Abstract
In Tetrahymena, HHT1 and HHT2 genes encode the same major histone H3; HHT3 and HHT4 encode similar minor H3 variants (H3s), H3.3 and H3.4. Green fluorescent protein (GFP)-tagged H3 is deposited onto chromatin through a DNA replication-coupled (RC) pathway. GFP-tagged H3.3 and H3.4 can be deposited both by a transcription-associated, replication-independent (RI) pathway and also weakly by an RC pathway. Although both types of H3s can be deposited by the RC pathway, DNA repair synthesis associated with meiotic recombination utilizes H3 specifically. The regions distinguishing H3 and H3.3 for their deposition pathways were identified. RC major H3 is not essential. Cells can grow without major H3 if the minor H3s are expressed at high levels. Surprisingly, cells lacking RI H3s are also viable and maintain normal nucleosome density at a highly transcribed region. The RC H3 is not detectably deposited by the RI pathway, even when there are no RI H3s available, indicating that transcription-associated RI H3 deposition is not essential for transcription. Minor H3s are also required to produce viable sexual progeny and play an unexpected role in the germ line micronuclei late in conjugation that is unrelated to transcription.
In eukaryotic cells, chromatin consists of repeating nucleosome cores in which 146 bp DNA is wrapped in ∼1.75 turns around a histone octamer containing two molecules each of the core histones H2A, H2B, H3, and H4. Usually, an additional 20 to 40 bp of DNA between the cores is associated with a linker histone, H1. The structural and functional diversity of nucleosomes is produced by covalent posttranslational modifications of the histones (19, 39) and by core histone variants, usually of H2A or H3 (17, 32).
The histone H3 multigene family has been well studied. Three important types of H3 variants (H3s) have been identified: major H3s; quantitatively minor H3.3s, which differ from the major H3s at four or five conserved sites in most organisms; and highly divergent CenH3s, which localize specifically to centromeres and are not dealt with in this paper. In Drosophila, deposition of the major H3 onto chromatin is exclusively DNA replication coupled (RC), while the minor H3.3 exhibits both RC deposition and replication-independent (RI) deposition (1). RI deposition requires transcription elongation (33). Three amino acids that differ between Drosophila H3 and H3.3 at the beginning of α-helix 2 (α2) are responsible for exclusive RC deposition of H3 (1). The DNA synthesis-dependent deposition of H3 and the DNA synthesis-independent deposition of H3.3 are mediated by different histone chaperones, CAF-1 and HIRA, respectively (31, 37). Mutations that eliminate the function of CAF-1 in Saccharomyces cerevisiae, Arabidopsis, or mammalian cells result in various phenotypes but not lethality. HIRA is essential for mouse embryonic development and is conserved among other eukaryotes (15, 17). Interestingly, S. cerevisiae and Schizosaccharomyces pombe contain only an H3.3-like variant, yet both contain CAF-1 and HIRA-related histone chaperones. Recent evidence also argues that S. pombe H3 is deposited through both an RC and a transcription-associated RI pathway (8). To date, no organism has been found containing only an H3-like variant, suggesting that minor, transcription-associated RI H3.3s may be essential (8, 25).
Tetrahymena thermophila is a well-studied protist (for a review, see reference 20). Like other ciliates, Tetrahymena has two types of nuclei (see Fig. S1 in the supplemental material): a small diploid germ line micronucleus (MIC) and a large polyploid (∼45C) somatic macronucleus (MAC). During vegetative (asexual) growth, the MIC divides mitotically and is transcriptionally inert while the MAC divides amitotically and is transcriptionally active. When Tetrahymena cells are starved, DNA replication and cell division stop, with the MAC in G1 phase and the MIC in G2, while the MAC remains transcriptionally active. The sexual reproduction process of Tetrahymena, conjugation, can be initiated by mixing starved Tetrahymena cells of different mating types. They first form conjugating pairs. In early conjugation, the MICs enter meiosis, adopting a highly elongated shape referred to as the crescent, in which chromosomes are arranged in parallel, with telomeres located at one end (22) and centromeres at the other end (9). Because micronuclei enter conjugation with the 4C amount of DNA and MIC DNA content does not change during meiosis (10), DNA synthesis detected in premeiotic crescents is associated with DNA repair following meiotic recombination between maternal and paternal chromosomes (2). MICs become transcriptionally active as they adopt the crescent shape (27, 36). After the crescent stage, MICs cease transcribing and undergo two meiotic divisions. One of the four meiotic products undergoes a prezygotic mitosis to produce two pronuclei, followed by the exchange of one pronucleus and fusion to produce a zygotic nucleus. Two postzygotic mitoses follow, producing four MICs. The two at the posterior end of each conjugant become the new MICs. The two at the anterior end enlarge to form the new MACs (NM, also known as anlagen), which initiate zygotic transcription, undergo genome rearrangement, and endoreplicate the MAC genome from 2C to 8C. When NM begin to develop, parental MACs (PMs) cease transcription and initiate an apoptosis-like degradation. The two cells of the conjugating pair then separate, become exconjugants, and resorb one MIC. If fed, each exconjugant divides and resumes vegetative growth. During growth, starvation, and conjugation, the nuclear activities, such as replication, recombination, and transcription, in the various nuclei at different stages have been well studied previously (2, 7, 10, 26, 27, 30, 35, 36, 41).
Three H3 genes were originally identified in Tetrahymena. HHT1 (accession no. M87304) and HHT2 (accession no. M87305) encode the same major H3 protein. HHT3 encodes H3.3 (38), a quantitatively minor H3 that differs from H3 at 16 residues (Fig. 1A), a much greater difference than that observed between the major and minor H3s of other organisms. The major H3 genes are expressed during vegetative growth and repressed during starvation, while the minor H3.3 gene is constitutively expressed in growing and starved cells (4). The major H3 localizes to both MACs and MICs, while the minor H3.3 was found only in MACs. H3.3, but not H3, is deposited in MACs in starved cells, when MACs are not replicating (3). Because the only process known to be associated constitutively and specifically with MACs in growing and starved cells is transcription, it was hypothesized that H3.3 is specifically associated with transcription in Tetrahymena (3, 43). Subsequent studies with multicellular eukaryotes demonstrated the association of H3.3 deposition with transcription (1, 18, 29, 33).
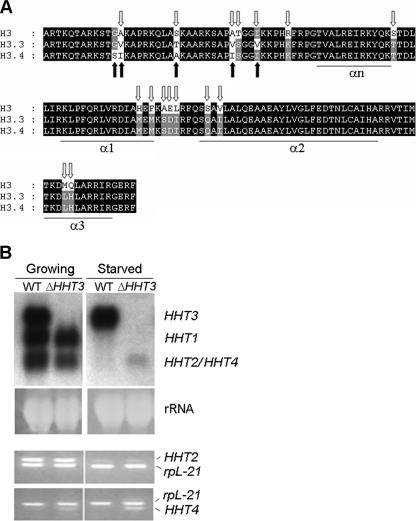
FIG. 1.
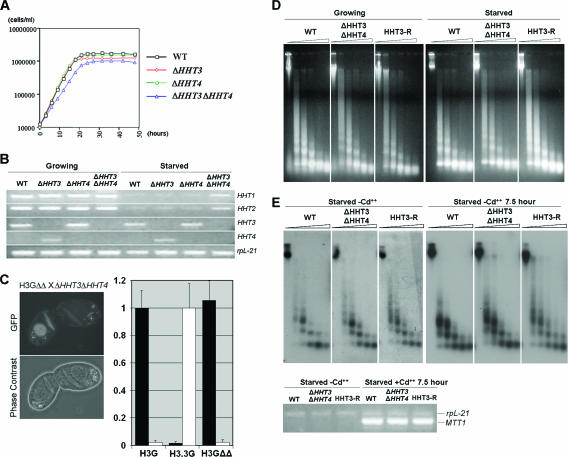
HHT4 is upregulated when HHT3 is knocked out. (A) Alignment of Tetrahymena histone H3, H3.3, and H3.4 sequences. The 16 residues that differ between H3 and H3.3 are indicated by open arrows, and the 5 that differ between H3.3 and H3.4 are indicated by filled arrows. (B) HHT4 is upregulated when HHT3 is knocked out. Upper panel, Northern blot using the HHT3 coding sequence as the probe, which detects all HHT genes due to their similarities in sequence but hybridizes most strongly to the homologous mRNA. The ethidium bromide staining of rRNA is shown as the loading control. Lower panel, RT-PCR using primers that amplify HHT2 or HHT4 specifically. The PCR products were sequenced to prove their identity. Negative controls lacking reverse transcriptase during the synthesis of cDNA resulted in blank lanes (data not shown). rpL21 encodes a ribosomal protein and was used as an RT-PCR control.
