Abstract
Metazoan genomes contain thousands of replication origins, but only a limited number have been characterized so far. We developed a two-step origin-trapping assay in which human chromatin fragments associated with origin recognition complex (ORC) in vivo were first enriched by chromatin immunoprecipitation. In a second step, these fragments were screened for transient replication competence in a plasmid-based assay utilizing the Epstein-Barr virus latent origin oriP. oriP contains two elements, an origin (dyad symmetry element [DS]) and the family of repeats, that when associated with the viral protein EBNA1 facilitate extrachromosomal stability. Insertion of the ORC-binding human DNA fragments in oriP plasmids in place of DS enabled us to screen functionally for their abilities to restore replication. Using the origin-trapping assay, we isolated and characterized five previously unknown human origins. The assay was validated with nascent strand abundance assays that confirm these origins as active initiation sites in their native chromosomal contexts. Furthermore, ORC and MCM2-7 components localized at these origins during G1 phase of the cell cycle but were not detected during mitosis. This finding extends the current understanding of origin-ORC dynamics by suggesting that replication origins must be reestablished during the early stages of each cell division cycle and that ORC itself participates in this process.
Transmission of genetic information from one cell generation to the next requires the accurate duplication of the genome. In bacteria, DNA replication initiates at a defined start site, the replicator, which is recognized and bound by an initiator protein (24). The first eukaryotic origins of replication were identified in Saccharomyces cerevisiae. In this unicellular organism, there are well-defined genetic elements that support the extrachromosomal replication of plasmids and were therefore called autonomously replicating sequences (ARS). These replication origins contain an 11-bp consensus sequence that is recognized by the origin recognition complex (ORC) (8, 52, 57).
However, in higher eukaryotes, replication origins are more difficult to define because of the increased complexity of these organisms and their genomes. Indeed, the existence of genetically defined replicator elements in higher eukaryotes is still debatable, since chromatin structure and epigenetic features are of fundamental importance for origin selection and activity (4, 9, 15, 19, 41). Many genomic sites can be used as replication origins when placed under appropriate conditions. In Xenopus and Drosophila embryos, any DNA sequence will efficiently replicate once per cell cycle up to the blastula stage of development when microinjected into embryos or introduced into egg extracts (20, 23, 67). We are only beginning to understand how, for example, higher-order chromatin organization influences origin organization and activity or how the genetic and epigenetic environment determines origin usage (37, 58). To elucidate how these processes are linked to and regulate replication initiation, it is necessary to identify and characterize DNA sequences that serve as replication origins in higher eukaryotes.
Results of biochemical fractionation, as well as genetic and physical studies, demonstrate that replication in higher eukaryotes initiates at specified start sites (10, 19). In support of genetically defined replicators in higher eukaryotic genomes, specific DNA fragments that contain the dihydrofolate reductase (DHFR), β-globin, c-myc, and lamin B2 start sites function as replicators at ectopic locations (15). As for that in budding yeast, ORC binding has been mapped to specific sites in mammalian chromosomes that colocalize with replication start sites (1, 6, 29, 35, 64). Mammalian genomes are thought to contain several thousand replication start sites (26, 39, 40, 56), some of which are hypothesized to colocalize with replicator elements, but the lack of a generally applicable and functional screening assay has hampered their identification and the definition of their characteristic features (63, 64).
Different types of general strategies to identify mammalian replicators have been developed. Early attempts to identify mammalian origins via their presumed ARS activity met with limited success due to poor autonomous replication activity and loss of small plasmids from the cells (19). To alleviate the problem of plasmid loss, Krysan and colleagues screened random mammalian DNA fragments for origin activity by cloning them into a replication-defective Epstein-Barr virus (EBV) oriP plasmid (33). Replication of wild-type oriP plasmids occurs once per cell cycle in human cells under the control of cellular replication-licensing factors and depends on two cis-acting components (11, 61, 68). One, the family of repeats (FR), is responsible for plasmid maintenance during cell division. The other, the dyad symmetry element (DS), serves as a replicator that through its interaction with the oriP-binding factor EBNA1 recruits ORC to DS (16, 61). Additional regulation of oriP replication occurs through nucleosome remodeling and covalent histone modifications (70, 71). Thus, random human fragments inserted into an oriP plasmid lacking DS were expected to replicate in human cells if the fragment served as a replicator. Indeed, Krysan and colleagues demonstrated autonomous replication of some plasmids in mammalian cells over a period of several months under selection (32, 34). Only human fragments of 6 kb or greater displayed replication activity in this assay, and activity-conferring features of the sequences other than their lengths were not identified (21). In recent years it has become clear that epigenetic processes, which are independent of replication competence, are involved in establishing long-term maintenance of plasmids (36). Currently, it is unknown to what extent DNA sequence contributes to the efficiency of establishment.
In a third general approach, a library of potential human DNA origins was prepared by cloning ORC-binding DNA fragments enriched by chromatin immunoprecipitation (ChIP). Origins located upstream of the Mcm4 and Top1 genes were identified using this approach (29). However, the effectiveness of this approach is currently uncertain because the efficiency and enrichment of the coprecipitated DNA fragments were not quantified (29). Moreover, ORC subunits may bind in vivo to regions of chromatin that serve other functions. For example, the Orc2 protein (Orc2p) has been shown to associate with nonorigin elements, such as heterochromatin, centrosomes, and centromeres (50).
