain of SRC-3 is in close proximity to the methylation region. Perhaps more likely, the mutation of the methylation site on SRC-3 leads to decreased CARM1 association with the p300-CARM1-SRC-3 coactivator complex, resulting in attenuated methylation of p300 by CARM1. Since methylation of p300 by CARM1 abrogates the p300-SRC-2/GRIP1 interaction (21), it is likely that less CARM1 association results in an increased association between SRC-3 and p300 and more transcriptional activity.
Acknowledgments
We thank Charles Foulds and Bin He for critical reading of the manuscript. We also thank Mark Bedford for CARM1 wild-type and knockout MEFs, Michael R. Stallcup for the pSG5-HA-CARM1 plasmid, Dean Edwards for ER alpha and PR-A proteins, Steven Nordeen for SRC-1 recombinant protein, W. Lee Kraus for ERE-E4 plasmid and p300 baculovirus, and Sophia Tsai and Ming-Jer Tsai for helpful discussion.
This work was supported by grants from NIH/NICHD, Welch Foundation, and NIDDK-NURSA (B.W.O.).
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 2.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 3.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21:4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, D., S. M. Huang, and M. R. Stallcup. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275:40810-40816. [DOI] [PubMed] [Google Scholar]
- 5.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation viaacetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 8.Chevillard-Briet, M., D. Trouche, and L. Vandel. 2002. Control of CBP co-activating activity by arginine methylation. EMBO J. 21:5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demarest, S. J., M. Martinez-Yamout, J. Chung, H. Chen, W. Xu, H. J. Dyson, R. M. Evans, and P. E. Wright. 2002. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415:549-553. [DOI] [PubMed] [Google Scholar]
- 10.Dickson, R. B., and G. M. Stancel. 2000. Estrogen receptor-mediated processes in normal and cancer cells. J. Natl. Cancer Inst. Monogr. 2000:135-145. [DOI] [PubMed] [Google Scholar]
- 11.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052-1058. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, B. C., and K. R. Yamamoto. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296:2232-2235. [DOI] [PubMed] [Google Scholar]
- 13.Fu, M., C. Wang, J. Wang, X. Zhang, T. Sakamaki, Y. G. Yeung, C. Chang, T. Hopp, S. A. Fuqua, E. Jaffray, R. T. Hay, J. J. Palvimo, O. A. Janne, and R. G. Pestell. 2002. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell. Biol. 22:3373-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 15.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 16.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassabov, S. R., B. Zhang, J. Persinger, and B. Bartholomew. 2003. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell 11:391-403. [DOI] [PubMed] [Google Scholar]
- 18.Katzenellenbogen, B. S., and J. A. Katzenellenbogen. 2000. Estrogen receptor transcription and transactivation: estrogen receptor alpha and estrogen receptor beta: regulation by selective estrogen receptor modulators and importance in breast cancer. Breast Cancer Res. 2:335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuang, S. Q., L. Liao, H. Zhang, A. V. Lee, B. W. O'Malley, and J. Xu. 2004. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 64:1875-1885. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. Y., C. Teyssier, B. D. Strahl, and M. R. Stallcup. 2005. Role of protein methylation in regulation of transcription. Endocr. Rev. 26:147-170. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y. H., S. A. Coonrod, W. L. Kraus, M. A. Jelinek, and M. R. Stallcup. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. USA 102:3611-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, Y. H., S. S. Koh, X. Zhang, X. Cheng, and M. R. Stallcup. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 22:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Z., J. Wong, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2001. Sequential recruitment of steroid receptor coactivator-1 (SRC-1) and p300 enhances progesterone receptor-dependent initiation and reinitiation of transcription from chromatin. Proc. Natl. Acad. Sci. USA 98:12426-12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Z., J. Wong, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1999. Steroid receptor coactivator-1 (SRC-1) enhances ligand-dependent and receptor-dependent cell-free transcription of chromatin. Proc. Natl. Acad. Sci. USA 96:9485-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 27.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 29.Onate, S. A., S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 30.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 31.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 32.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 33.Thackray, V. G., D. O. Toft, and S. K. Nordeen. 2003. Novel activation step required for transcriptional competence of progesterone receptor on chromatin templates. Mol. Endocrinol. 17:2543-2553. [DOI] [PubMed] [Google Scholar]
- 34.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 35.Torres-Arzayus, M. I., J. Font de Mora, J. Yuan, F. Vazquez, R. Bronson, M. Rue, W. R. Sellers, and M. Brown. 2004. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell 6:263-274. [DOI] [PubMed] [Google Scholar]
- 36.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, C., M. Fu, R. H. Angeletti, L. Siconolfi-Baez, A. T. Reutens, C. Albanese, M. P. Lisanti, B. S. Katzenellenbogen, S. Kato, T. Hopp, S. A. Fuqua, G. N. Lopez, P. J. Kushner, and R. G. Pestell. 2001. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 276:18375-18383. [DOI] [PubMed] [Google Scholar]
- 38.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 39.Wu, R. C., J. Qin, Y. Hashimoto, J. Wong, J. Xu, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2002. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by I kappa B kinase. Mol. Cell. Biol. 22:3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, R. C., J. Qin, P. Yi, J. Wong, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2004. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic responses to multiple cellular signaling pathways. Mol. Cell 15:937-949. [DOI] [PubMed] [Google Scholar]
- 41.Xu, J., Y. Qiu, F. J. DeMayo, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1998. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922-1925. [DOI] [PubMed] [Google Scholar]
- 42.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 43.Yadav, N., J. Lee, J. Kim, J. Shen, M. C. Hu, C. M. Aldaz, and M. T. Bedford. 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. USA 100:6464-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi, P., R. C. Wu, J. Sandquist, J. Wong, S. Y. Tsai, M.-J. Tsai, A. R. Means, and B. W. O'Malley. 2005. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol. Cell. Biol. 25:9687-9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, F. F., R. C. Wu, C. L. Smith, and B. W. O'Malley. 2005. Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor. Mol. Cell. Biol. 25:8273-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]