HHT3 is not essential for cell growth in Tetrahymena. The deletion of HHT3 resulted in the induced expression of an mRNA in starved cells whose electrophoretic mobility was indistinguishable from that of the HHT2 mRNA found in growing cells, leading to the suggestion that HHT2, encoding a major H3, is expressed constitutively in starved cells in the absence of H3.3 and that it was the replication-independent, constitutive (basal) expression, not the primary sequence differences, that was important in distinguishing the function of the minor H3.3 from that of the major H3 in Tetrahymena (43).
The recently completed Tetrahymena macronuclear genome sequence (http://www.tigr.org/tdb/e2k1/ttg/index.shtml) revealed two new H3 genes, CNA1 and HHT4. CNA1 (accession no. DQ126145) encodes the centromeric H3, which is described elsewhere (6, 9). HHT4 (accession no. CH445775) encodes H3.4, a minor H3, similar to H3.3. Here, we describe this new gene and analyze the deposition and function of the entire HHT gene family. We show that, in Tetrahymena, H3-green fluorescent protein (GFP) is deposited onto chromatin when DNA is synthesized, both during DNA replication and during meiotic recombination. In contrast, H3.3-GFP and H3.4-GFP depositions can be both DNA replication coupled and (transcription associated) DNA replication independent. HHT4 is expressed little, if at all, in wild-type (WT) cells. When HHT3 is knocked out, HHT4, not HHT2, is upregulated, showing that H3.3 and H3.4 function redundantly as RI minor variants. Domain-swapping experiments indicate that most of the regions that differ between H3 and H3.3 contribute to the efficiency of H3 RC deposition, while the residues in Loop1 and at the beginning of α-helix 2 are responsible for excluding H3 from RI deposition. Cells in which both major H3 genes were knocked out are viable but grow very slowly unless either HHT3 or HHT4 is overexpressed. However, the MICs in these cells are hypodiploid, likely due to the inefficient RC deposition of the RI H3s. Surprisingly, although no known organisms lack transcription-associated RI H3s, Tetrahymena cells that have both minor RI H3s knocked out are viable. Moreover, in those cells, the major H3-GFP is not detectably deposited through the transcription-associated RI pathway, and nucleosome density is maintained normally on a highly transcribed gene. Thus, deposition of nucleosomes containing newly synthesized H3 is not required for transcription. However, RI H3s are essential for producing viable sexual progeny. In matings between cells lacking minor H3s, MICs appear abnormally decondensed at the early NM stage and some cells lose their MICs at the exconjugant stage. Correspondingly, H3.3-GFP and H3.4-GFP appear transiently in MICs at the NM stage in WT cells, and this specific deposition is not associated with either replication or transcription, indicating that the minor H3s may have an unexpected novel function during the development of the sexual progeny.
MATERIALS AND METHODS
Tetrahymena strains and culture conditions.
Wild-type CU428 and B2086 strains of Tetrahymena thermophila were provided by Peter J. Bruns (Cornell University). Cells were grown in 1× SPP medium with 1% proteose peptone (14) at 30°C. To starve cells, mid-log-phase growing cells were washed twice with 10 mM Tris (pH 7.5) and incubated for 22 to 24 h at 30°C. To induce conjugation, starved cells with different mating types were mixed together.
Creation of the endogenous HHT2-GFP and HHT3-GFP strains.
A 6-amino-acid linker encoded by TCTAGGCCTGTTGCTACT and the GFP-coding sequence were inserted into the endogenous HHT2 or HHT3 gene right before the stop codon. A neo2 cassette (12) was inserted into the flanking sequence ∼0.6 kb downstream of the HHT2 stop codon or ∼1.6 kb upstream of the HHT3 start codon, as a selectable marker. The constructs were transformed into the micronuclear HHT2 or HHT3 locus of mating B2086 or CU428 cells, respectively, by biolistic transformation (5). Progeny that were resistant to paromomycin were obtained, and standard procedures (16) were used to obtain homozygous HHT2-GFP/HHT2-GFP or HHT3-GFP/HHT3-GFP heterokaryons (homozygous for the tagged gene in the MIC, WT in the MAC). Two such heterokaryons were then mated to obtain homozygous homokaryons (tagged gene in both the MIC and the MAC).
Creation of MTT1-HHT-GFP strains.
The pTTMet construct (34) containing the 489-bp MTT1 coding, ∼2-kb 5′ flanking, and ∼400-bp 3′ flanking sequences was used to create the MTT1-HHT-GFP construct. The neo2 cassette was inserted into the 5′ flanking sequence of the pTTMet construct, ∼900 bp upstream of the start codon. The MTT1 coding sequence in the resulting construct was replaced by the HHT2-GFP or HHT3-GFP sequence. The constructs were transformed into the macronuclear MTT1 locus of CU428 or B2086 cells by biolistic transformation and completely assorted to give MTT1-HHT2-GFP and MTT1-HHT3-GFP cell strains.
To induce H3-GFP (or H3.3-GFP) expression in growing cells, MTT1-HHT2-GFP or MTT1-HHT3-GFP cells were grown to mid-log phase. Cd2+ was then added to 0.75 μg/ml, and 2 h later, cells were fixed and photographed (see below). To induce H3-GFP or H3.3-GFP expression in starved cells, cells were grown to mid-log phase and starved in 10 mM Tris for 24 h, and then Cd2+ was added to 0.06 μg/ml; 2 h later, cells were fixed and photographed.
Cloning the HHT4 gene.
The HHT4 gene was identified in the Tetrahymena thermophila macronuclear genome database (http://tigrblast.tigr.org/er-blast/index.cgi?project=ttg). The gene accession no. is CH445775. The protein accession no. is EAR88155. A PCR product containing a 2.4-kb 5′ flanking region, a 408-bp coding region, and a 0.77-kb 3′ flanking region amplified from genomic DNA was digested by HindIII and EcoRI and cloned into Bluescript KS vector (Stratagene). The construct was named pHHT4 and verified by sequencing.
Creation of ΔHHT4 cells.
The HHT4 knockout construct was created by replacing the 408-bp HHT4 coding sequence in pHHT4 with the neo2 cassette. The construct was transformed into the micronuclear HHT4 locus of mating B2086 and CU428 cells. The hht4::neo2/hht4::neo2 homozygous heterokaryons and subsequently the ΔHHT4 homokaryons were obtained by following the procedure described above.
Creation of ΔHHT3 ΔHHT4 cells.
A homozygous ΔHHT3 heterokaryon using the neo2 cassette as the selection marker was first created as described above for the ΔHHT4 heterokaryon. An HHT4 knockout construct, in which the HHT4 coding sequence was replaced by a bsr1 cassette (conferring blasticidin resistance), which consists of the HHF1 promoter, the bsr gene, and a ∼0.3-kb BTU2 3′ flanking sequence (42), was transformed into the micronucleus of mating B2086 and the ΔHHT3 heterokaryon. The ΔHHT3 and ΔHHT4 heterokaryons were mated, and progeny that were resistant to both paromomycin and blasticidin were processed by following the procedure described above to obtain first the hht3::neo2, hht4::bsr1/hht3::neo2, hht4::bsr1 homozygous heterokaryons and subsequently the ΔHHT3 ΔHHT4 homokaryons.
Creation of HHT2-GFP; ΔHHT3 ΔHHT4 cells.
HHT2-GFP/HHT2-GFP and hht3::neo2, hht4::bsr1/hht3::neo2, hht4::bsr1 homozygous heterokaryons were mated, and the resulting paromomycin-resistant progeny were processed by following the procedure described above to obtain first the HHT2-GFP; hht3::neo2, hht4::bsr1/HHT2-GFP; hht3::neo2, hht4::bsr1 homozygous heterokaryons and subsequently the HHT2-GFP; ΔHHT3 ΔHHT4 homokaryons.
Rescuing the progeny of mating ΔHHT1ΔHHT2ΔHHF2ΔHHF1 heterokaryons.
