Abstract
The physiological and pathological manifestations of Sonic hedgehog (Shh) signaling arise from the specification of unique transcriptional programs dependent upon key nuclear effectors of the Ci/Gli family of transcription factors. However, the underlying mechanism by which Gli proteins regulate target gene transcription in the nucleus remains poorly understood. Here, we identify and characterize a physical and functional interaction between Gli3 and the MED12 subunit within the RNA polymerase II transcriptional Mediator. We show that Gli3 binds to MED12 and intact Mediator both in vitro and in vivo through a Gli3 transactivation domain (MBD; MED12/Mediator-binding domain) whose activity derives from concerted functional interactions with both Mediator and the histone acetyltransferase CBP. Analysis of MBD truncation mutants revealed an excellent correlation between the in vivo activation strength of an MBD derivative and its ability to bind MED12 and intact Mediator in vitro, indicative of a critical functional interaction between the Gli3 MBD and the MED12 interface in Mediator. Disruption of the Gli3-MED12 interaction through dominant-negative interference inhibited, while RNA interference-mediated MED12 depletion enhanced, both MBD transactivation function and Gli3 target gene induction in response to Shh signaling. We propose that activated Gli3 physically targets the MED12 interface within Mediator in order to functionally reverse Mediator-dependent suppression of Shh target gene transcription. These findings thus link MED12 to the modulation of Gli3-dependent Shh signaling and further implicate Mediator in a broad range of developmental and pathological processes driven by Shh signal transduction.
The proper specification and maintenance of cell fate during embryonic development and postnatal homeostasis are driven by signal transduction pathways that specify unique programs of protein-encoding gene transcription. Among these, the Hedgehog (Hh) signaling pathway plays a particularly important role in embryonic pattern formation and maintenance of adult stem cell number (27, 32, 90). Accordingly, dysregulated Hh signaling has been linked to a variety of pathologies, including developmental anomalies and cancer (43, 63, 65, 90).
Our current knowledge regarding Hh signaling at the molecular level derives from studies of a broad range of model metazoan organisms, including Drosophila melanogaster, where Hh was first identified as a segment polarity gene product (8, 51, 68, 72, 88, 89). Drosophila Hh occupies a central role in pattern formation of the anterior-posterior (A-P) axis through its ability to modulate the functional status of Cubitus interruptus (Ci), a DNA-binding transcription factor and bifunctional regulator of Hh target gene transcription (1, 3, 4, 8, 13, 23, 26, 51, 58, 64, 89, 95). Because Ci is targeted for Hh-dependent proteolytic cleavage, its functional status in the anterior compartment is determined by a diffusible gradient of Hh concentration that decreases anteriorly from the compartment border. Thus, in anterior cells distant from the A-P border where Hh is low or absent, cleavage of Ci produces a truncated DNA-binding-competent derivative that silences Hh target gene transcription through an N-terminal repression domain (4, 21, 64). By contrast, in anterior cells at the A-P border where Hh is present in high concentrations, suppression of Ci cleavage leads to accumulation of full-length Ci that stimulates border-specific gene transcription through a C-terminal transactivation domain (4, 21, 64, 98). In this manner, a morphogenetic gradient of Hh controls the pattern of expressed genes that function in turn to specify cell fate.
The intracellular signaling events leading to Hh-induced suppression of Ci cleavage, while not fully clarified, have nonetheless been deciphered in considerable detail. The active signaling form of Hh corresponds to a 19-kDa dually lipidated species derived from autocatalytic processing of the full-length Hh translation product (60). This Hh derivative binds to the 12-pass transmembrane protein Patched (Ptc), thereby precluding Ptc-mediated inhibition of Smoothened (Smo), a seven-pass transmembrane protein with distant homology to G-protein-coupled receptors (15, 24, 41, 61, 85, 91). Activated Smo in turn promotes the accumulation and activation of latent full-length Ci in the cytoplasm through a complex series of events including suppression of Ci repressor processing and possible posttranslational modification (38, 40, 57, 73). Ultimately, full-length Ci activator translocates into the nucleus, whereupon it binds to and stimulates Hh target genes that function further in cell fate specification.
Fundamental aspects of invertebrate Hh signaling have been conserved in vertebrates, and the enhanced complexity of the latter is generally reflected in a combinatorial expansion in the number of Hh pathway components. Thus, mammals encode three distinct Hh family members (Sonic [Shh], Indian [Ihh], and Desert [Dhh]), two Ptch homologues (Ptch1 and Ptch2), and three Ci-related transcription factors (Gli1, Gli2, and Gli3) (12, 22, 25, 70, 81, 85). Among the mammalian Hh protein family, Shh is the best characterized and functions to regulate cell fate specification, proliferation, and/or differentiation in a wide variety of target tissues and organ systems. For example, Shh controls dorsoventral patterning of the neural tube and the somites, anteroposterior patterning of the limb buds, and morphogenesis of the hindgut (32, 71). As obligate effectors of Shh signaling, the three Gli proteins possess distinct transcriptional properties. Whereas Gli1 functions primarily as an activator, Gli2 and Gli3 more closely resemble Ci structurally and functionally as bipartite transcriptional regulators of Shh target genes (6, 18, 20, 29, 30, 50, 54, 62, 69, 75, 80, 82). With specific respect to Gli3, Shh suppresses the proteolytic production of a Gli3 repressor, thereby promoting the accumulation of full-length Gli3 with activator potential (18, 55, 96). While the Gli3 repressor has clearly been shown to play a broad and essential role in antagonizing Shh activity in a variety of tissues, the relative importance of the Gli3 activator as an effector of Shh signaling has been more controversial (29, 54, 55, 79, 93, 99). However, recent gene targeting studies of mice have clearly established the importance of a Gli3 activator function in proper patterning of the ventral spinal cord (7, 53). In addition, a Gli3 activator function has been implicated in both sclerotome formation and regulation of gastric glandular growth in mice (10, 45).
Reflective of its role as an effector of Shh signaling, mutations and translocations involving the Gli3 locus cause a variety of developmental disorders, including Greig cephalopolysyndactyly syndrome and Pallister-Hall syndrome (39, 42). Mutations associated with these polysyndactylous disorders are predicted to affect the repressor and/or the activator functions of Gli3. Surprisingly, little is currently known regarding the mechanism by which Gli3 functions as a bipartite regulator of gene transcription in the nucleus. However, a more detailed description of the transcriptional regulatory properties of Gli3 will be essential to fully understand the molecular basis for its important roles in development and disease.
With respect to the activation function of Gli3, previous studies have revealed a physical and functional interaction between mammalian Gli3 and the histone acetyltransferase CBP (18). The observation that the CBP-binding domain on Gli3 (CBD; amino acids 827 to 1132 of the 1,596-residue protein) can function as an independent transactivation domain supports the idea that CBP is a transcriptional coactivator of Gli3 (18). However, more recent studies have identified Gli3 sequences outside the CBD with autonomous transactivation function, suggesting the involvement of additional unidentified activities in Gli3-directed transcription (42).
In this regard, recent genetic studies of Drosophila have revealed that mutations in Mediator subunits MED12 and MED13 variously affect developmental pathways regulated by Hh signaling. In the eye disc, loss of MED12 or MED13 results in diminished expression of the Hh target genes decapentaplegic (dpp) and atonal (ato) in the morphogenetic furrow and persistent expression of these genes posterior to the morphogenetic furrow, where cells are normally unresponsive to Hh (94). As a result, eye disc cells mutant for either MED12 or MED13 fail to differentiate (94). In the wing disc, loss of MED12 or MED13 results in migration of anterior cells into the posterior compartment at the A-P border, suggesting a requirement for MED12 and MED13 for the expression of cell adhesion molecules normally activated by Ci (36). However, whether these effects reflect a direct or indirect role for Mediator in Ci target gene transcription has heretofore remained unknown.
Mediator is an evolutionarily conserved multiprotein interface between gene-specific transcription factors and the RNA polymerase II general transcription machinery (16, 47, 59, 86). In this capacity, Mediator serves to promote the assembly, activation, and regeneration of transcription complexes on core promoters during the initiation and reinitiation phases of transcription (5, 11, 34, 48, 56, 66, 76, 77, 97, 103). Because of its direct association with both signal-activated transcription factors and the RNA polymerase II transcription machinery, Mediator has been proposed to function as a general conduit and integrator of regulatory signals that converge on the promoters of protein-encoding genes (100). Consistent with this idea, several Mediator subunits are functionally required for activated transcription in response to diverse cell signaling pathways. These include MED1 for nuclear receptor, MED14 for gamma interferon, MED23 for RAS/mitogen-activated protein kinase, and MED15 for transforming growth factor β signaling pathways (9, 44, 49, 84, 102).
To clarify the role of Mediator in transcription control, we undertook a broad-base yeast two-hybrid screening approach to identify interaction partners for different human Mediator subunits. Among MED12-interacting proteins, we identified human Gli3. Herein, we document a direct physical and functional interaction between Gli3 and the MED12 subunit in Mediator. We show that Gli3 binds to isolated MED12 and intact Mediator both in vitro and in vivo through a novel transactivation domain (MBD; MED12/Mediator-binding domain) whose activity derives from concerted functional interactions with both Mediator and CBP. Analysis of MBD truncation mutants revealed an excellent correlation between the in vivo activation strength of an MBD derivative and its ability to bind MED12 and intact with Mediator in vitro, indicative of a critical functional interaction between the Gli3 MBD and the MED12 interface in Mediator. Disruption of the Gli3-MED12 interaction through dominant-negative interference inhibited while MED12 depletion enhanced both MBD transactivation activity and Gli3-dependent gene induction in response to Shh signaling. We propose that activated Gli3 physically targets the MED12 interface within Mediator in order to functionally reverse Mediator-dependent suppression of Shh target gene transcription. These findings thus link MED12 to the modulation of Gli3-dependent Shh signaling and suggest that MED12 could be important to ensure an appropriate transcriptional response to a graded Shh signal.
MATERIALS AND METHODS
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed following the manufacturer's recommended procedures for the Matchmaker GAL4 Two-Hybrid System 3 (Clontech, Mountain View, CA). Briefly, a human MED12 cDNA fragment encoding amino acids 1750 to 2212 was expressed as a GAL4 DNA-binding domain fusion protein from the bait plasmid pGBKT7. For screening purposes, the MED12 bait plasmid was cotransformed into the Saccharomyces cerevisiae strain AH109 along with a library of human embryonic brain cDNAs, expressed from the prey plasmid pACT2 as GAL4 activation domain fusion proteins (Clontech, Mountain View, CA). After two successive rounds of selection on Leu-Trp-His-Ade-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates, positive clones were inoculated into selective yeast culture medium, and plasmid DNA purified from cultures was transformed into Escherichia coli to amplify the isolated prey plasmids under ampicillin selection. Individual prey plasmids were subsequently cotransformed along with the MED12 bait plasmid back into AH109 yeast cells to confirm their positive status by selection on Leu-Trp-His-Ade-X-Gal plates. Subsequently, positive prey plasmids were sequenced and the BLAST (GenBank) alignment program was used to determine protein similarity indices.