ΔHHT1ΔHHT2 cells are viable; therefore, efficient rescue of them is not possible. To create a system for facilitating rescue with genes encoding an H3, ΔmTF knockout heterokaryons were created (21) in which all major H3 and H4 genes (HHT1, colinear HHT2 and HHF2, and HHF1) were disrupted in the MIC, and WT genes were present in the MAC. When ΔmTF heterokaryons were mated, the progeny were not viable due to the lack of major H3 and H4. The progeny can then be rescued by simultaneous transformation with functional, linked H3 and H4 genes targeted to the HHT2-HHF2 locus. In the experiments described in this study, rescues were performed with constructs in which an HHT gene coding region was inserted into the HHT2 locus, which was linked with a WT HHF2 gene. Cells rescued with a wild-type HHT2-HHF2 gene pair are indistinguishable from WT cells; therefore, any phenotype exhibited by the rescued cells is due to the mutated HHT gene used for the rescue.
Microscopy of the GFP-labeled cells.
For images shown in Fig. 2B and 3C and Fig. S3B and S4 in the supplemental material, cells were fixed in 0.5% formaldehyde at room temperature for 5 min and resuspended in phosphate-buffered saline with the DNA-specific dye DAPI (4′,6′-diamidino-2-phenylindole) at 0.1 μg/ml (Roche). GFP fluorescence was examined by confocal microscopy (Leica TCS SP). The images shown in Fig. 2D and Fig. S3A in the supplemental material were Triton extracted (11, 24) to remove the cytoplasm and nucleoplasm while retaining cytoskeletal and chromosomal elements, enabling visualization of chromosomal GFP-tagged H3s. This experiment was repeated with formaldehyde-fixed cells, and similar results were obtained (data not shown). Living cells were also observed in all cases to confirm the results obtained with fixed cells. Figures 5B and 6C show living-cell images obtained from a conventional fluorescence microscope to achieve more focal depth. The intensities of the crescent (Wc) and parental MAC (Wpm) for the WT mating partner, the parental MAC for the GFP-expressing cells (Gpm), and the background (b) were obtained by ImageJ software. The relative intensity of the WT crescent (RWc) in each mating was calculated as (Wc − b)/(Gpm − b). The RWc in the crossing of H3-GFP with the WT was arbitrarily set as 1, and the RWcs in other crossings were normalized to it. The relative intensity of the parental MAC (RWpm) in each mating was calculated as (Wpm − b)/(Gpm − b). The RWpm in the crossing of H3.3-GFP with the WT was arbitrarily set as 1, and the RWpms in other crossings were normalized to it.
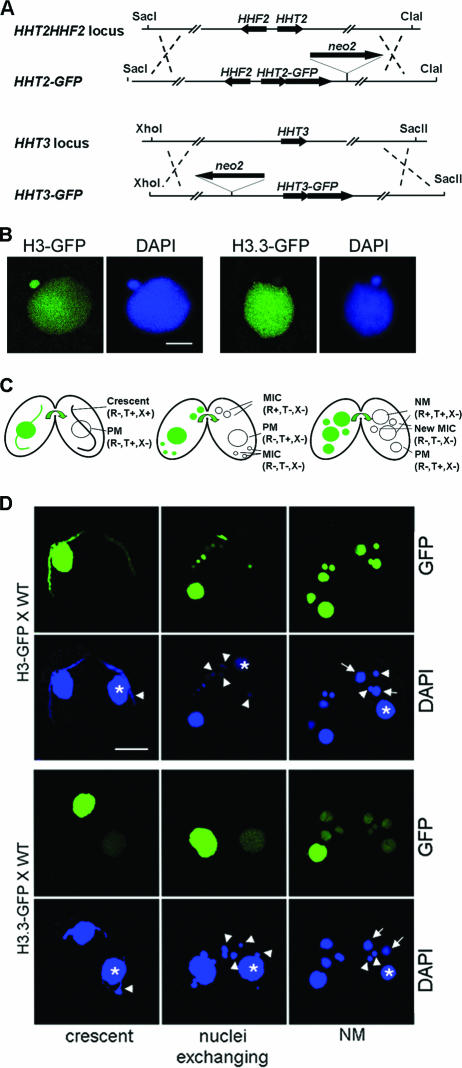
FIG. 2.
H3-GFP deposition is DNA replication coupled, while H3.3-GFP can be deposited independently of DNA replication. (A) The constructs of GFP-tagged H3 and H3.3 targeting their endogenous loci. (B) Direct GFP and DAPI (a DNA-specific dye) fluorescence microscopy showing that H3-GFP localizes in both MICs and MACs in vegetative growing cells. H3.3-GFP localizes mainly in MACs and faintly in MICs. Scale bar, 8 μm. (C) Diagrams of crescent, nucleus-exchanging, and NM stages during conjugation. The nuclei are indicated with their names and status in parentheses. PM, parental MAC; NM, new MAC; R, replication; T, transcription; X, recombination. Arrows indicate direction of transfer of the GFP label. (D) Deposition of H3-GFP is coupled to DNA replication, while H3.3-GFP can be deposited independently of DNA replication. H3-GFP-expressing cells (upper panel) or H3.3-GFP-expressing cells (lower panel) were crossed with WT cells (shown on the right in each mating pair). Asterisks, parental MAC; arrowheads, MIC; arrows, NM; scale bar, 16 μm.
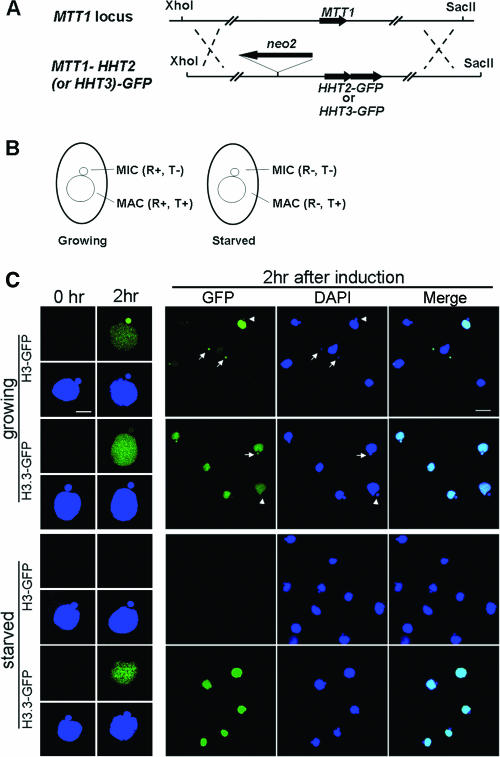
FIG. 3.
H3.3-GFP can be deposited through the DNA replication-coupled pathway. (A) HHT2-GFP or HHT3-GFP construct targeting to the MTT1 locus. (B) Diagram of growing and starved cells, with the replication (R) and transcription (T) statuses of the nuclei indicated. (C) H3-GFP and H3.3-GFP localization 2 h after Cd2+ induction in growing or starved cells. Individual cells are shown in the left panel at high magnification. Scale bar, 8 μm; first column, cells before induction; second column, cells 2 h after induction; green, GFP; blue, DAPI. Right panel, fields of cells after 2 h of induction at lower magnification. Scale bar, 24 μm; arrows, MICs after S phase that have strong H3-GFP or H3.3-GFP signal; arrowheads, MICs that have little GFP signal.
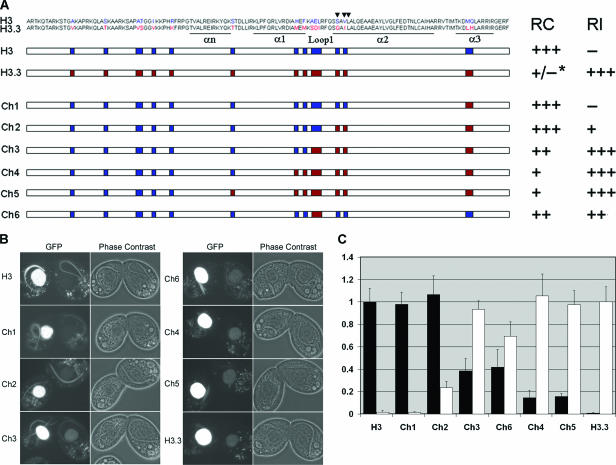
FIG. 5.
Regions that distinguish H3 and H3.3 function. GFP-tagged chimeric H3s that contain different domains of H3 and H3.3 were used to rescue the ΔmTF cells and crossed with WT cells. The localizations of the chimeric H3-GFP proteins in the crescents (RC pathway) and parental MAC (RI pathway) of WT cells were examined. (A) Chimeric H3s. White bars, sequences common between H3 and H3.3; blue bars, amino acids specific for H3; red bars, amino acids specific for H3.3. The positions of the three amino acids that are important for RC deposition of Drosophila H3 are indicated by arrowheads. *, since the RC deposition of H3.3 during conjugation is not detectable, the RC deposition capacity of H3.3 is noted as “+/−.” (B) Images of cells expressing GFP-tagged chimeric H3s crossed with WT cells. (C) Quantification of the relative GFP intensities of the crescent MIC (black bar) and parental MAC (white bar) in the WT mating partner (see Materials and Methods).
FIG. 6.
Minor H3s are not essential for transcription or for growth. (A) Growth curves for WT cells, ΔHHT3, ΔHHT4, and ΔHHT3 ΔHHT4. The doubling times are as follows: for WT cells, 2.9 h; for ΔHHT3, 2.9 h; for ΔHHT4, 2.8 h; and for ΔHHT3 ΔHHT4, 3.8 h. (B) Major HHT1 and HHT2 are upregulated in starved cells when both minor H3 genes are knocked out. Primers that amplify different HHT genes were used for RT-PCR. rpL21 was used as a control for RT-PCR. Negative controls were blank and are not shown. (C) H3-GFP is not deposited through the RI pathway when there is no minor H3 available in cells. Left panel, HHT2-GFP ΔHHT3 ΔHHT4 cell (H3GΔΔ, expressing H3-GFP in a ΔHHT3 ΔHHT4 background) mating with ΔHHT3 ΔHHT4 cell; right panel, relative GFP intensities of the crescents (black bars) and the parental MAC (white bars) in the non-GFP-expressing ΔHHT3 ΔHHT4 cells, which were mated with H3-GFP cells (H3G), H3.3-GFP cells (H3.3G) (images not shown), or H3GΔΔ cells. (D) Global chromatin structure is not affected in ΔHHT3 ΔHHT4 and HHT3 rescue (HHT3-R) cells. DNA extracted from isolated nuclei that had been incubated with MNase for increasing periods of time was separated on agarose gel and visualized by ethidium bromide staining. (E) Nucleosome density on a transcriptionally active region is normal in ΔHHT3 ΔHHT4 and HHT3-R cells. Upper panel, nuclei from starved WT, ΔHHT3 ΔHHT4, or HHT3-R cells that have been incubated without (left) or with (right) Cd2+ at 0.1 μg/ml for 7.5 h were digested as described above. A Southern blot of the extracted DNA was hybridized with an MTT1 coding sequence probe. Lower panel, RT-PCR showing that MTT1 gene expression is induced after 7.5 h in 0.1 μg/ml Cd2+. rpL21 was used as a quantity control.
MNase digestion.
Macronuclei were isolated as described previously (14), washed twice with reticulocyte standard buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.2 mM phenylmethylsulfonyl fluoride), and resuspended with reticulocyte standard buffer with 0.1 mM CaCl2 at 1 × 107 MACs/ml. Micrococcal nuclease (MNase; Worthington) was added to 50 U/ml, and the nuclei were digested for 0 and 40 seconds and 1, 2, 4, and 7 min at 37°C. Digestion was stopped by addition of 3.5 volumes of stop buffer (1% sodium dodecyl sulfate, 0.5 M EDTA, 18 mM Tris, pH 9.5, prewarmed to 65°C), followed by incubation at 65°C for at least 20 min. Proteinase K was then added to 1 mg/ml, and incubation continued at 55°C for 4 h. An equal volume of water was added to the digestion buffer, and the DNA was extracted with phenol-chloroform, precipitated, and separated on a 1.2% agarose gel.
RESULTS
HHT4 encodes a new H3 variant in Tetrahymena.
We identified a new HHT family gene, HHT4, in the recently sequenced Tetrahymena macronuclear genome. It encodes a protein, referred to as H3.4, which differs from H3.3 by 5 amino acids near the N terminus (Fig. 1A), a region that is unimportant for H3.3 RI deposition in Drosophila (1). HHT3 and HHT4 are located only 10 kb apart on the same MAC chromosome. About 1 kb each of the sequences in the HHT3 and HHT4 5′ flanking regions share 84.7% identity, and there is an identical 70-bp region in their 3′ flanking regions immediately after the stop codons. These observations suggest that HHT3 and HHT4 are products of a recent duplication event and may have similar functions.
HHT4, not HHT2, is upregulated in HHT3 knockout cells.
As reported earlier (43), HHT3 knockout (ΔHHT3) cells grew normally. When ΔHHT3 cells were starved, increased amounts of a message comigrating with the HHT2 mRNA on a Northern blot were detected using an HHT3 probe (Fig. 1B, upper panel, ΔHHT3 lane), which detects the entire HHT family due to the sequence similarities among them. This upregulated message was thought to be derived from the HHT2 gene, but the discovery of HHT4 raised the possibility that the gene encoding the upregulated mRNA had been misidentified. To clarify this issue, primers that amplify either HHT2 or HHT4 were used for reverse transcriptase (RT)-PCR to analyze their expression. HHT2 was highly expressed in growing cells and expressed at similar low levels in starved cells regardless of the presence of HHT3. HHT4 was barely detected in WT cells but was upregulated in both growing and starved ΔHHT3 cells (Fig. 1B, lower panel). Therefore, HHT4, not HHT2, was upregulated when no H3.3 was available in starved cells, suggesting that both H3.3 and H3.4 are replacement variants that function redundantly.
H3 deposition is DNA replication dependent, and H3.3 deposition is mainly DNA replication independent.
To study the deposition of histone H3s in Tetrahymena, the proteins were tagged with GFP at their C termini. HHT2 and HHT3 genes were studied initially, because HHT1 and HHT2 encode the same major H3 and are coexpressed, while HHT3 and HHT4 are likely redundant and HHT3 is expressed at a higher level than HHT4. HHT2 or HHT3 genes were replaced in both MICs and MACs with HHT2-GFP (encoding H3-GFP) or HHT3-GFP (encoding H3.3-GFP), respectively (Fig. 2A). The GFP-tagged H3 and H3.3 were chromosomal because they were retained in Triton-extracted cells (see Materials and Methods) and, during the closed micronuclear division of Tetrahymena, they were not observed in the region which connects the daughter nuclei and lacks DNA staining. The tagged proteins were also functional because they could rescue cells whose major H3 genes had been disrupted (see Fig. 5B and below). In vegetative cells, H3-GFP localized in both MACs and MICs, while H3.3-GFP localized mainly in MACs and very faintly in MICs (Fig. 2B). The presence of H3.3 in MICs was not observed previously when histones isolated from MICs were examined (3), likely because the biochemical methods used were not as sensitive as the GFP tagging used here. Transcription has not been detected in vegetative-cell MICs, suggesting that the small amounts of H3.3 observed in the MIC reflect the ability of this variant to inefficiently enter the RC pathway (see below).
The major approach we took to study the deposition properties of H3s was to mate cells expressing GFP-tagged H3 or H3.3 with WT cells and examine the localization of the GFP-tagged proteins in WT cells. In these experiments, the GFP-expressing cell functions as a carrier of the tagged protein and/or RNA, both of which can pass between the mating partners during conjugation when their cytoplasms are connected (28). As noted above, different nuclei have distinct activities of replication, transcription, and recombination at different stages of conjugation (see Fig. S1 in the supplemental material; Fig. 2C) that have been very well established by previous studies. By observing the timing and nuclear localization of the GFP protein into the untagged WT cell, the deposition properties of the GFP-tagged H3s can be associated with the nuclear activities of the corresponding nuclei.
In mating pairs of H3-GFP and WT cells (Fig. 2D, upper panel), H3-GFP was detected in the WT meiotic MICs at the crescent stage but not in the transcribing MACs of the same cells. As MICs are 4C when they enter conjugation, this H3-GFP deposition is not associated with genome duplication and therefore must be related to DNA repair synthesis associated with meiotic homologous recombination that occurs at this stage (2). The association of H3-GFP deposition with DNA synthesis was further confirmed by the later stages. When the mating pairs reached the stage when nuclei are exchanged between the mating partners, the MIC had gone through a DNA replication and the prezygotic mitosis, and the H3-GFP signal in MIC had increased. By the NM stage, when the MIC had gone through two more replications and mitoses and the NM were amplifying their genomes, there was very strong H3-GFP signal in the MIC and NM. Importantly, no H3-GFP signal was ever detected in the parental MAC, which is the only nucleus in these cells that does not undergo DNA synthesis in the WT cell at any stage but is transcriptionally active throughout the conjugation process until NM are formed. These observations strongly argue that H3-GFP is deposited exclusively through a DNA RC pathway that can be associated with either DNA duplication or repair of breakage that occurs during meiotic recombination.