Expression and reporter plasmids.
A detailed description of all expression and reporter plasmids used in this study will be furnished upon request. Full-length MED12 and truncation fragments thereof used in glutathione S-transferase (GST) pull-down experiments have been described previously (46) and were expressed by coupled in vitro transcription-translation reactions (TNT SP6/T7 Quickcoupled transcription-translation system; Promega, Madison, WI). Human Gli3 cDNA sequences encoding Gli3 amino acids 1006 to 1596, 1046 to 1596, 1090 to 1596, 1006 to 1376, and 1006 to 1260 were subcloned by PCR-based methods into the E. coli expression plasmid pGEX-6P-1 for production of GST fusion proteins used in GST pull-down experiments or mammalian expression plasmid pM1 for production of GAL4 fusion proteins used in transient reporter-based transcription assays. Plasmid pact-FLAG-Gli3 for mammalian expression of FLAG epitope-tagged human Gli3 has been described previously (18). Plasmid pCS2+E1A-12S for mammalian expression of the adenovirus 12S E1A protein was generated by PCR-based subcloning of the adenovirus 12S cDNA into pCS2+. Plasmid pRc/RSV-CBP for mammalian expression of human CBP was a kind gift of Paul M. Lieberman (The Wistar Institute, Philadelphia, PA). Plasmids pCMV-GAL4-β-Cat for mammalian expression of a GAL4 DNA-binding domain-human β-catenin chimera and pGEX4T-β-Cat for bacterial expression of a GST-β-catenin activation domain chimera were generous gifts of Andreas Hecht (Universitaet Freiburg, Freiburg, Germany). The pGAL4-E1A and pGAL4-VP16 expression plasmids as well as the pG5E1B-Luc reporter plasmid used in transient reporter-based transcription assays were a kind gift of Arnold J. Berk (University of California, Los Angeles). Reporter plasmid 8X3′Gli-BS Luc bearing eight copies of a Gli-binding site derived from the murine HNF3β floor plate enhancer was a generous gift of Hiroshi Sasaki (Osaka University, Osaka, Japan). Plasmid pJT4-Shh for mammalian expression of Shh has been described previously (82).
Cell culture.
HeLa human cervical carcinoma, 293 human embryonic kidney, and CH310T1/2 mouse fibroblast cell lines were routinely cultured at 37°C and 10% CO2 in Dulbecco's modified Eagle's medium (Invitrogen/Gibco BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and penicillin-streptomycin-l-glutamine (Invitrogen/Gibco BRL, Gaithersburg, MD).
GST pull-down assays.
GST-Gli3 and GST-β-Cat derivatives were expressed in E. coli strain BL21 CodonPlus (Stratagene, La Jolla, CA), and soluble lysates were prepared in Lysis 250 buffer (50 mM Tris-HCl, 250 mM NaCl, 5 mM EDTA, 0.1% NP-40) supplemented with protease inhibitors (20 μM antipain, 2 mM pepstatin, 20 μM leupeptin, 2 μg/ml aprotinin). Resuspended cells were subjected to one freeze-thaw cycle followed by sonication and clarification by centrifugation at 35,000 × g for 30 min at 4°C. HeLa/S3 nuclear extracts were prepared as described previously (19). For GST pull-down assays using radiolabeled recombinant MED12 or HeLa nuclear extract, 20 μg of each GST derivative was immobilized on glutathione-Sepharose beads (GE Healthcare Life Sciences, Piscataway, NJ) and washed extensively with Lysis 250 buffer containing 0.2% NP-40. Bead-immobilized GST-Gli3 derivatives were washed once with 0.1 M KCl D buffer (20 mM HEPES, pH 7.9, 0.2 mM EDTA, 20% glycerol) containing 0.1% NP-40 and subsequently incubated with either radiolabeled recombinant MED12 (40 μl of a standard in vitro-coupled transcription-translation reaction), HeLa/S3 nuclear extract (1 mg; previously dialyzed against 0.1 M KCl D buffer), or nuclear extract derived from HeLa cells transfected with 20 μM control or MED-specific short interfering RNA (siRNA; 1 mg; previously dialyzed against 0.1 M KCl D buffer). After incubation overnight at 4°C, beads were washed five times with 0.15 M KCl D buffer (0.2% NP-40). Bound proteins were eluted with Laemmli sample buffer. Ten percent of each eluate was resolved by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE) and analyzed by Coomassie brilliant blue staining to ensure that equivalent amounts of GST-Gli3 fusion proteins were immobilized. The remaining 90% of each eluate was resolved by SDS-10% PAGE and processed by either PhosphorImager analysis in the case of recombinant MED12 pull-downs or, alternatively, immunoblot analysis using Mediator subunit-specific antibodies for intact Mediator pull-downs.
GST pull-down assays using recombinant CBP and/or purified Mediator were performed as described above with minor modifications. Briefly, recombinant CBP, purified Mediator, or both were incubated with immobilized GST derivatives in 0.15 M KCl D buffer (0.1% NP-40) for 2 h at 4°C and washed three times with the same buffer. Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-PAGE, and either visualized by silver staining or detected by immunoblot analysis using CBP-specific and Mediator subunit-specific antibodies.
Recombinant full-length CBP was expressed in and purified from Sf9 insect cells as described previously (14). Mediator purification from a HeLa/S3-derived clonal cell line expressing a hemagglutinin epitope-tagged MED6 Mediator subunit has been described previously (46).
Transfections, reporter assays, and RNAi.
HeLa or CH310T1/2 cells were seeded into 12-well cell culture plates 24 h prior to transfection. Cells (50 to 60% confluence) were transfected with reporter, activator, and effector plasmids as indicated in the figure legends using Fugene 6 (Roche, Indianapolis, IN) following the manufacturer's instructions. At 48 h posttransfection, transfected cells were harvested and assayed for luciferase (Promega, Madison, WI) and β-galactosidase (Applied Biosystems, Foster City, CA) activities as described previously (92). All transfections were performed a minimum of three times in duplicate. For coupled transient reporter/RNA interference (RNAi) experiments in HeLa cells, cells were transfected with 20 nM CBP-specific (sc-29244; Santa Cruz Biotechnology, Santa Cruz, CA), MED12-specific (M-009092-00), MED23-specific (M-013220-00), or control (D-001210-01-05) siRNAs (Dharmacon, Chicago, IL) using Transit SiQuest transfection reagent (Mirus Bio Corporation, Madison, WI) 48 h prior to transfection with reporter and activator plasmids using Fugene 6 as described above. Reporter assays were performed 36 h posttransfection as described above.
For coupled transient reporter/RNAi experiments in CH310T1/2 cells, cells were electroporated with 2.2 mM Gli3-specific (M-045798-00), MED12-specific (M-048744-00), or control (D-001210-01-20) siRNAs (Dharmacon, Chicago, IL) using a Nucleofector II instrument and Nucleofector Kit R (Amaxa Biosystems, Koeln, Germany) according to the manufacturer's instructions. Electroporated cells were seeded at 1.5 × 105 cells/well in 12-well cell culture plates 24 h prior to transfection with the 8X3′Gli-BS Luc reporter plasmid (0.4 μg) and the internal control pact-β-galactosidase expression plasmid (0.1 μg) using Fugene 6 (Roche, Indianapolis, IN). Twenty-fours hours following plasmid transfections, transfected cell culture medium was replaced with control or Shh-conditioned medium, and cells were cultured for an additional 24 h prior to cell harvest and assay of reporter gene activity as described above.
Antibodies.
Antibodies used for immunoblot analyses correspond to the following: MED1 (sc-8998), MED6 (sc-9433), MED12 (sc-5372), CDK8 (sc-13155, sc-1521), CBP (sc-369), and GAL4 (sc-510) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); MED23 (551175) was purchased from BD Pharmingen (San Diego, CA). Purified rabbit antibody raised against murine Gli3 was a generous gift from Susan Mackem (Laboratory of Pathology, National Cancer Institute, Bethesda, MD). MED14 antibody was a kind gift from Michael J. Garabedian (New York University School of Medicine, New York, NY). Antibody used for standard immunoprecipitation analysis corresponds to the following: CDK8 (sc-1521). Antibodies used for chromatin immunoprecipitation (ChIP) analysis correspond to the following: Gli3 (sc-20688) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA); CDK8 (RB-018-P1) was purchased from Lab Vision Corporation (Fremont, CA).
Immunoprecipitation assays.
HeLa cells transfected with pact-FLAG-Gli3 (6 μg) were harvested at 48 h and nuclear extracts prepared as described previously (22). Nuclear extract (1 mg; adjusted to 0.1 M KCl and 0.2% NP-40) was incubated for 12 h at 4°C with 10 μg of either CDK8-specific goat polyclonal antibodies or goat immunoglobulin G (IgG) as a negative control. Following addition of protein A-Sepharose beads (20 μl) and incubation for 2 h at 4°C, immune complexes were washed five times for 5 min each at 4°C with 0.1 M KCl D buffer (0.2% NP-40). Immunoprecipitated proteins were subsequently eluted with Laemmli sample buffer, resolved by SDS-10% PAGE, and processed by immunoblot analysis for the presence of individual Mediator subunits and FLAG epitope-tagged Gli3.
Shh-conditioned medium production.
Shh-conditioned medium was prepared as described previously (74). Briefly, 293 cells were transfected with the empty pJT4 expression plasmid or the pJ.T4-shh expression plasmid using Fugene 6 (Roche, Indianapolis, IN). Seventy-two hours posttransfection, control-conditioned and Shh-conditioned media were collected, filtered through 0.2-μm cellulose acetate filters, and stored at −80°C.
Shh signaling assays and quantitative real-time RT-PCR.
CH310T1/2 cells (50 to 60% confluence) were either untransfected or transfected with 100 nM Gli3-specific (M-045798-00), MED12-specific (M-048744-00), or control (D-001210-01-20) siRNAs (Dharmacon, Chicago, IL) using Oligofectamine transfection reagent (Invitrogen, Carlsbad, CA). At 48 h posttransfection, transfected cell culture medium was replaced with control-conditioned or Shh-conditioned medium, and cells were cultured for an additional 24 h prior to harvest and isolation of RNA using Trizol reagent (Invitrogen, Carlsbad, CA). RNA was reverse transcribed using oligo(dT) and Superscript III (Invitrogen, Carlsbad, CA) following standard procedures and used in quantitative reverse transcription-PCR (RT-PCR). Primers used for quantitative real-time PCR were as follows: Gli1 (5′-GAG CCC TTC TTT AGG ATT CCC A-3′ and 5′-ACC CCG AGT AGA GTC ATG TGG-3′), cyclin D1 (5′-TTG TGC ATC TAC ACT GAC AAC TC-3′ and 5′-AGG GTG GGT TGG AAA TGA ACT-3′), and β-actin (5′-CAA AGA CCT GTA CGC CAA CAC AGT-3′ and 5′-ACT CCT GCT TGC TGA TCC ACA TCT-3′). Quantitative RT-PCR was performed using an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) and Absolute SYBR Green ROX Mix (ABgene, Epsom, United Kingdom). Gli1 and cyclin D1 RNA levels were normalized to β-actin levels.