In the mating between H3.3-GFP and WT cells (Fig. 2D, lower panel), GFP signal was detected in the WT PM of crescent stage cells and the signal intensity increased as conjugation proceeded. As noted, there is transcription but no DNA replication in the PM at any time during conjugation. Therefore, this H3.3-GFP deposition is DNA replication independent and is correlated with transcription. H3.3-GFP was also found in the NM of the WT cells when they initiated zygotic transcription. We conclude that H3.3-GFP is deposited through a transcription-associated RI pathway.
Interestingly, H3.3-GFP was not detected in crescents when they were transcriptionally active, indicating that, although H3.3 RI deposition is coupled to transcription, it is not essential for transcription. This is consistent with our later observations that cells can grow without the H3.3 and H3.4 RI variants (see Fig. 6). Surprisingly, H3.3-GFP was detected in the MIC at the NM stage (Fig. 2D, lower panel, NM), persisted in exconjugants, and then decreased markedly after a few vegetative divisions (data not shown). This deposition of H3.3 in MICs is not associated with the transcription-associated RI pathway since no transcription has been detected in MICs at this stage. The absence of H3.3-GFP in micronuclei right after they undergo replications associated with prezygotic mitosis (Fig. 2D, lower panel, nuclei exchanging) and two postzygotic mitoses (data not shown) also makes RC deposition unlikely. How H3.3 is deposited into MICs at the NM stage is not clear, although it appears to have a function, since cells lacking H3.3 and H3.4 have conjugation-specific phenotypes (see Fig. 7).
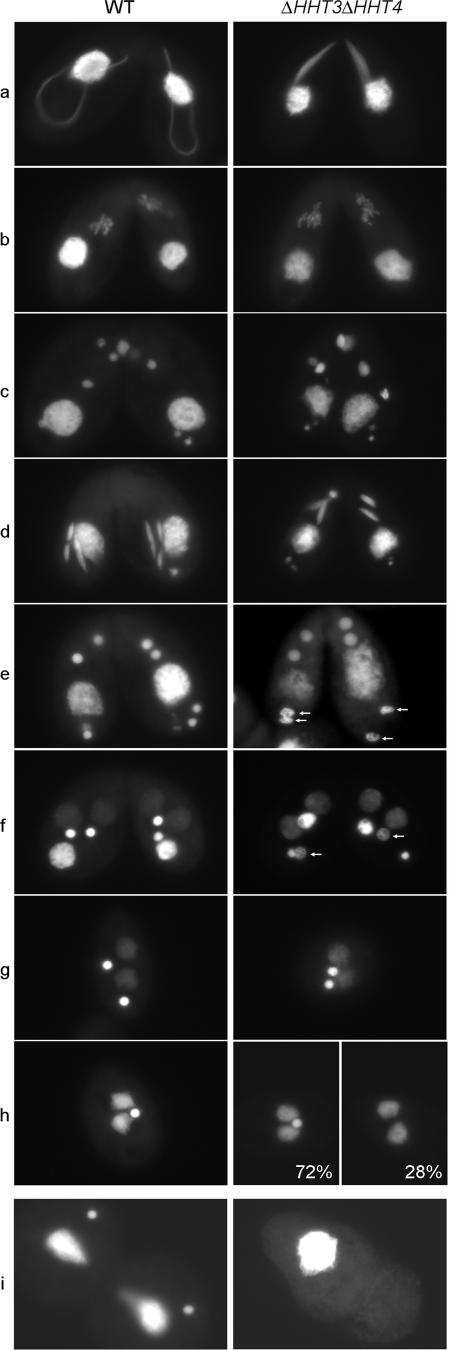
FIG. 7.
Minor H3s are required for full crescent extension, proper MIC condensation at the NM stage, and production of viable progeny. WT or ΔHHT3 ΔHHT4 homozygous homokaryons were mated, and their conjugation was followed. a, crescent stage; b, meiosis I; c, nuclear exchange; d, postzygotic mitosis II; e and f, NM stage; g, two-MIC exconjugant; h, all WT exconjugants have 1 MIC, ∼72% of ΔHHT3 ΔHHT4 cells have 1 MIC, and ∼28% of ΔHHT3 ΔHHT4 cells have no MIC; i, the exconjugants were transferred into growth medium, where WT cells resumed vegetative growth, while the ΔHHT3 ΔHHT4 cells could not divide and died.
H3.3-GFP is also deposited through a DNA replication-coupled pathway.
In Drosophila, H3.3 is deposited through both RC and RI pathways. In Tetrahymena, RC deposition was not observed in conjugating cells when H3.3 was expressed at its endogenous level but was weakly detected in MICs of vegetative cells. One possible explanation for these observations is that H3.3 can enter the RC pathway very inefficiently compared to major H3 and therefore its detection depends on its concentration in cells relative to that of the major H3. If this is the case, overexpression of H3.3-GFP should lead to greater incorporation into replicating nuclei. To test this, the coding sequence of a Cd2+-inducible metallothionein gene, MTT1 (34), was replaced by either HHT2-GFP or HHT3-GFP (Fig. 3A), so that the H3-GFP or H3.3-GFP can be overexpressed from the MTT1 locus. In growing cells, MACs are the only transcriptionally active nuclei, but both MACs and MICs replicate their DNA (Fig. 3B) and the timing of their nonoverlapping S phases during the cell cycle has been well studied by [3H]thymidine labeling (41). Two hours after Cd2+ addition, H3-GFP was deposited into both nuclei, but some cells had strong GFP-labeled MICs and faint MACs while others had strong MACs and faint MICs (Fig. 3C), consistent with their nonoverlapping S phases (40, 41). Thus, only nuclei that had been through S phase contain H3-GFP, confirming that H3-GFP deposition is RC. In contrast, H3.3-GFP was deposited into MACs to similar extents in all cells, consistent with constitutive, transcription-associated RI deposition. Importantly, when overexpressed, the pattern of H3.3-GFP deposition into MICs was like that of H3-GFP, strong in some cells and very faint in others, arguing that the H3.3-GFP deposition in MICs was RC. In starved cells, where transcription occurs in MACs and no DNA replication occurs in either MICs or MACs, H3-GFP could not be detected in either nucleus 2 h after induction by Cd2+. H3.3-GFP, in contrast, was deposited efficiently into the MAC, confirming that H3.3 is deposited through a transcription-associated RI pathway. Importantly, when induced in starved cells, H3.3-GFP was never observed in MICs, arguing that the H3.3-GFP deposition observed in the MICs of growing cells occurs through an RC pathway. These observations argue strongly that H3.3 can be deposited by an RC pathway, although less efficiently than H3, explaining the low levels of endogenously expressed H3.3-GFP observed in MICs of growing cells (Fig. 2B).
Minor H3s alone can support cell growth.
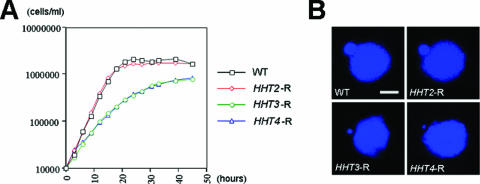
The above-described results show that H3 is deposited exclusively through an RC pathway, and H3.3 can be deposited through both RC and RI pathways. In yeasts, H3.3 must be responsible for both RC and RI nucleosome assembly since it is the only H3 in the cells. To determine whether minor H3s alone can also support cell growth in Tetrahymena, both genes encoding the major H3 (HHT1 and HHT2) were knocked out. ΔHHT1 ΔHHT2 cells were viable but grew very slowly, with a doubling time of ∼10 days (data not shown). To determine whether the slow-growth phenotype was caused by low expression of the remaining endogenous minor H3s, the HHT3 gene, along with a colinear HHF2 gene (encoding H4.2), was used to rescue the progeny of mating ΔHHT1 ΔHHT2 ΔHHF2 ΔHHF1 (ΔmTF) knockout heterokaryon cells, in which both major H3 genes (HHT1 and HHT2) and H4 genes (HHF1 and HHF2) were knocked out in the micronuclei. The progeny of these cells die if they are not rescued with both functional H3 and H4 genes (21 [see Materials and Methods for details]). In the rescuing construct, the HHT3 coding region replaced the HHT2 coding region and was targeted to the HHT2-HHF2 locus so that it would be expressed under the control of the HHT2 promoter. The rescued cells grew well, albeit slightly slower than WT cells (Fig. 4A, HHT3-R), but their MICs were significantly smaller and contained less DNA than wild-type cells (Fig. 4B). Control cells that were rescued with HHT2 and HHF2 genes (Fig. 4A, HHT2-R) were indistinguishable from wild-type cells in both growth rate and micronuclear morphology. Thus, the extreme slow growth of cells containing only HHT3 and HHT4 at their endogenous loci is largely due to the inadequate amount of H3 proteins. However, rescue with HHT3 expressed at a similar level as HHT2 still resulted in slight growth reduction and a small MIC phenotype. To test whether these are caused by inefficient deposition of H3.3 through the RC pathway, ΔmTF cells were rescued with HHT3, colinear HHF2, and an HHT3-GFP fusion gene that was overexpressed from the MTT1 locus. Although cells with normal-sized MICs were obtained and strong H3.3-GFP was observed in both the MAC and MIC, these strains were not stable in terms of their MIC sizes and their growth rate was not improved (see Fig. S2 in the supplemental material). These observations suggest that the MIC phenotype is mostly due to the inefficient entry of the RI variants into the RC pathway, but there are some other functions that H3.3 cannot fulfill as efficiently as H3 even when highly overexpressed.
FIG. 4.
Cells can grow without major RC H3. (A) Growth curves of WT and rescued (R) cells. ΔmTF cells, in which all major H3 and H4 genes are disrupted, were rescued with different HHT genes expressed from the HHT2 locus and an HHF2 gene (encoding H4.2). The doubling times are as follows: for WT cells, 2.5 h; for HHT2-R, 2.5 h; for HHT3-R, 3.8 h; and for HHT4-R, 4.0 h. (B) Small MICs are observed in HHT3-R and HHT4-R but not in HHT2-R cells. Scale bar, 8 μm.
H3.4 functions similarly to H3.3.
To see whether H3.4 functions similarly to H3.3, H3.4 was tagged with Flag or GFP at its endogenous locus. No tagged protein was detectable by either Western blot assays or fluorescence microscopy (data not shown), consistent with our previous observations that HHT4 messages were barely detectable in WT cells (Fig. 1B). Similar to HHT3, HHT4 coupled with HHF2 could rescue the ΔmTF progeny when expressed from the HHT2 locus and the rescued cells had small MICs and grew slightly more slowly than WT cells (Fig. 4, HHT4-R). When the HHT4-GFP construct was used to rescue the ΔmTF cells, H3.4-GFP signal localized in both the MAC and the small MIC. When HHT4-GFP rescued cells were crossed with WT cells, their behavior also mimicked that of H3.3. H3.4-GFP was deposited into the WT MAC at the crescent stage, and in the few mating cells that proceeded to NM stage, H3.4-GFP was observed in the new MIC and MAC (see Fig. S3A in the supplemental material).
To determine whether H3.4 can be deposited through the RC pathway, HHT4-GFP was used to replace the MTT1 gene coding sequence. H3.4-GFP overexpressed by Cd2+ induction localized in both MACs and MICs in growing cells and only in MACs in starved cells (see Fig. S3B in the supplemental material), as seen with H3.3-GFP. All of the above-described results show that H3.4 functions similarly to H3.3 and can be deposited through both the RI and the RC pathways.
Protein regions that distinguish H3 and H3.3 function.
In Drosophila, the 3 amino acids at the beginning of α-helix 2 that differ between H3 and H3.3 are responsible for the exclusive RC deposition that distinguishes H3 from H3.3, and mutating these residues allows the mutated H3 to be deposited through the RI pathway (1). In Tetrahymena, there are 16 amino acid differences between H3 and H3.3 (Fig. 5A). To identify the region(s) that distinguishes major and minor H3s for the RC and RI pathways, a series of chimeric HHT-GFP genes that swap the domains of H3 and H3.3 were used to rescue the ΔmTF cells. The GFP-expressing cells were then mated with WT cells, and the deposition of GFP-tagged protein was quantified (Fig. 5B and C). Because H3.3-GFP and some of the chimeric H3-GFP rescue cells have small MICs and cannot proceed to the later stages of conjugation, only the crescent stage of each mating was studied. RC deposition was detected by the appearance of GFP-tagged protein in crescents and RI deposition by its appearance in parental MACs.
The results show that GFP-tagged Ch1, in which two residues in α3 of H3 were mutated to the corresponding H3.3 residues, was exclusively and efficiently deposited like H3-GFP into crescents through the RC pathway, indicating that these residues have no detectable effect on H3 deposition. Ch2-GFP, with two more residues in the beginning of α2 mutated to the corresponding H3.3 residues, was observed in crescents as intensely as H3-GFP but was also detected faintly in PM, indicating that it can be weakly deposited by the RI pathway. Ch3-GFP, with three more residues in Loop1 mutated to the corresponding H3.3 residues, was deposited into crescents through the RC pathway ∼40% as efficiently as H3-GFP but was deposited into PM through the RI pathway almost as efficiently as H3.3-GFP. Ch6-GFP, in which the three residues in Loop1 alone were mutated to the corresponding H3.3 residues, was deposited into crescents similarly to Ch3-GFP and was deposited into PM ∼70% as efficiently as H3.3. Ch4-GFP, with mutations in two more residues at the end of α1, or Ch5-GFP, with mutations in one more residue in the LoopN connecting αn and α1, was deposited into crescents even less efficiently but could be deposited through the RI pathway as efficiently as H3.3-GFP. From these results, we conclude that the Loop1 region and the beginning of α2 are both required for excluding H3 from RI deposition, while all the different regions except for those in α2 and α3 contribute to efficient H3 RC deposition. Small MICs were also observed in cells rescued by Ch3, Ch4, Ch5, and Ch6 (data not shown), which have less-efficient RC deposition, consistent with the H3.3-GFP overexpression rescue results (see Fig. S2 in the supplemental material), suggesting that the small MIC phenotype was due to the less efficient RC deposition of the minor variants.
Minor RI H3s are not essential for Tetrahymena cell growth.
Both H3.3 and H3.4 can be deposited through both the RI and the RC pathways and appear to function redundantly, and as expected, cells containing only these variants are viable. The major H3, on the other hand, can be deposited only by the RC pathway, and no organism with only RC H3 has been described to date, suggesting that transcription-associated RI deposition is an essential process (8) and that cells lacking an RI variant would not be viable. To test whether this was the case, we knocked out the HHT3 and HHT4 genes (ΔHHT3 ΔHHT4). Surprisingly, the ΔHHT3 ΔHHT4 cells were viable and grew only slightly slower than WT cells (Fig. 6A). Therefore, the RI minor H3s are not essential for vegetative growth of Tetrahymena.
The transcription-associated RI deposition of H3 proteins is not essential.
A possible explanation for the viability of ΔHHT3 ΔHHT4 cells is that, in the absence of the RI variants, the major H3 can be incorporated into the transcription-associated RI pathway. Consistent with this, both Northern blot assays (data not shown) and RT-PCR analyses (Fig. 6B) showed that the HHT1 and HHT2 genes were slightly upregulated when the ΔHHT3 ΔHHT4 cells were starved. To determine whether major H3 was actually deposited through the transcription-associated RI pathway in ΔHHT3 ΔHHT4 cells, we created cells in which the endogenous HHT2 gene was replaced by HHT2-GFP and both HHT3 and HHT4 were knocked out. These cells were then mated with ΔHHT3 ΔHHT4 cells, and the deposition of H3-GFP into the transcriptionally active, nonreplicating PM of the non-GFP-expressing partner was determined. No incorporation of H3-GFP into the PM was detected at any stage of conjugation (Fig. 6C; also see Fig. S4 in the supplemental material), indicating that H3 is not deposited by the transcription-associated RI pathway in the absence of H3.3 and H3.4. These results suggest that the upregulation of the major H3 genes in starved cells in the absence of RI variants simply reflects the mechanism by which Tetrahymena cells sense the need for more H3 and induce H3 gene expression and that transcription-associated RI deposition of H3s is not essential. The observation that no GFP-tagged H3.3 or H3.4 was deposited in crescent micronuclei (Fig. 2D), where a significant level of transcription was occurring, also argues that transcription can occur without the incorporation of new RI H3s.
Chromatin structure is maintained properly in ΔHHT3 ΔHHT4 and HHT3 rescue cells.