ChIP assays.
CH310T1/2 cells cultured for 6 h with control or Shh-conditioned medium were treated with 1% formaldehyde for 15 min at room temperature. Cross-linking reactions were quenched with glycine, and soluble chromatin was obtained by sonication of pelleted cells in cell lysis buffer {50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 8.0, 85 mM KCl, 1% NP-40} prior to processing as described previously (28). DNA isolated with a QIAquick PCR purification kit was subsequently used in amplification reactions using primers specific for the Gli-binding region within the Gli1 gene (18).
RESULTS
Gli3 binds directly to MED12 both in vitro and vivo.
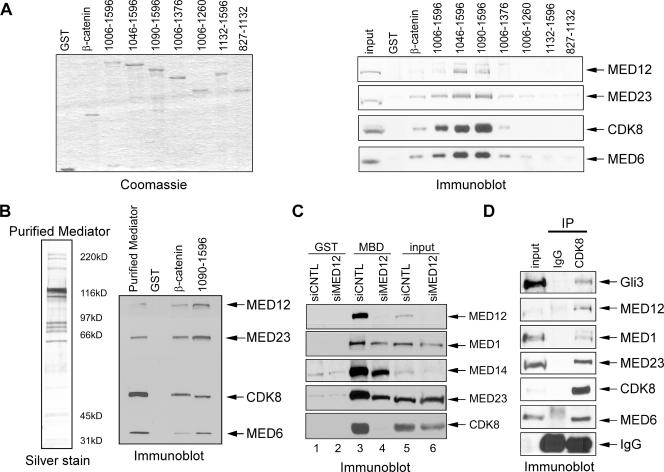
To identify MED12-interacting proteins, we used the MED12 C terminus (amino acids 1750 to 2212) as bait to screen a human fetal brain cDNA library by the yeast two-hybrid assay (Fig. 1A). This region of MED12 encompasses part of a PQL domain rich in Pro, Gln, and Leu residues and a Gln-rich OPA domain. From among ∼2 × 106 independent clones screened, we identified a partial cDNA for Gli3 (encoding amino acids 1006 to 1596; Fig. 1B).
FIG. 1.
Gli3 binds specifically to MED12 in vitro. (A and B) Schematic diagrams of MED12 (A) and Gli3 (B) derivatives used in binding reactions. The MED12 PQL and OPA domains and the Gli3 DNA- and CBP-binding domains (DBD and CBD, respectively) are highlighted. Gli3 amino acids (1006 to 1596) encoded by the partial Gli3 cDNA recovered in the yeast two-hybrid screen are indicated in boldface. Numbers refer to amino acid coordinates. (C and D) Recombinant full-length MED12 or its indicated truncation derivatives were expressed and radiolabeled with [35S]methionine by translation in vitro prior to incubation with glutathione-Sepharose-immobilized GST or the indicated GST-Gli3 truncation derivatives. Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-10% PAGE, and visualized by phosphorimager analysis. In panel D, 10% of each eluate was resolved by SDS-10% PAGE and visualized by Coomassie brilliant blue staining to ensure that roughly equivalent amounts of GST-Gli3 derivatives were used in binding reactions. Input corresponds to 10% of each in vitro-translated product used in binding reactions. Numbers at left of panel C are in thousands.
To confirm the physical interaction between MED12 and Gli3 and also to map the Gli3-binding domain on MED12, we expressed Gli3 amino acids 1006 to 1596 as a GST fusion protein and tested the ability of GST-Gli3 1006-1596 to bind to various MED12 fragments produced by transcription and translation in vitro (Fig. 1A). GST-Gli3 1006-1596 bound strongly to full-length MED12 and the MED12 PQL domain (amino acids 1651 to 2086) but failed to bind to MED12 fragments corresponding to the N terminus, an internal region, and the OPA domain at the C terminus (Fig. 1C). Interestingly, GST-Gli3 1006-1596 bound only weakly to the MED12 yeast two-hybrid bait fragment (amino acids 1750 to 2212), thus indicating that the amino-terminal region of the PQL domain is particularly important for the interaction between MED12 and Gli3 (Fig. 1C). Apparently, the weak interaction between Gli3 and the MED12 bait fragment was nonetheless sufficient to support yeast colony outgrowth in the two-hybrid assay. In fact, we confirmed by quantitative β-galactosidase assay that Gli3 1006-1596 and the MED12 bait fragment interact weakly, but specifically, in yeast (data not shown). In summary, the Gli3-binding domain on MED12 corresponds to the MED12 PQL domain (amino acids 1651 to 2086).
To reciprocally map the MED12-binding domain on Gli3, we screened a panel of GST-Gli3 1006-1596 amino- and carboxyl-terminal truncation derivatives (Fig. 1B) for their respective abilities to bind to full-length MED12 produced by transcription and translation in vitro. Analysis of amino-terminal truncation derivatives revealed comparably strong MED12 binding to GST-Gli3 1006-1596 and its derivatives 1046-1596 and 1090-1596 and a significant reduction in MED12 binding to GST-Gli3 1132-1596 (Fig. 1D). Analysis of GST-Gli3 1006-1596 carboxyl-terminal truncation derivatives revealed a significant loss of MED12 binding to derivatives 1006-1376 and 1006-1260 (Fig. 1D). Therefore, the MED12-binding domain on Gli3 corresponds to amino acids 1090 to 1596. Although this domain partially overlaps the previously identified Gli3 CBD (amino acids 827 to 1132) (18), MED12 does not bind to the CBD (Fig. 1D). Significantly, the MED12-binding domain on Gli3 corresponds almost perfectly to a region (amino acids 1044 to 1580) shown previously to harbor autonomous transactivation function (42), raising the possibility that Mediator, through its MED12 subunit, might represent a functionally important target of Gli3.
Gli3 binds directly to intact Mediator both in vitro and in vivo.
Because Gli3 sequence 1006 to 1596 interacted specifically with MED12 by yeast two-hybrid and GST pull-down assays, we next asked whether GST-Gli3 1006-1596 could pull down intact Mediator present in a HeLa cell nuclear extract. A GST-β-catenin activation domain fusion protein was included as a positive control for these experiments, since we recently documented a direct physical and functional interaction between the β-catenin activation domain and the MED12 interface in Mediator (46). Similarly to GST-β-catenin, GST-Gli3 1006-1596, but not GST alone, bound intact Mediator, as revealed by the presence in pull-down eluates of individual Mediator subunits MED12, MED23, CDK8, and MED6 (Fig. 2A). To identify the minimal Mediator-binding domain on Gli3, we also examined the same panel of Gli3 1006-1596 amino- and carboxyl-terminal truncation derivatives that were tested for MED12 binding for their respective abilities to bind to intact Mediator. In general, we observed an excellent correlation between the ability of a particular Gli3 truncation derivative to bind to isolated MED12 and intact Mediator. Thus, GST-Gli3 1006-1596 and its amino-terminal truncation derivatives 1046-1596 and 1090-1596 all bound intact Mediator strongly, while GST-Gli3 1132-1596 did not (Fig. 2A). Interestingly, the level of Mediator binding was significantly enhanced by incremental truncation of Gli3 sequences between 1006 and 1090, although the basis for this effect is presently unknown. Analysis of GST-Gli3 1006-1596 carboxyl-terminal truncation derivatives revealed a significant reduction in Mediator binding to GST-Gli3 derivatives 1006-1376 and 1006-1260 (Fig. 2A). Similarly to MED12, intact Mediator did not bind to the Gli3 CBD (Fig. 2A). Therefore, the Mediator-binding domain on Gli3 coincides with the MED12-binding domain and corresponds to amino acids 1090 to 1596. We hereafter refer to this region on Gli3 as the MBD.
FIG. 2.
Gli3 binds specifically to Mediator in vitro and in vivo. (A and B) Immobilized GST, GST-β-catenin, or GST-Gli3 derivatives as indicated were incubated with HeLa nuclear extract (A) or purified Mediator (B). Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-10% PAGE, and processed by immunoblot analysis for the presence of individual Mediator subunits as indicated. Input represents 5% of total nuclear extract (A) or 30% of purified Mediator (B) used in binding reactions. In panel A, 10% of each eluate was resolved by SDS-10% PAGE and visualized by Coomassie brilliant blue staining to ensure that equivalent amounts of GST-Gli3 derivatives were used in binding reactions. In panel B, purified Mediator used in binding reactions was resolved by SDS-4 to 12% gradient PAGE and visualized by silver staining. (C) Immobilized GST or GST-MBD derivatives were incubated with nuclear extracts derived from HeLa cells transfected with control (siCNTL) or MED12-specific (siMED12) siRNA as indicated. Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-10% PAGE, and processed by immunoblot analysis for the presence of individual Mediator subunits as indicated. Input represents 5% of total nuclear extract used in binding reactions. (D) Nuclear extracts derived from HeLa cells transfected with FLAG epitope-tagged Gli3 were subjected to immunoprecipitation using CDK8-specific goat polyclonal antibodies or goat IgG as a negative control. Immunoprecipitates were resolved by SDS-10% PAGE and processed by immunoblot analysis for the presence of individual Mediator subunits and FLAG-tagged Gli3 as indicated. Note that FLAG-tagged Gli3 was detected using Gli3-specific antibodies. Input corresponds to 10% of total nuclear extract used in immunoprecipitation reactions.
To determine if Gli3 binds Mediator directly, we examined the ability of GST-Gli3 MBD to bind Mediator purified from a HeLa/S3-derived cell line expressing a hemagglutinin epitope-tagged MED6 Mediator subunit (46). This analysis revealed that GST-Gli3 MBD bound specifically and efficiently to intact purified Mediator, demonstrating that the interaction between Gli3 and Mediator is indeed direct (Fig. 2B).
To determine if the Gli3 MBD binds Mediator exclusively through its MED12 interface, we examined the ability of GST-Gli3 MBD to bind Mediator present in nuclear extracts derived from HeLa cells transfected with control or MED12-specific siRNA. MED12 knockdown (≥80%) was accompanied by a corresponding reduction (≥65%) in the steady-state level of CDK8 protein (Fig. 2C). This result is consistent with our recent finding that RNAi-mediated MED12 depletion in HeLa cells led to a corresponding reduction in the steady-state levels of CDK8 and CycC proteins as well as their stable incorporation into Mediator and further suggests that MED12 is critical to ensure the integrity of a MED12/13/CDK8/CycC module (46). Surprisingly, despite significant depletion of the MED12/13/CDK8/CyC module in MED12 siRNA-transfected cell extracts, GST-Gli3 MBD nonetheless still bound Mediator, as revealed by the presence in pull-down eluates of individual Mediator subunits MED1, MED14, and MED23 (Fig. 2C, lane 4). This observation suggests that the Gli3 MBD interacts with Mediator through at least two different subunits, MED12 within the MED12/13/CDK8/CycC module and an unidentified subunit(s) within the core Mediator lacking this module. Notably, the Gli3 MBD bound MED12-deficient Mediator with apparently lower affinity than MED12-proficient Mediator, since reduced levels of MED1, MED14, and MED23 were recovered from GST-MBD matrices loaded with MED12 knockdown versus control extracts (Fig. 2C; compare lanes 3 and 4). Thus, both MED12 and the unidentified MBD target subunit(s) within core Mediator apparently contribute to maximal binding between Mediator and the Gli3 MBD.