To determine whether ΔHHT3 ΔHHT4 cells and HHT3 rescued cells, which grow more slowly than WT cells, exhibit any global alteration in chromatin structure, MNase digestion was done on nuclei isolated from these cells. No obvious differences were detected in the kinetics or pattern of chromatin fragments, indicating that the overall structure of bulk chromatin was maintained properly without either major H3 (HHT3-R cells) or minor H3s (ΔHHT3 ΔHHT4 cells) (Fig. 6D). Because no transcription-associated deposition of H3s was detected in ΔHHT3 ΔHHT4 cells, we then investigated whether the chromatin structure of the MTT1 locus, which can be highly induced by Cd2+ in these cells, was maintained properly (Fig. 6E). Starved cells were used to eliminate RC nucleosome deposition. The MNase digestion pattern of the MTT1 gene became less distinct in all strains when its expression was induced. This difference between the transcriptionally active and inactive states of the gene resembles the differences between the overall nucleosome ladders obtained from transcriptionally active MACs and transcriptionally inactive MICs (13) and likely reflects changes in nucleosomes accompanying movement of the transcription machinery along the gene. Importantly, the kinetics of MNase digestion at the MTT1 locus in ΔHHT3 ΔHHT4 cells were very similar to those in WT cells, regardless of the activity of the gene. Thus, both the pattern and the kinetics of digestion indicate that the chromatin structure of the highly transcribed MTT1 gene was properly maintained in the absence of transcription-associated RI deposition of H3s.
RI H3 variants are required for producing viable conjugation progeny.
To determine whether minor H3s have any function during conjugation, ΔHHT3 ΔHHT4 cells were mated and the conjugation process was analyzed. In early conjugation, fully extended crescents were rarely observed, most MICs adopted a partially extended shape (Fig. 7a), and conjugation was delayed (data not shown). During the crescent stage, the MIC is transcriptionally active and meiotic recombination occurs. However, the abnormal shape of the crescent nuclei is probably not caused directly by lack of RI H3s in the MIC, since neither H3.3-GFP nor H3.4-GFP was detected in the WT crescents in the analyses described above. It is more likely an indirect effect of the absence of RI H3s in the parental MACs, which are responsible for expressing most gene products, including the ones utilized by MICs. Consistent with this, we have observed changes in the expression of some genes in ΔHHT3 ΔHHT4 cells (unpublished observations).
After the crescent stage, the mating of ΔHHT3 ΔHHT4 cells proceeded as in WT cells (Fig. 7b to d). At the stage when new MACs were formed, the MICs in ΔHHT3 ΔHHT4 cells appeared decondensed (Fig. 7e to f). This decondensation was transient since, at the two-MIC exconjugant stage, all cells had two condensed MICs (Fig. 7g). However, at the following one-MIC exconjugant stage, instead of all cells having one MIC, ∼28% of exconjugants lost both of their MICs (Fig. 7h). When put into growth medium, all exconjugants lost their MICs and were unable to undergo MAC division and died (Fig. 7i). These results suggest that the minor H3s have specific functions during the late developmental stages of conjugation and are essential for producing viable progeny. The unexpected appearance of H3.3-GFP and H3.4-GFP in MICs at the NM stage (Fig. 2D; also see Fig. S3A in the supplemental material) suggests that these defects reflect a direct function for these variants in the MIC itself.
RI H3 variants affect the developmental process of sexual maturation in Tetrahymena.
In wild-type Tetrahymena, cells that have just completed conjugation are immature and cannot mate again until they have undergone ∼65 divisions (20). When mating was performed between two ΔHHT3 ΔHHT4 knockout heterokaryons (disrupted HHT3 and HHT4 genes in the MIC, WT genes in the MAC), their homokaryon progeny (complete knockout in both MIC and MAC) were mature immediately after finishing conjugation, indicating that the minor H3s may function to regulate genes controlling the sexual maturation process of Tetrahymena. Interestingly, when mated cells expressed HHT3-GFP from the zygotic NM, bright GFP spots were observed in NM 27 h after mixing (see Fig. S5 in the supplemental material). Such spots were never observed in HHT2-GFP cells, indicating that H3.3 was deposited into specific loci in late exconjugants. The deposition of H3.3-GFP probably reflects a high expression level at these loci and is not mediated by the RC pathway, since replication ceases much earlier (∼14 h postmixing). While the nature of these specific loci is not known, it is plausible to speculate that they involve genes that function in sexual maturation.
DISCUSSION
Major H3 is deposited exclusively by the RC pathway, while minor H3.3 and H3.4 are deposited by both RC and transcription-associated RI pathways.
In both conjugating and growing cells, H3-GFP is deposited only into nuclei that have gone through either replication- or recombination-associated DNA synthesis, demonstrating that in Tetrahymena, as in other eukaryotes, H3 deposition occurs exclusively through the RC pathway. H3.3-GFP and H3.4-GFP can be deposited through the transcription-associated RI pathway, as shown by their deposition into the MAC in starved cells and into PM during conjugation. H3.3-GFP and H3.4-GFP can also be deposited through the RC pathway, as demonstrated by their deposition into the MIC in growing cells but not in starved cells, even when overexpressed. H3.3 and H3.4 appear to function redundantly. Importantly, our observations show that H3, but not the RI H3 variants, is incorporated into crescent nuclei undergoing meiotic recombination, indicating that the major H3 is utilized for repair synthesis associated with meiotic recombination, while H3.3 is not detectable in this process even though it is being synthesized and deposited in the old MACs in the same cells.
We also demonstrated that the gene previously reported to be induced in starved cells in the absence of HHT3 (43) was HHT4, a newly identified RI variant, instead of HHT2. Nonetheless, it is true that, in the absence of both HHT3 and HHT4, expression of HHT1 and HHT2 is induced in starved cells. However, we conclude that the major H3 cannot serve a transcription-associated RI function, since, unlike H3.3 and H3.4, it was not incorporated into nonreplicating, transcribing PM in ΔHHT3 ΔHHT4 cells. Thus, for Tetrahymena, the important functional distinction between RC and RI H3s does not reside in the constitutive expression of the RI variant as previously concluded, but in the differences in their protein sequences, which target them specifically into their appropriate deposition pathways, as in other organisms.
Transcription-associated RI deposition of H3s is not essential for life or for transcription.
It is not surprising to find that RC H3 is not essential in Tetrahymena and that RI H3s alone can support cell growth when expressed at a level similar to that for a major H3, since they can be deposited through both RI and RC pathways and organisms (yeasts) containing only the H3.3 variant exist. However, the fact that no organism has been found lacking minor H3.3 (25) and the association of H3.3 deposition with transcription have led to the suggestion that H3.3 RI deposition is essential for maintaining transcriptionally active chromatin (8), which is likely to be essential for viability. However, we found that Tetrahymena cells can grow without RI H3s. Moreover, in starved ΔHHT3 ΔHHT4 cells, where no DNA replication occurs, the chromatin structure of a highly transcribed region is not obviously different from that in wild-type cells in the absence of detectable RI deposition of H3s. These observations show that transcription-associated RI deposition of H3s is not essential for transcription or for cell viability.
The regions that distinguish H3 and H3.3 functions.
We mapped the regions that exclude H3 from RI deposition to two residues in α-helix 2 and three in Loop1. Mutating them causes H3 deposition through the RI pathway. The two residues in α-helix 2 reside in the same region that excludes H3 from RI deposition in Drosophila, which is proposed to be the region that interacts with histone chaperones associated with the different deposition pathways (1). Tetrahymena H3s appear to have evolved to include the adjacent Loop1 region in this recognition process. We have also found that amino acids that differ between H3 and H3.3 in the N-terminal tail, LoopN, α1, and Loop1 also contribute to H3 RC deposition efficiency. It is interesting that, in most eukaryotes, H3 and H3.3 differ by only 4 or 5 amino acids while Tetrahymena H3 and H3.3 differ at more than 10 other residues. A likely explanation for these additional differences may be the complete physical separation of the transcriptionally silent, mitotically dividing MIC and the transcriptionally active, amitotically dividing MAC. This compartmentalization of functions (mitosis and transcription), usually found in the same nucleus in other organisms, causes the RI variants of Tetrahymena to function mainly in the amitotic macronuclei of vegetative cells. The major H3s, in contrast, comprise most of the H3 and function in both MACs and MICs. Because the RI variants are present in very low amounts in mitotic micronuclei, it could be argued that, in Tetrahymena, they are under little selection pressure to retain residues that are required for mitosis.
Evolution of RC and RI H3 variants.
It seems highly likely that both RC and RI H3 variants had a common ancestor at some time in eukaryotic evolution and that the genes encoding the two types of proteins arose by duplication of that ancestral gene. Our finding that Tetrahymena cells can grow with only RC or RI H3s makes it difficult to determine which H3 was the ancestral state or whether the ancestral gene had already acquired both RC and RI functions prior to its duplication. In different organisms, the two types of H3s appear to have specialized for RC or RI deposition to different degrees. In yeasts, one type of H3, more similar to the RI type, must accomplish both functions efficiently. Two alternative scenarios can explain the existence of a single type of H3 in yeasts. Because organisms both higher (metazoans) and lower (Tetrahymena) than fungi on the evolutionary tree still contain both types of H3, one possibility is that yeasts lost one of the two types of H3. However, phylogenetic analyses indicate that these two types of H3 arose independently in multiple lineages during eukaryotic evolution (25, 38). This suggests that the existence of a single, bifunctional H3 (as in yeast) might in fact be the ancestral state and that duplication and divergence into two more-distinct types has been a frequent but later event. In this case, it would be the strictly RC type of H3 that arose late. What remains unclear in this scenario is why the RI H3.3s studied to date still retain the capacity to enter the RC pathway, while the RC H3s have lost the capacity to enter the RI pathway.
Minor H3s function in development during late conjugation in Tetrahymena.
H3.3 and H3.4 have unexpected functions in Tetrahymena in the development of sexual progeny. Both H3.3-GFP and H3.4-GFP appear in germ line MICs at the NM stage. Since MICs are neither replicating nor transcribing at this time, the deposition of H3.3 into micronuclei appears to be a specific process mediated by a replication-independent pathway that is not associated with transcription. The progeny of ΔHHT3 ΔHHT4 mating cells have decondensed MICs at the NM stage, and some lose their MICs at the exconjugant stage. These results suggest that the transient appearance of minor H3s in MICs may have an important function in regulating the chromatin status of germ line micronuclei. The absence of expression of HHT3 and HHT4 in zygotic nuclei also has a remarkable effect on the number of vegetative fissions required for exconjugant progeny to attain sexual maturity. Again, there is a particular localization of RI H3 in developing MACs that may correlate with this effect. Perhaps the reason that no organism has been found lacking RI H3s is their important developmental functions instead of their transcription-associated RI function, which we have shown is not essential. H3.3 deposition into male pronuclei through a HIRA-mediated pathway in Drosophila (23) might be another instance where H3.3 regulates chromatin during development in addition to its “traditional” role of association with transcriptionally active chromatin.
Supplementary Material
Acknowledgments
We thank Josephine Bowen for critical reading of the manuscript.
This work is supported by NIH grant GM21793.
Footnotes
Published ahead of print on 14 August 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 2.Allis, C. D., M. Colavito-Shepanski, and M. A. Gorovsky. 1987. Scheduled and unscheduled DNA synthesis during development in conjugating Tetrahymena. Dev. Biol. 124:469-480. [DOI] [PubMed] [Google Scholar]
- 3.Allis, C. D., C. V. Glover, J. K. Bowen, and M. A. Gorovsky. 1980. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell 20:609-617. [DOI] [PubMed] [Google Scholar]
- 4.Bannon, G. A., F. J. Calzone, J. K. Bowen, C. D. Allis, and M. A. Gorovsky. 1983. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 11:3903-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassidy-Hanley, D., J. Bowen, J. H. Lee, E. Cole, L. A. VerPlank, J. Gaertig, M. A. Gorovsky, and P. J. Bruns. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervantes, M. D., X. Xi, D. Vermaak, M. C. Yao, and H. S. Malik. 2006. The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus. Mol. Biol. Cell 17:485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalker, D. L., and M. C. Yao. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, E. S., J. A. Shin, H. S. Kim, and Y. K. Jang. 2005. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res. 33:7102-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui, B., and M. A. Gorovsky. 2006. Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila. Mol. Cell. Biol. 26:4499-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doerder, F. P., and L. E. Debault. 1975. Cytofluorimetric analysis of nuclear DNA during meiosis, fertilization and macronuclear development in the ciliate Tetrahymena pyriformis, syngen 1. J. Cell Sci. 17:471-493. [DOI] [PubMed] [Google Scholar]
- 11.Dou, Y., J. Bowen, Y. Liu, and M. A. Gorovsky. 2002. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaertig, J., L. Gu, B. Hai, and M. A. Gorovsky. 1994. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 22:5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorovsky, M. A., C. Glover, C. A. Johmann, J. B. Keevert, D. J. Mathis, and M. Samuelson. 1978. Histones and chromatin structure in Tetrahymena macro- and micronuclei. Cold Spring Harb. Symp. Quant. Biol. 42:493-503. [DOI] [PubMed] [Google Scholar]
- 14.Gorovsky, M. A., M. C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 9:311-327. [DOI] [PubMed] [Google Scholar]
- 15.Gunjan, A., J. Paik, and A. Verreault. 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie 87:625-635. [DOI] [PubMed] [Google Scholar]
- 16.Hai, B., and M. A. Gorovsky. 1997. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 94:1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henikoff, S., and K. Ahmad. 2005. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21:133-153. [DOI] [PubMed] [Google Scholar]
- 18.Janicki, S. M., T. Tsukamoto, S. E. Salghetti, W. P. Tansey, R. Sachidanandam, K. V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R. H. Singer, and D. L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Karrer, K. M. 2000. Tetrahymena genetics: two nuclei are better than one. Methods Cell Biol. 62:127-186. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y., K. Mochizuki, and M. A. Gorovsky. 2004. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 101:1679-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loidl, J., and H. Scherthan. 2004. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J. Cell Sci. 117:5791-5801. [DOI] [PubMed] [Google Scholar]
- 23.Loppin, B., E. Bonnefoy, C. Anselme, A. Laurencon, T. L. Karr, and P. Couble. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437:1386-1390. [DOI] [PubMed] [Google Scholar]
- 24.Love, H. D., Jr., A. Allen-Nash, Q. A. Zhao, and G. A. Bannon. 1988. mRNA stability plays a major role in regulating the temperature-specific expression of a Tetrahymena thermophila surface protein. Mol. Cell. Biol. 8:427-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik, H. S., and S. Henikoff. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882-891. [DOI] [PubMed] [Google Scholar]
- 26.Marsh, T. C., E. S. Cole, K. R. Stuart, C. Campbell, and D. P. Romero. 2000. RAD51 is required for propagation of the germinal nucleus in Tetrahymena thermophila. Genetics 154:1587-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martindale, D. W., C. D. Allis, and P. J. Bruns. 1985. RNA and protein synthesis during meiotic prophase in Tetrahymena thermophila. J. Protozool. 32:644-649. [DOI] [PubMed] [Google Scholar]
- 28.McDonald, B. B. 1966. The exchange of RNA and protein during conjugation in Tetrahymena. J. Protozool. 13:277-285. [DOI] [PubMed] [Google Scholar]
- 29.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mochizuki, K., and M. A. Gorovsky. 2004. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot. Cell 3:1233-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray-Gallet, D., J. P. Quivy, C. Scamps, E. M. Martini, M. Lipinski, and G. Almouzni. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091-1100. [DOI] [PubMed] [Google Scholar]
- 32.Sarma, K., and D. Reinberg. 2005. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 6:139-149. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang, Y., X. Song, J. Bowen, R. Corstanje, Y. Gao, J. Gaertig, and M. A. Gorovsky. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 99:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stargell, L. A., and M. A. Gorovsky. 1994. TATA-binding protein and nuclear differentiation in Tetrahymena thermophila. Mol. Cell. Biol. 14:723-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugai, T., and K. Hiwatashi. 1974. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J. Protozool. 21:542-548. [DOI] [PubMed] [Google Scholar]
- 37.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 38.Thatcher, T. H., J. MacGaffey, J. Bowen, S. Horowitz, D. L. Shapiro, and M. A. Gorovsky. 1994. Independent evolutionary origin of histone H3.3-like variants of animals and Tetrahymena. Nucleic Acids Res. 22:180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 40.Woodard, J., E. Kaneshiro, and M. A. Gorovsky. 1972. Cytochemical studies on the problem of macronuclear subnuclei in Tetrahymena. Genetics 70:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, M., C. D. Allis, and M. A. Gorovsky. 1988. Cell-cycle regulation as a mechanism for targeting proteins to specific DNA sequences in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 85:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia, L., B. Hai, Y. Gao, D. Burnette, R. Thazhath, J. Duan, M. H. Bre, N. Levilliers, M. A. Gorovsky, and J. Gaertig. 2000. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 149:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, L., and M. A. Gorovsky. 1997. Constitutive expression, not a particular primary sequence, is the important feature of the H3 replacement variant hv2 in Tetrahymena thermophila. Mol. Cell. Biol. 17:6303-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.