To examine the physical interaction between Gli3 and Mediator in vivo, we employed coimmunoprecipitation analysis using nuclear extracts from HeLa cells transfected with FLAG epitope-tagged Gli3. Immunoprecipitation of intact Mediator with CDK8-specific antibodies resulted in the specific and efficient coimmunoprecipitation of FLAG-tagged Gli3, confirming an association between Gli3 and MED12-proficient Mediator in mammalian cells (Fig. 2D).
The Gli3 MBD is a potent transactivation domain.
Because Gli3 bound directly to isolated MED12 and intact Mediator, we considered it likely that the Gli3 MBD might represent a functional transactivation domain within Gli3. To test this possibility, we expressed the Gli3 MBD as a GAL4 DNA-binding domain fusion protein in HeLa cells and tested the GAL4-MBD chimera for its ability to activate transcription from a GAL4-responsive reporter plasmid. We also tested a subset of the Gli3 1006-1596 amino- and carboxyl-terminal truncation derivatives used in the MED12/Mediator binding experiments for their respective transactivation activities when expressed as GAL4 derivatives (Fig. 3A).
FIG. 3.
The Gli3 MBD is a potent transactivation domain in vivo. (A) Schematic diagram of full-length Gli3 and GAL4-Gli3 truncation derivatives used in transient-transactivation assays. (B) HeLa cells were transfected with expression plasmids for the each of the indicated GAL4-Gli3 derivatives, a GAL4-reponsive luciferase reporter template (pG5E1B-Luc) harboring 5XGAL4 DNA-binding sites upstream of the adenovirus E1B core promoter, and an internal control β-galactosidase expression vector (pact-β-galactosidase). Forty-eight hours posttransfection, harvested whole-cell lysates were processed for luciferase and β-galactosidase activities. Luciferase activities were normalized against β-galactosidase values and expressed relative to the level of luciferase activity obtained in cells transfected without activator, which was arbitrarily assigned a value of 1. In this and all subsequent transient-transactivation assays, error bars represent the standard deviations from the averages of at least three independent transfection experiments performed in duplicate. (C) Transfected whole-cell lysates were resolved by SDS-10% PAGE and processed by immunoblot analysis with GAL4 DNA-binding domain-specific antibodies for the expression levels of each GAL4-Gli3 derivative used in transactivation assays. Note that differences in transactivation activities between GAL4-Gli3 derivatives (B) cannot be explained by differences in their relative expression levels.
Several observations can be made from these functional analyses. First, the Gli3 MBD is a potent transactivator in vivo, with an activity at least 2 orders of magnitude stronger than that of the Gli3 CBD that has previously been shown to function as a physiologically relevant Gli3 transactivation domain (Fig. 3B) (18). Second, an excellent correlation exists between the ability of a particular Gli3 truncation derivative to bind MED12 and intact Mediator in vitro and its ability to function as a potent transactivation domain in vivo. Thus, Gli3 1006-1596 and the Gli3 MBD (1090 to 1596) are both potent transactivation domains in vivo (Fig. 3B) and also bind strongly to MED12 and Mediator in vitro, with the MBD stronger in all respects (Fig. 1D and 2A). By contrast, Gli3 amino-terminal truncation derivative 1132-1596 was significantly reduced in its transactivation capacity in vivo (Fig. 3B) and binds MED12 and Mediator comparatively poorly in vitro (Fig. 1D and 2A). Finally, the Gli3 CBD (827 to 1132) was the weakest transactivator in vivo (Fig. 3B) and did not bind MED12 or Mediator to an appreciable extent in vitro (Fig. 1D and 2A). In summary, Gli3 truncation derivatives within the MBD exhibit the same rank order and relative strength with respect to MED12/Mediator binding activity in vitro and transactivation function in vivo. This observation supports the notion that Mediator, through its MED12 subunit, might represent a functionally important target of Gli3.
The Gli3 MBD interacts functionally with both Mediator and CBP.
Should Gli3 target Mediator for a functional interaction through its MED12 interface, the Gli3-binding domain on MED12 (PQL domain) might be expected to inhibit the transactivation function of the Gli3 MBD through dominant-negative interference. To test this possibility, we examined in HeLa cells the effect of MED12-PQL expression on the transactivation function of GAL4-MBD. This analysis revealed that ectopic expression of the MED12-PQL domain, but not the neighboring MED12-OPA domain to which the Gli3 MBD does not bind, inhibited the transcriptional activity of the MBD in a dose-dependent manner (Fig. 4A). By contrast, the MED12-PQL domain had no effect on the activity of the Gli3 CBD, which binds to CBP, but not to MED12 or Mediator (Fig. 4A). Because the Gli3 MBD partially overlaps the CBD, we also examined a possible CBP requirement for MBD function. To this end, we examined the influence of adenovirus 12S E1A, a functional inhibitor of CBP, on MBD transactivation activity. As expected, ectopic expression of 12S E1A inhibited the transactivation function of GAL4-CBD (Fig. 4B). Strikingly, however, 12S E1A also proved to be a potent dose-dependent inhibitor of GAL4-MBD-directed transactivation, thus implicating CBP in Gli3 MBD function (Fig. 4B). Consistent with this possibility, ectopic overexpression of CBP stimulated, while RNAi-mediated CBP depletion inhibited, Gli3 MBD transactivation activity (Fig. 4C and D). Taken together, these findings reveal the Gli3 MBD to be a composite transactivation domain whose activity likely derives from concerted functional interactions with both Mediator and CBP.
FIG. 4.
The Gli3 MBD interacts functionally with both MED12 and CBP. (A to C) HeLa cells were transfected with expression plasmids for GAL4-Gli3 MBD (G4-MBD) or GAL4-Gli3 CBD (G4-CBD), as indicated, along with the GAL4-reponsive luciferase reporter plasmid pG5E1B-Luc and the internal control pact-β-galactosidase expression plasmid. Transfections additionally included the indicated nanogram quantities of expression plasmids for the MED12 PQL and OPA domains (A), the adenovirus 12S E1A protein (B), or human CBP (C). Forty-eight hours posttransfection, harvested whole-cell lysates were processed for luciferase and β-galactosidase activities. Luciferase activities were calculated as described in the legend to Fig. 3. For comparative purposes on the same plot, the relative luciferase activity of the MBD or CBD alone was arbitrarily assigned a transactivation level of 100%; their corresponding activities in the presence of increasing amounts of other expression plasmids are expressed relative to this value. (D) Top panel: HeLa cells were transfected with control (siCNTL) or CBP-specific (siCBP) siRNA as indicated 48 h prior to transfection with expression plasmids for GAL4-Gli3 MBD (G4-MBD), the pG5E1B-Luc reporter plasmid, and the internal control pact-β-galactosidase expression plasmid. Thirty-six hours following DNA transfections, harvested whole-cell lysates were processed for luciferase and β-galactosidase activities. Relative transactivation levels were calculated as described for panels A to C. Bottom panel: lysates from an aliquot of cells used in a representative transient-transactivation assay were resolved by SDS-10% PAGE and processed by immunoblot analysis with the indicated antibodies specific for CBP or the p89 subunit of transcription factor IIH (TFIIH), the latter of which served as a loading control.
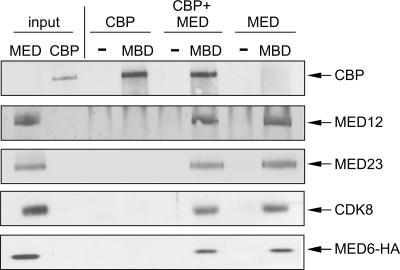
To explore a possible physical basis to explain the functional requirement for both Mediator and CBP in MBD-directed transactivation, we examined the ability of CBP to bind to the Gli3 MBD as a function of Mediator using highly purified proteins. This analysis revealed that purified recombinant CBP bound to GST-Gli3 MBD efficiently and comparably in the absence and presence of purified Mediator (Fig. 5). Collectively, these results reveal the Gli3 MBD to be a platform for the independent physical and functional association of CBP and Mediator.
FIG. 5.
The Gli3 MBD interacts physically with both Mediator and CBP. Immobilized GST (−) or GST-Gli3 MBD (MBD) was incubated with purified CBP, purified Mediator (MED), or both as indicated. Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-9% PAGE, and processed by immunoblot analysis for the presence of CBP and individual Mediator subunits as indicated. Input represents 33% of recombinant CBP or purified Mediator used in binding reactions.
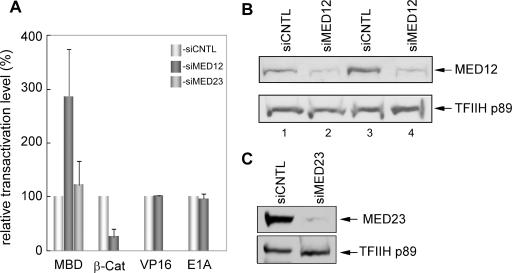
MED12 negatively modulates Gli3 MBD activation domain function.
The observation that the Gli3-binding domain on MED12 (PQL domain) is a dominant-negative inhibitor of the Gli3 MBD suggests that MED12 is likely to be an important functional target of Gli3 within Mediator. To more rigorously examine the functional interaction between the Gli3 MBD and the MED12 interface in Mediator, we monitored the influence of RNAi-mediated MED12 depletion on the transactivation activity of GAL4-MBD. In HeLa cells, MED12-specific siRNA reduced the steady-state level of MED12 protein by ≥60% and, unexpectedly, enhanced MBD transactivation activity by nearly threefold relative to control siRNA (Fig. 6A and B). This surprising observation suggests that MED12 is a negative regulator of Gli3 MBD activation domain function. By contrast, RNAi-mediated MED23 knockdown (≥85%) had no appreciable effect on Gli3 MBD function, demonstrating the specificity of the effect of MED12 knockdown on Gli3 MBD transactivation activity (Fig. 6A and C). Further specificity of the effect of MED12 knockdown on enhancement of Gli3 MBD activity was revealed by analysis of the influence of MED12 knockdown on the activity of the β-catenin transactivation domain. In this case, RNAi-mediated MED12 knockdown (≥60%) inhibited β-catenin transactivation activity by ∼5-fold (Fig. 6A and B), consistent with our recent finding that MED12 is a direct physical and functional target of the β-catenin transactivation domain (46). The fact that MED12 depletion has opposing functional consequences for the Gli3 and β-catenin transactivation domains suggests a mechanistic distinction between these two transactivators that share a common target subunit within Mediator.
FIG. 6.
MED12 depletion enhances Gli3 MBD transactivation activity. HeLa cells were transfected with control (siCNTL), MED12-specific (siMED12), or MED23-specific (siMED23) siRNA as indicated 48 h prior to transfection with expression plasmids for either GAL4-Gli3 MBD (MBD), GAL4-β-catenin (β-Cat), GAL4-VP16 (VP16), or GAL4-E1A (E1A) along with the pG5E1B-Luc reporter plasmid and the internal control pact-β-galactosidase expression plasmid. Thirty-six hours following DNA transfections, harvested whole-cell lysates were processed for luciferase and β-galactosidase activities. Luciferase activities were calculated as described in the legend to Fig. 3. For comparative purposes on the same plot, the relative luciferase activity of each activation domain in control siRNA-transfected cells was arbitrarily assigned a transactivation level of 100%; their corresponding activities in MED12-specific or MED23-specific siRNA-transfected cells are expressed relative to this value. (B and C) Cell lysates from representative transient-transactivation assays in panel A were resolved by SDS-10% PAGE and processed by immunoblot analysis with the indicated antibodies specific for MED12, MED23, or the p89 subunit of TFIIH, the last of which served as a loading control. Note that GAL4-MBD, GAL4-VP16, and GAL4-E1A transfections were performed in parallel using the same pool of control or MED12 knockdown cells. Hence, for the immunoblots shown in panel B, lanes 1 and 2 corresponding to control and MED12 knockdown lysates are representative for transfections involving the MBD, VP16, and E1A transactivation domains. Lanes 3 and 4 are representative of transfections with the β-catenin transactivation domain, which involved an independent pool of control or MED12 knockdown cells.
To confirm the specificity of the effect of MED12 knockdown on both the Gli3 and β-catenin transactivation domains, we also monitored the influence of RNAi-mediated MED12 depletion on the activities of the herpes simplex virus VP16 and adenovirus E1A transactivation domains, whose established targets in Mediator are MED17/25 and MED23, respectively (9, 33, 67, 101). This analysis revealed that MED12 knockdown (≥60%) had no appreciable influence on the activities of either the VP16 or the E1A transactivation domains (Fig. 6A and B). Thus, the influence of MED12 knockdown appears to be restricted functionally to the Gli3 and β-catenin transactivation domains, both of which physically target Mediator for direct interaction through its MED12 interface.
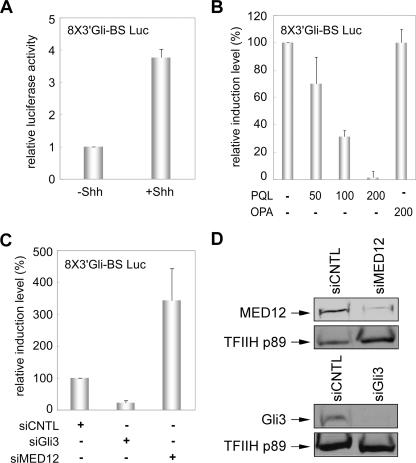
MED12 negatively modulates Gli3-dependent Shh signal transduction.
To characterize the functional interaction between Gli3 and MED12 during Shh signal transduction, we initially established an assay for Gli3-dependent stimulation of a Gli-binding site reporter plasmid in response to Shh signaling. Accordingly, murine CH310T1/2 cells were transfected with a Gli-responsive luciferase reporter plasmid (8X3′Gli-BS Luc) bearing eight copies of a Gli-binding site derived from the murine HNF3β floor plate enhancer (82), and 24 h later transfected cells were treated with conditioned medium from control or Shh-expressing 293 cells. Relative to control-conditioned medium, Shh-conditioned medium induced reporter gene activity by ∼4-fold, in a manner largely reversible by concurrent Gli3 knockdown (Fig. 7A and C). Thus, in CH310T1/2 cells, the 8X3′Gli-BS Luc reporter gene is induced by Shh in a Gli3-dependent manner. Next, we monitored the influence of MED12 PQL expression and MED12 knockdown on Gli3-dependent reporter gene activity in response to Shh signaling. Reporter gene activity induced by Shh was largely abrogated by ectopic expression of the MED12 PQL domain (Fig. 7B) but enhanced over threefold by RNAi-mediated MED12 knockdown (≥65%) (Fig. 7C and D). These results are congruent with our earlier finding that MED12 suppresses the transactivation function of the isolated MBD and further support the notion that MED12 is a negative regulator of Shh-induced Gli3 activator function.
FIG. 7.
Functional interaction between Gli3 and MED12 during induction of an episomal Shh-responsive reporter gene. (A) CH310T1/2 cells were transfected with the 8X3′Gli-BS Luc reporter plasmid bearing eight copies of a Gli-binding site from the murine HNF3β floor plate enhancer along with the internal control pact-β-galactosidase expression plasmid. Twenty-four hours posttransfection, cells were treated with conditioned medium from control (−Shh) or Shh-expressing (+Shh) 293 cells as indicated for an additional 24 h prior to processing of harvested whole-cell lysates for luciferase and β-galactosidase activities. Luciferase activities were normalized against β-galactosidase values and expressed relative to the level of luciferase activity obtained in transfected cells treated with control-conditioned medium, which was arbitrarily assigned a value of 1. (B) CH310T1/2 cells were transfected with the 8X3′Gli-BS Luc reporter plasmid and the internal control pact-β-galactosidase expression plasmid without or with the indicated nanogram quantities of expression plasmids for the MED12 PQL and OPA domains. Twenty-four hours posttransfection, cells were treated with conditioned medium from control or Shh-expressing 293 cells for an additional 24 h prior to processing of harvested whole-cell lysates for luciferase and β-galactosidase activities. Relative luciferase activities were calculated as described for panel A. For comparative purposes on the same plot, the level of Shh-induced reporter gene activity obtained in the absence of cotransfected PQL or OPA expression plasmids was arbitrarily assigned a value of 100%; corresponding induction levels in the presence of increasing amounts of the PQL or OPA expression plasmids are expressed relative to this value. (C) CH310T1/2 cells were electroporated with control (siCNTL), Gli3-specific (siGli3), or MED12-specific (siMED12) siRNA as indicated 24 h prior to transfection with the 8X3′Gli-BS Luc reporter plasmid and the internal control pact-β-galactosidase expression plasmid. Twenty-four hours following DNA transfections, cells were treated with conditioned medium from control or Shh-expressing 293 cells for an additional 24 h prior to processing of harvested whole-cell lysates for luciferase and β-galactosidase activities. Relative luciferase activities were calculated as described for panel A. For comparative purposes on the same plot, the level of Shh-induced reporter gene activity obtained in control siRNA-transfected cells was arbitrarily assigned a value of 100%; corresponding induction levels in Gli3-specific and MED12-specific siRNA-transfected cells are expressed relative to this value. (D) Cell lysates from a representative transient transactivation assay in panel C were resolved by SDS-10% PAGE and processed by immunoblot analysis with the indicated antibodies specific for MED12, Gli3, or the p89 subunit of TFIIH, the last of which served as a loading control.
To examine the functional interaction between Gli3 and MED12 under more biologically relevant conditions, we monitored the influence of MED12 PQL expression and MED12 knockdown on Gli3-dependent activation of endogenous Gli3 target genes in response to Shh signaling. For these experiments, we monitored expression of the Gli1 and cyclin D1 genes, both of which have been reported to be direct transcriptional targets of Gli3 (18, 28). Treatment of CH310T1/2 cells for 24 hours with conditioned medium from Shh-expressing 293 cells, but not control 293 cells, induced Gli1 and cyclin D1 mRNAs by ∼6-fold and ∼4.5-fold, respectively, as revealed by quantitative RT-PCR analysis (Fig. 8A). Induction of both Gli1 and cyclin D1 was completely blocked by cyclopamine, a pharmacological inhibitor of Smo (17, 31, 91), thus confirming that Gli1 and cyclin D1 activation in this system derives from Shh signal transduction (Fig. 8A). Furthermore, RNAi-mediated Gli3 knockdown (≥70%) reduced Gli1 and cyclin D1 induction by 50% and 60%, respectively, thereby implicating Gli3 in the activation of Gli1 and cyclin D1 gene transcription in response to Shh signaling (Fig. 8B).
FIG. 8.
Functional interaction between Gli3 and MED12 during induction of endogenous Shh target genes. (A) CH310T1/2 cells cultured for 24 h in control (−Shh) or Shh-conditioned (+Shh) medium with or without cyclopamine (5 μM) as indicated were harvested, and RNA was processed by quantitative RT-PCR analysis for the level of Gli1, cyclin D1, and β-actin mRNAs. Gli1 and cyclin D1 RNA levels were normalized to β-actin levels and expressed relative to the level of Gli1 and cyclin D1 RNAs in control cells without cyclopamine, which was arbitrarily assigned a value of 1. Error bars represent the standard deviations from the averages of three independent experiments. (B and D) CH310T1/2 cells were transfected with control (siCNTL), Gli3-specific (siGli3), or MED12-specific (siMED12) siRNA as indicated, and 48 h later, cells were treated with control or Shh-conditioned medium for an additional 24 h. Top panels: extracted RNA was processed by quantitative RT-PCR analysis for the level of Gli, cyclin D1, and β-actin mRNAs. Relative RNA levels were calculated as described in for panel A. For comparative purposes on the same plot, Gli1 and cyclin D1 induction levels in control siRNA-transfected cells were arbitrarily assigned a value of 100%; the corresponding induction level for each gene in Gli3-specific (B) or MED12-specific (D) siRNA-transfected cells is expressed relative to this value. Error bars represent the standard deviations from the averages of three independent experiments. Bottom panels: cell lysates from representative quantitative RT-PCR analyses in panels B and D were resolved by SDS-10% PAGE and processed by immunoblot analysis with the indicated antibodies specific for Gli3, MED12, or the p89 subunit of TFIIH, the last of which served as a loading control. (C) CH310T1/2 cells were transfected without or with 2 μg of an expression plasmid for the MED12 PQL domain as indicated. Twenty-four hours posttransfection, cells were treated with conditioned medium from control or Shh-expressing 293 cells for an additional 24 h. Extracted RNA was processed by quantitative RT-PCR analysis for the level of Gli1 and β-actin mRNAs. Relative RNA levels were calculated as described for panel A. For comparative purposes on the same plot, the Gli1 induction level in cells transfected without the PQL domain was arbitrarily assigned a value of 100%; the corresponding Gli1 induction level cells transfected with the PQL domain is expressed relative to this value.
Having thus established that Gli1 and cyclin D1 gene transcription in CH310T1/2 cells is induced by Shh in a Gli3-dependent manner, we next examined the role of MED12 in this process. Ectopic expression of the MED12 PQL domain inhibited Gli3-dependent induction of Gli1 in response to Shh by ∼70% (Fig. 8C). On the other hand, RNAi-mediated MED12 knockdown (≥60%) enhanced Shh-mediated Gli3-dependent induction of Gli1 and cyclin D1 by ∼3-fold and ∼2-fold, respectively (Fig. 8D). In terms of actual induction levels, Shh induced Gli1 and cyclin D1 expression by an average of ∼8-fold and ∼3-fold, respectively, in control siRNA-treated cells and by an average of ∼22-fold and ∼6-fold, respectively, in MED12-depleted cells. These results are consistent with our earlier findings that MED12 suppresses the transactivation function of the Gli3 MBD on both artificial and natural binding site reporter plasmids and further reveals MED12 to be a negative regulator of Gli3 activator function.
MED12-proficient Mediator is recruited to the Gli1 promoter in a Shh-dependent manner.
To further examine the functional interaction between Gli3 and Mediator, we employed ChIP analysis to monitor occupancy of Gli-binding sites within the Gli1 promoter by Gli3 and Mediator as a function of Shh signaling. To this end, CH310T1/2 cells were treated with control or Shh-conditioned medium for 6 h prior to ChIP analysis. We confirmed by quantitative RT-PCR analysis that Gli1 gene expression was induced by Shh under these conditions; relative to control medium, Shh-conditioned medium stimulated Gli1 expression by ∼4-fold (Fig. 9A). As expected, Gli3 was detected by ChIP on the Gli1 promoter in both the absence and the presence of Shh signaling (Fig. 9B), an observation concordant with the fact that the Gli3 N-terminal specific antibodies used in this ChIP analysis recognize both the processed Gli3 repressor and full-length Gli3 activator species, and the former is predicted to occupy Gli-binding sites in the absence of Shh signaling (29, 54, 55, 79, 82, 93, 99). Notably, however, occupancy of the Gli1 promoter by the Mediator subunit CDK8 was observed only in Shh-treated cells (Fig. 9B). Because CDK8 is a reliable indicator of the MED12/13/CDK8/CycC Mediator module and MED12 is essential for incorporation of CDK8 into Mediator (46), this observation suggests that the Mediator species recruited to the Gli1 promoter in response to Shh is Mediator containing the MED12/13/CDK8/CycC module.
FIG. 9.
MED12-proficient Mediator is recruited to the Gli1 promoter in a Shh-dependent manner. (A and B) CH310T1/2 cells were cultured for 6 h in control (−Shh) or Shh-conditioned (+Shh) medium as indicated. (A) Total RNA recovered from CH310T1/2 cells cultured in control or Shh-conditioned medium was processed by quantitative RT-PCR analysis for the levels of Gli1 and β-actin mRNAs. Relative RNA levels were calculated as described in the legend to Fig. 8A. (B) Soluble chromatin prepared from CH310T1/2 cells cultured in control or Shh-conditioned medium was subjected to immunoprecipitation with rabbit IgG as a negative control or rabbit polyclonal antibodies specific for Gli3 or CDK8 as indicated. Immunoprecipitated DNA was PCR amplified using primers that span Gli-binding sites within the Gli1 gene (positions +91 to +413 relative to the transcription start site). Input corresponds to 0.25% of the soluble chromatin that was subjected to immunoprecipitation.
DISCUSSION
The underlying mechanisms by which Gli proteins regulate target gene transcription in response to Shh signaling remain to be fully clarified. Herein, we have investigated the molecular and cellular requirements for human Gli3 activator function. Of principal import, we identify and characterize a carboxyl-terminal Gli3 transactivation domain (MBD) whose activity derives from concerted functional interactions with both Mediator and CBP. This finding extends a previous report of autonomous transactivation function within this region of Gli3 by providing a molecular basis to explain its transactivation activity (42). A physical basis to explain the functional requirement for both Mediator and CBP in Gli3 transactivation derives from our observation that both Mediator and CBP can independently bind to the Gli3 MBD in vitro. Currently, we do not know whether Gli3 might target CBP and Mediator sequentially or simultaneously in vivo. Nonetheless, our collective findings reveal the Gli3 MBD to be a composite transactivation domain that may function at two distinct steps in the transcriptional activation process—CBP-dependent chromatin modification and Mediator-dependent RNA polymerase II transcription complex activation.
While previous studies have directly implicated CBP in Gli3-dependent Shh signaling, the contribution of Mediator to this process has been less clear. Thus, while previous genetic studies of Drosophila have implicated Mediator subunits MED12 and MED13 in Hh signal transduction (36, 94), it has heretofore remained unknown whether and how Mediator plays a direct or indirect role in Shh signaling. Our work extends these studies and provides the first evidence to implicate Mediator directly in Shh signal transduction through a demonstrated physical and functional interaction between Gli3 and the MED12 interface in Mediator. First, we show that Gli3 binds specifically and directly to isolated MED12 and intact Mediator both in vitro and in vivo. Second, we observe that mutant derivatives of the Gli3 MBD exhibit the same rank order and relative strength with respect to MED12/Mediator binding activity in vitro and transactivation function in vivo. Third, we demonstrate that disruption of the Gli3-MED12 interaction through dominant-negative interference or RNAi-mediated MED12 depletion has profound functional consequences for induction of Gli3 target genes in response to Shh signaling. Finally, we observed recruitment of MED12-containing Mediator to a Gli3-target gene in response to Shh signaling. Collectively, these findings suggest that MED12 is an important functional target of Gli3 within Mediator.
In an effort to investigate the mechanistic consequence of the Gli3-MED12 interaction, we examined the influence of RNAi-mediated MED12 depletion on Gli3 transactivation activity. Unexpectedly, we observed that MED12 depletion enhanced Gli3 MBD transactivation function as well as Gli3-dependent induction of Gli1 and cyclin D1 gene transcription in response to Shh signaling. How might these observations be reconciled with the observation that the Gli3-binding domain on MED12 (PQL domain) inhibits Gli3 transactivation in a dominant-negative manner? We propose a model whereby a Mediator-imposed constraint on Gli3 transactivation is relieved only by direct interaction of the Gli3 MBD with the MED12 interface in Mediator (Fig. 10). Hence, ectopic expression of the MED12 PQL domain would preclude a functional interaction between Gli3 and MED12 necessary to reverse a Mediator-dependent constraint on Gli3 transactivation, one revealed only by RNAi-mediated MED12 depletion. Interestingly, unlike most other signaling pathways, Hh signal transduction proceeds largely by sequential repressive interactions, including effects of Ptch on Smo (90). Our findings indicate that this theme extends to the endpoint of Hh signaling in the nucleus, wherein MED12 is targeted by Gli3 to reverse Mediator-dependent inhibition of Shh signaling.
FIG. 10.
Schematic model to describe the functional interaction between Gli3, CBP, and Mediator. Solid and broken lines represent established and hypothetical protein interactions, respectively. Arrowheads and capped lines represent cooperative and antagonistic protein interactions, respectively. We propose that activated Gli3, through its MBD, physically targets both CBP and the MED12 interface in Mediator, the latter to reverse Mediator-dependent suppression of Shh target gene (i.e., Gli1 or cyclin D1) transcription. An established interaction between Gli3 and a presently unidentified target subunit(s) within core Mediator could account for MED12-independent recruitment of Mediator to Gli3 target genes. In this model, the MED12/13/CDK8/CycC module within Mediator is shown to be responsible for suppression of transcription, consistent with its established repression function. However, the actual basis for Mediator-dependent suppression of Gli3 target gene transcription remains to be established.
Our findings are consistent with a model whereby Gli3 targets MED12 in order to reverse Mediator-dependent suppression of Gli3 transactivation activity; however, whether and how MED12 contributes directly to such repression remain to be established. A role for MED12 in the negative regulation of Gli3 transactivation is consistent with a large body of evidence linking MED12 to transcriptional repression. For example, MED12, along with Mediator subunits MED13, CDK8, and CycC, marks a transcriptionally repressive form of Mediator (Arc-L) that is biochemically and structurally distinguishable from an activating form of Mediator (CRSP) lacking this module and additionally containing MED26 (Crsp70) (87). Within the repressive MED12/13/CDK8/CycC module, CDK8/CycC has previously been shown to directly repress transcription through phosphorylation and consequent inactivation of the RNA polymerase II CTD kinase activity of TFIIH (2). Thus, disruption of MED12, which anchors CDK8/CycC within Mediator (46), could abrogate CDK8/CyC-mediated repression. Alternatively, MED12 itself might contribute directly to repression through presently unknown mechanisms. Further studies will be required to distinguish among these possibilities.
Regarding the switch between transcriptionally repressive and active Mediator species, current data suggest a model in which the MED12/13/CDK8/CycC module is displaced upon activation of transcription. The mechanism and regulation relevant to this switch have only recently begun to be clarified. Recent work suggests that gene-specific cofactors (i.e., PARP-1) may be important to facilitate an activator-induced reconfiguration in Mediator composition (78). Our work suggests a potential additional mechanism for this switch whereby an activation domain (Gli3 MBD) directly targets the MED12/13/CDK8/CycC module to overcome repression. Whether additional cofactors are also required to reconfigure Mediator during Gli3 transactivation will require further evaluation.
Interestingly, we observed that while MED12 depletion enhanced Gli3 MBD activity, it nonetheless reduced β-catenin transactivation domain function, consistent with our recent report that MED12 is a direct functional target of the β-catenin transactivation domain (46). Thus, depletion of a common target subunit within Mediator has opposing functional consequences for two distinct transactivation domains. One simple interpretation is that the β-catenin-MED12 interaction facilitates Mediator recruitment, while the Gli3-MED12 interaction triggers a postrecruitment step in the activation process (97), one that concomitantly reconfigures Mediator and reverses negative regulation imposed by the MED12/13/CDK8/CycC module in Mediator. According to this model, Mediator recruitment to Gli3 target promoters would likely occur in a MED12-independent manner. While chromatin immunoprecipitation analysis revealed Shh-induced recruitment of MED12-proficient Mediator to the Gli1 promoter, our data cannot presently establish whether such recruitment is dependent on MED12. Probable MED12-independent recruitment of Mediator to Gli3 target genes could be achieved through any of several alternative possibilities (Fig. 10). First, the Gli3 MBD could recruit Mediator directly through a subunit other than MED12. Second, the MBD could recruit Mediator indirectly through CBP or another component of the transcriptional machinery that is assembled in a Gli3-dependent manner. Our finding that the Gli3 MBD can bind to MED12-deficient Mediator leads us to favor the former possibility. Nonetheless, further experiments will be required to establish the means by which Mediator is recruited to Gli3 target genes.
Our conclusion that MED12 is a negative regulator of Gli3-dependent Shh signaling is not wholly unsupported by genetic studies in model metazoan organisms. For example in the Drosophila eye disc, mutational inactivation of MED12 results in persistent expression of the Hh response genes decapentaplegic and atonal posterior to the morphogenetic furrow, where cells are normally unresponsive to Hh, as well as reduced expression of these genes in the morphogenetic furrow where they are normally expressed (94). The former observation would be consistent with a role for MED12 in the negative regulation of Hh signaling if, for example, MED12 was important to establish a threshold response to a gradient of Hh. Reduced expression of Hh response genes in certain cell contexts could derive from the combinatorial input of other developmental signaling pathways in which MED12 plays a positive role. In fact, the phenotypes of MED12 mutations in model metazoan organisms are quite complex and do not clearly phenocopy mutations in any one developmental signaling pathway. It therefore seems likely that the MED12 interface within Mediator functions to process and integrate a diverse range of developmental signals, of which Hh is but one. In support of this possibility, MED12 was recently identified as one of six highly connected “hub” genes linked to multiple developmental signaling pathways during a systematic genetic mapping exercise in Caenorhabditis elegans (52). Our identification herein of MED12 as a negative regulator of Gli3-dependent Shh signaling, coupled with our recent finding that MED12 is a transducer of Wnt/β-catenin signaling (46), further supports the idea that MED12 may function as a global integrator of diverse developmental signaling networks.
Finally, the role of MED12 as a negative regulator of Shh signal transduction could have important biological implications for the proper physiological response to a graded Shh signal. A central issue in developmental biology concerns the means by which morphogenetic gradients elicit differential gene expression and alter cell fates. In this regard, the influence of Shh on the specification of neuronal cell fates in the developing spinal cord has served as a paradigm for the study of vertebrate morphogenetic gradients. In the developing neural tube, a long-range gradient of Shh signaling is sufficient to direct the emergence of distinct neuronal subtypes at precise positions along the dorsal-ventral axis (35, 37). Recent studies suggest that this Shh gradient is transduced into a gradient of Gli transcription factor activity that, in turn, controls the differential expression of homeodomain and helix-loop-helix transcription factors that define distinct neuronal progenitor domains (83). Although the precise mechanism(s) responsible for differential control of gene expression through a gradient of Gli transcription factor activity remains to be elucidated, several strategies have been invoked, each of which functions to effectively alter the threshold response of target genes to a gradient of Gli transcription factor activity. These include differential binding affinities among Gli response elements as well as the presence of additional regulatory elements that bind synergistic activators and/or repressor proteins (83). Our data suggest the possibility of an additional mechanism that involves intrinsic modulation of Gli transcription factor activity by Mediator. In this regard, cell-type-specific changes in MED12 expression and/or activity could alter the concentration threshold of Shh required to induce Gli3-dependent gene activation. Future studies should clarify this issue and further reveal the contribution of Mediator to the entire spectrum of developmental and pathological processes driven by Shh signaling.
Acknowledgments
We thank the following individuals for their generous gifts of reagents used in this study: Susan Mackem for Gli3 antibody, Michael Garabedian for MED14 antibody, Paul Lieberman for mammalian and baculovirus expression vectors for human CBP, Hiroshi Sasaki for the 8X3′Gli-BS Luc reporter plasmid, Andreas Hecht for GAL4- and GST-β-catenin expression plasmids, and Arnold Berk for the GAL4-E1A and GAL4-VP16 expression plasmids. We are grateful to Pao-Tien Chuang and Alan Brooks for advice regarding production of Shh-conditioned medium. We also thank Amy Trauernicht, Ning Ding, Xuan Xu, Sejin Kim, Paula Garza, Wei Tan, and P. Renee Yew for advice, discussion, and comments.
This work was supported by Public Health Service grant CA-0908301 from the National Cancer Institute (T.G.B.) and by U.S. Army Department of Defense BCRP grants DAMD17-03-1-0272 and DAMD17-02-1-0584 (T.G.B.).
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Akimaru, H., Y. Chen, P. Dai, D. X. Hou, M. Nonaka, S. M. Smolik, S. Armstrong, R. H. Goodman, and S. Ishii. 1997. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386:735-738. [DOI] [PubMed] [Google Scholar]
- 2.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 3.Alexandre, C., A. Jacinto, and P. W. Ingham. 1996. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10:2003-2013. [DOI] [PubMed] [Google Scholar]
- 4.Aza-Blanc, P., F. A. Ramirez-Weber, M. P. Laget, C. Schwartz, and T. B. Kornberg. 1997. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89:1043-1053. [DOI] [PubMed] [Google Scholar]
- 5.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol. Cell. Biol. 22:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai, C. B., W. Auerbach, J. S. Lee, D. Stephen, and A. L. Joyner. 2002. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129:4753-4761. [DOI] [PubMed] [Google Scholar]
- 7.Bai, C. B., D. Stephen, and A. L. Joyner. 2004. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6:103-115. [DOI] [PubMed] [Google Scholar]
- 8.Basler, K., and G. Struhl. 1994. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368:208-214. [DOI] [PubMed] [Google Scholar]
- 9.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 10.Buttitta, L., R. Mo, C. C. Hui, and C. M. Fan. 2003. Interplays of Gli2 and Gli3 and their requirement in mediating Shh-dependent sclerotome induction. Development 130:6233-6243. [DOI] [PubMed] [Google Scholar]
- 11.Cantin, G. T., J. L. Stevens, and A. J. Berk. 2003. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. USA 100:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpenter, D., D. M. Stone, J. Brush, A. Ryan, M. Armanini, G. Frantz, A. Rosenthal, and F. J. de Sauvage. 1998. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc. Natl. Acad. Sci. USA 95:13630-13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C. H., D. P. von Kessler, W. Park, B. Wang, Y. Ma, and P. A. Beachy. 1999. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98:305-316. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C. J., Z. Deng, A. Y. Kim, G. A. Blobel, and P. M. Lieberman. 2001. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21:476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Y., and G. Struhl. 1996. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87:553-563. [DOI] [PubMed] [Google Scholar]
- 16.Conaway, R. C., S. Sato, C. Tomomori-Sato, T. Yao, and J. W. Conaway. 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30:250-255. [DOI] [PubMed] [Google Scholar]
- 17.Cooper, M. K., J. A. Porter, K. E. Young, and P. A. Beachy. 1998. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 280:1603-1607. [DOI] [PubMed] [Google Scholar]
- 18.Dai, P., H. Akimaru, Y. Tanaka, T. Maekawa, M. Nakafuku, and S. Ishii. 1999. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274:8143-8152. [DOI] [PubMed] [Google Scholar]
- 19.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding, Q., J. Motoyama, S. Gasca, R. Mo, H. Sasaki, J. Rossant, and C. C. Hui. 1998. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125:2533-2543. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez, M., M. Brunner, E. Hafen, and K. Basler. 1996. Sending and receiving the hedgehog signal: control by the Drosophila Gli protein Cubitus interruptus. Science 272:1621-1625. [DOI] [PubMed] [Google Scholar]
- 22.Echelard, Y., D. J. Epstein, B. St.-Jacques, L. Shen, J. Mohler, J. A. McMahon, and A. P. McMahon. 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75:1417-1430. [DOI] [PubMed] [Google Scholar]
- 23.Forbes, A. J., Y. Nakano, A. M. Taylor, and P. W. Ingham. 1993. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev. Suppl. 1993:115-124. [PubMed] [Google Scholar]
- 24.Fuse, N., T. Maiti, B. Wang, J. A. Porter, T. M. Hall, D. J. Leahy, and P. A. Beachy. 1999. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc. Natl. Acad. Sci. USA 96:10992-10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodrich, L. V., R. L. Johnson, L. Milenkovic, J. A. McMahon, and M. P. Scott. 1996. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 10:301-312. [DOI] [PubMed] [Google Scholar]
- 26.Heberlein, U., T. Wolff, and G. M. Rubin. 1993. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75:913-926. [DOI] [PubMed] [Google Scholar]
- 27.Hooper, J. E., and M. P. Scott. 2005. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6:306-317. [DOI] [PubMed] [Google Scholar]
- 28.Hu, M. C., R. Mo, S. Bhella, C. W. Wilson, P. T. Chuang, C. C. Hui, and N. D. Rosenblum. 2006. GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development 133:569-578. [DOI] [PubMed] [Google Scholar]
- 29.Hui, C. C., and A. L. Joyner. 1993. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 3:241-246. [DOI] [PubMed] [Google Scholar]
- 30.Hynes, M., D. M. Stone, M. Dowd, S. Pitts-Meek, A. Goddard, A. Gurney, and A. Rosenthal. 1997. Control of cell pattern in the neural tube by the zinc finger transcription factor and oncogene Gli-1. Neuron 19:15-26. [DOI] [PubMed] [Google Scholar]
- 31.Incardona, J. P., W. Gaffield, R. P. Kapur, and H. Roelink. 1998. The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development 125:3553-3562. [DOI] [PubMed] [Google Scholar]
- 32.Ingham, P. W., and A. P. McMahon. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15:3059-3087. [DOI] [PubMed] [Google Scholar]
- 33.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 34.Ito, M., C. X. Yuan, H. J. Okano, R. B. Darnell, and R. G. Roeder. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5:683-693. [DOI] [PubMed] [Google Scholar]
- 35.Jacob, J., and J. Briscoe. 2003. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 4:761-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janody, F., Z. Martirosyan, A. Benlali, and J. E. Treisman. 2003. Two subunits of the Drosophila mediator complex act together to control cell affinity. Development 130:3691-3701. [DOI] [PubMed] [Google Scholar]
- 37.Jessell, T. M. 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1:20-29. [DOI] [PubMed] [Google Scholar]
- 38.Jiang, J. 2002. Degrading Ci: who is Cul-pable? Genes Dev. 16:2315-2321. [DOI] [PubMed] [Google Scholar]
- 39.Johnston, J. J., I. Olivos-Glander, C. Killoran, E. Elson, J. T. Turner, K. F. Peters, M. H. Abbott, D. J. Aughton, A. S. Aylsworth, M. J. Bamshad, C. Booth, C. J. Curry, A. David, M. B. Dinulos, D. B. Flannery, M. A. Fox, J. M. Graham, D. K. Grange, A. E. Guttmacher, M. C. Hannibal, W. Henn, R. C. Hennekam, L. B. Holmes, H. E. Hoyme, K. A. Leppig, A. E. Lin, P. Macleod, D. K. Manchester, C. Marcelis, L. Mazzanti, E. McCann, M. T. McDonald, N. J. Mendelsohn, J. B. Moeschler, B. Moghaddam, G. Neri, R. Newbury-Ecob, R. A. Pagon, J. A. Phillips, L. S. Sadler, J. M. Stoler, D. Tilstra, C. M. Walsh Vockley, E. H. Zackai, T. M. Zadeh, L. Brueton, G. C. Black, and L. G. Biesecker. 2005. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of GLI3 mutations. Am. J. Hum. Genet. 76:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalderon, D. 2005. The mechanism of hedgehog signal transduction. Biochem. Soc. Trans. 33:1509-1512. [DOI] [PubMed] [Google Scholar]
- 41.Kalderon, D. 2000. Transducing the hedgehog signal. Cell 103:371-374. [DOI] [PubMed] [Google Scholar]
- 42.Kalff-Suske, M., A. Wild, J. Topp, M. Wessling, E. M. Jacobsen, D. Bornholdt, H. Engel, H. Heuer, C. M. Aalfs, M. G. Ausems, R. Barone, A. Herzog, P. Heutink, T. Homfray, G. Gillessen-Kaesbach, R. Konig, J. Kunze, P. Meinecke, D. Muller, R. Rizzo, S. Strenge, A. Superti-Furga, and K. H. Grzeschik. 1999. Point mutations throughout the GLI3 gene cause Greig cephalopolysyndactyly syndrome. Hum. Mol. Genet. 8:1769-1777. [DOI] [PubMed] [Google Scholar]
- 43.Kasper, M., G. Regl, A. M. Frischauf, and F. Aberger. 2006. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur. J. Cancer 42:437-445. [DOI] [PubMed] [Google Scholar]
- 44.Kato, Y., R. Habas, Y. Katsuyama, A. M. Naar, and X. He. 2002. A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature 418:641-646. [DOI] [PubMed] [Google Scholar]
- 45.Kim, J. H., Z. Huang, and R. Mo. 2005. Gli3 null mice display glandular overgrowth of the developing stomach. Dev. Dyn. 234:984-991. [DOI] [PubMed] [Google Scholar]
- 46.Kim, S., X. Xu, A. Hecht, and T. G. Boyer. 2006. Mediator is a transducer of Wnt/beta-catenin signaling. J. Biol. Chem. 281:14066-14075. [DOI] [PubMed] [Google Scholar]
- 47.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30:235-239. [DOI] [PubMed] [Google Scholar]
- 48.Kuras, L., T. Borggrefe, and R. D. Kornberg. 2003. Association of the Mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. USA 100:13887-13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau, J. F., I. Nusinzon, D. Burakov, L. P. Freedman, and C. M. Horvath. 2003. Role of metazoan mediator proteins in interferon-responsive transcription. Mol. Cell. Biol. 23:620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, J., K. A. Platt, P. Censullo, and A. Ruiz i Altaba. 1997. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 124:2537-2552. [DOI] [PubMed] [Google Scholar]
- 51.Lee, J. J., D. P. von Kessler, S. Parks, and P. A. Beachy. 1992. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71:33-50. [DOI] [PubMed] [Google Scholar]
- 52.Lehner, B., C. Crombie, J. Tischler, A. Fortunato, and A. G. Fraser. 2006. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 38:896-903. [DOI] [PubMed] [Google Scholar]
- 53.Lei, Q., A. K. Zelman, E. Kuang, S. Li, and M. P. Matise. 2004. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development 131:3593-3604. [DOI] [PubMed] [Google Scholar]
- 54.Litingtung, Y., and C. Chiang. 2000. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 3:979-985. [DOI] [PubMed] [Google Scholar]
- 55.Litingtung, Y., R. D. Dahn, Y. Li, J. F. Fallon, and C. Chiang. 2002. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418:979-983. [DOI] [PubMed] [Google Scholar]
- 56.Liu, Y., C. Kung, J. Fishburn, A. Z. Ansari, K. M. Shokat, and S. Hahn. 2004. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 24:1721-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lum, L., and P. A. Beachy. 2004. The Hedgehog response network: sensors, switches, and routers. Science 304:1755-1759. [DOI] [PubMed] [Google Scholar]
- 58.Ma, C., Y. Zhou, P. A. Beachy, and K. Moses. 1993. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75:927-938. [DOI] [PubMed] [Google Scholar]
- 59.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30:256-263. [DOI] [PubMed] [Google Scholar]
- 60.Mann, R. K., and P. A. Beachy. 2004. Novel lipid modifications of secreted protein signals. Annu. Rev. Biochem. 73:891-923. [DOI] [PubMed] [Google Scholar]
- 61.Marigo, V., R. A. Davey, Y. Zuo, J. M. Cunningham, and C. J. Tabin. 1996. Biochemical evidence that patched is the Hedgehog receptor. Nature 384:176-179. [DOI] [PubMed] [Google Scholar]
- 62.Matise, M. P., D. J. Epstein, H. L. Park, K. A. Platt, and A. L. Joyner. 1998. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development 125:2759-2770. [DOI] [PubMed] [Google Scholar]
- 63.McMahon, A. P., P. W. Ingham, and C. J. Tabin. 2003. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53:1-114. [DOI] [PubMed] [Google Scholar]
- 64.Methot, N., and K. Basler. 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96:819-831. [DOI] [PubMed] [Google Scholar]
- 65.Ming, J. E., E. Roessler, and M. Muenke. 1998. Human developmental disorders and the Sonic hedgehog pathway. Mol. Med. Today 4:343-349. [DOI] [PubMed] [Google Scholar]
- 66.Mittler, G., E. Kremmer, H. T. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mittler, G., T. Stuhler, L. Santolin, T. Uhlmann, E. Kremmer, F. Lottspeich, L. Berti, and M. Meisterernst. 2003. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 22:6494-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mohler, J., and K. Vani. 1992. Molecular organization and embryonic expression of the hedgehog gene involved in cell-cell communication in segmental patterning of Drosophila. Development 115:957-971. [DOI] [PubMed] [Google Scholar]
- 69.Motoyama, J., L. Milenkovic, M. Iwama, Y. Shikata, M. P. Scott, and C. C. Hui. 2003. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev. Biol. 259:150-161. [DOI] [PubMed] [Google Scholar]
- 70.Motoyama, J., T. Takabatake, K. Takeshima, and C. Hui. 1998. Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat. Genet. 18:104-106. [DOI] [PubMed] [Google Scholar]
- 71.Murone, M., A. Rosenthal, and F. J. de Sauvage. 1999. Sonic hedgehog signaling by the patched-smoothened receptor complex. Curr. Biol. 9:76-84. [DOI] [PubMed] [Google Scholar]
- 72.Nusslein-Volhard, C., and E. Wieschaus. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287:795-801. [DOI] [PubMed] [Google Scholar]
- 73.Ogden, S. K., M. Ascano, Jr., M. A. Stegman, and D. J. Robbins. 2004. Regulation of Hedgehog signaling: a complex story. Biochem. Pharmacol. 67:805-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olsen, C. L., P. P. Hsu, J. Glienke, G. M. Rubanyi, and A. R. Brooks. 2004. Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors. BMC Cancer 4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park, H. L., C. Bai, K. A. Platt, M. P. Matise, A. Beeghly, C. C. Hui, M. Nakashima, and A. L. Joyner. 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127:1593-1605. [DOI] [PubMed] [Google Scholar]
- 76.Park, J. M., H. S. Kim, S. J. Han, M. S. Hwang, Y. C. Lee, and Y. J. Kim. 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y. J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8:9-19. [DOI] [PubMed] [Google Scholar]
- 78.Pavri, R., B. Lewis, T. K. Kim, F. J. Dilworth, H. Erdjument-Bromage, P. Tempst, G. de Murcia, R. Evans, P. Chambon, and D. Reinberg. 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 18:83-96. [DOI] [PubMed] [Google Scholar]
- 79.Persson, M., D. Stamataki, P. te Welscher, E. Andersson, J. Bose, U. Ruther, J. Ericson, and J. Briscoe. 2002. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 16:2865-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruiz i Altaba, A. 1999. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development 126:3205-3216. [DOI] [PubMed] [Google Scholar]
- 81.Ruppert, J. M., K. W. Kinzler, A. J. Wong, S. H. Bigner, F. T. Kao, M. L. Law, H. N. Seuanez, S. J. O'Brien, and B. Vogelstein. 1988. The GLI-Kruppel family of human genes. Mol. Cell. Biol. 8:3104-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sasaki, H., Y. Nishizaki, C. Hui, M. Nakafuku, and H. Kondoh. 1999. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126:3915-3924. [DOI] [PubMed] [Google Scholar]
- 83.Stamataki, D., F. Ulloa, S. V. Tsoni, A. Mynett, and J. Briscoe. 2005. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 19:626-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 85.Stone, D. M., M. Hynes, M. Armanini, T. A. Swanson, Q. Gu, R. L. Johnson, M. P. Scott, D. Pennica, A. Goddard, H. Phillips, M. Noll, J. E. Hooper, F. de Sauvage, and A. Rosenthal. 1996. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384:129-134. [DOI] [PubMed] [Google Scholar]
- 86.Taatjes, D. J., M. T. Marr, and R. Tjian. 2004. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5:403-410. [DOI] [PubMed] [Google Scholar]
- 87.Taatjes, D. J., A. M. Naar, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295:1058-1062. [DOI] [PubMed] [Google Scholar]
- 88.Tabata, T., S. Eaton, and T. B. Kornberg. 1992. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 6:2635-2645. [DOI] [PubMed] [Google Scholar]
- 89.Tabata, T., and T. B. Kornberg. 1994. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76:89-102. [DOI] [PubMed] [Google Scholar]
- 90.Taipale, J., and P. A. Beachy. 2001. The Hedgehog and Wnt signalling pathways in cancer. Nature 411:349-354. [DOI] [PubMed] [Google Scholar]
- 91.Taipale, J., M. K. Cooper, T. Maiti, and P. A. Beachy. 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418:892-897. [DOI] [PubMed] [Google Scholar]
- 92.Tan, W., L. Zheng, W. H. Lee, and T. G. Boyer. 2004. Functional dissection of transcription factor ZBRK1 reveals zinc fingers with dual roles in DNA-binding and BRCA1-dependent transcriptional repression. J. Biol. Chem. 279:6576-6587. [DOI] [PubMed] [Google Scholar]
- 93.te Welscher, P., A. Zuniga, S. Kuijper, T. Drenth, H. J. Goedemans, F. Meijlink, and R. Zeller. 2002. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science 298:827-830. [DOI] [PubMed] [Google Scholar]
- 94.Treisman, J. 2001. Drosophila homologues of the transcriptional coactivation complex subunits TRAP240 and TRAP230 are required for identical processes in eye-antennal disc development. Development 128:603-615. [DOI] [PubMed] [Google Scholar]
- 95.Von Ohlen, T., D. Lessing, R. Nusse, and J. E. Hooper. 1997. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc. Natl. Acad. Sci. USA 94:2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang, B., J. F. Fallon, and P. A. Beachy. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100:423-434. [DOI] [PubMed] [Google Scholar]
- 97.Wang, G., M. A. Balamotis, J. L. Stevens, Y. Yamaguchi, H. Handa, and A. J. Berk. 2005. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17:683-694. [DOI] [PubMed] [Google Scholar]
- 98.Wang, Q. T., and R. A. Holmgren. 1999. The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development 126:5097-5106. [DOI] [PubMed] [Google Scholar]
- 99.Wijgerde, M., J. A. McMahon, M. Rule, and A. P. McMahon. 2002. A direct requirement for Hedgehog signaling for normal specification of all ventral progenitor domains in the presumptive mammalian spinal cord. Genes Dev. 16:2849-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Woychik, N. A., and M. Hampsey. 2002. The RNA polymerase II machinery: structure illuminates function. Cell 108:453-463. [DOI] [PubMed] [Google Scholar]
- 101.Yang, F., R. DeBeaumont, S. Zhou, and A. M. Naar. 2004. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 101:2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]