To isolate new origins, we developed a two-step origin-trapping assay suitable to screen for potential replicators in human chromatin. We first used ChIP to enrich for DNA sequences bound in vivo by a subunit of ORC. In the second step, we selected for their ability to rescue transient replication of a replication-defective EBV oriP plasmid. This two-step origin-trapping assay differs from that used in the early oriP-based studies (32, 34) in that the human DNA fragments cloned into oriP plasmids are not random but rather are enriched for their ability to bind ORC in vivo. To minimize the contributions of epigenetic processes, the period of selection for oriP plasmid maintenance was comparatively brief. This new origin-trapping strategy also enhances the effectiveness of the ORC-binding library approach (29) by following it with a plasmid-based replication screen.
In this initial report we describe five new human origins of replication that were identified by the origin-trapping assay. We present evidence that the assay is a stringent screen that allows the unbiased identification of sequences that serve as replication start sites in their native chromosomal context. Finally, we show that the association of replication-licensing proteins with these sites is cell cycle regulated and that Orc subunits are largely absent from origins during mitosis.
MATERIALS AND METHODS
Plasmids.
p2832 is a 12.1-kb oriP plasmid containing oriP, enhanced green fluorescent protein (GFP), and the puromycin selection marker. p2932 (ΔDS) was obtained by deleting the DS element between BsaBI and HpaI sites and replacing it with an AscI site containing a linker element. In addition, a 2.4-kb BamHI fragment containing a 3′ long terminal repeat was removed, resulting in a 9.4-kb plasmid. DNA fragments isolated by ChIP were ligated to the AscI linker, amplified, and cloned into plasmid p2932. The lamin B2 fragment was cloned by PCR from plasmid IRALp962B012Q2 (RZPD) with an AscI-linked primer and cloned into p2932. All plasmid DNA sequences are available upon request.
Cell culture.
The Burkitts lymphoma cell line A39 (61) and the semiadherent embryonic kidney cell line HEK293/EBNA1 (36) were cultured in RPMI medium (Invitrogen) supplemented with 10% fetal calf serum and 220 μg/ml G418 (HEK293/EBNA1). HEK293 cells were drug selected with 80 μg/ml hygromycin or 200 ng/ml puromycin. The 143/EBNA1 line was generated from the fibroblast cell line 143 (ATCC CRL-7092) (69) and maintained in Dulbecco's modified Eagle's minimal medium (Invitrogen) supplemented with 10% fetal calf serum and 600 μg/ml G418. Adherent HeLa cells (ATCC CCL116) were grown in Dulbecco's modified Eagle's minimal medium (Invitrogen) containing 10% fetal calf serum. For G1/S synchronization, the cells were subjected to a double thymidine block (54). They were first arrested by a single thymidine block for 15.5 h and then released for 9 h and blocked again with 2.2 mM thymidine for 15 h. For synchronization in mitosis, cells were treated with 40 ng/ml nocodazole for 9 h and isolated by mitotic shake off. Synchronization was controlled by flow cytometry using standard procedures.
ChIP cloning.
For cloning of Orc2p-coprecipitated DNA fragments, 10 immunoprecipitations with 1 × 108 cells were performed in parallel from A39 cells and isolated as described below. To generate blunt ends, the purified ChIP sequences were pooled, dissolved in 100 μl of buffer (50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris-HCl [pH 7.9]), and treated with 2 U Klenow enzyme for 1 h. After enzyme inactivation and ethanol precipitation, the DNA fragments were ligated to 10 pmol of annealed AscI linker with 5 U T4 DNA ligase (50-μl volume, overnight at 14°C). For PCR amplification, the ligated DNA was ethanol precipitated and dissolved in 20 μl water. PCR was performed with 10 pmol AscI PCR oligonucleotides by use of 1 U TaqPlus precision polymerase (Stratagene) in 100 μl by adding 2% dimethyl sulfoxide (25 cycles of 1 min at 94°C, 1 min at 64°C, and 2 min at 72°C). PCR products of between 200 and 1,000 bp were isolated from a 0.8% Tris-acetate-EDTA agarose gel and purified (gel extraction kit; QIAGEN). One microgram of amplified DNA fragments was digested with 2 U AscI (100 μl) and separated from the nucleotides (PCR purification kit; QIAGEN). These inserts were ligated with 1 U T4 DNA ligase in five parallel samples of 20 μl overnight into AscI-digested p2932.
Chromatin immunoprecipitation.
Chromatin was prepared for immunoprecipitation exactly as described previously (55). For cloning of Orc2p-coprecipitated DNA fragments, 10 immunoprecipitations with 1 × 108 cells were performed in parallel from A39 cells. The enrichment of specifically Orc2p-bound chromatin was determined on the mini-EBV episome by use of the PCR fragment p4.2 (sc6) and the reference I3 (55).
ChIP experiments with HeLa cells were performed essentially as described previously (60). Briefly, 1 × 108 HeLa cells were washed with phosphate-buffered saline (PBS) and treated with 1% formaldehyde in prewarmed medium for 4 min at 37°C. To prepare nuclei, cells were resuspended in hypotonic RSB buffer (10 mM Tris-HCl [pH 8.0], 3 mM MgCl2), incubated for 10 min on ice, and disrupted by Dounce homogenization. Nucleoprotein was washed and resuspended in NSB buffer (10 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.1% NP-40, 1 mM EDTA). After centrifugation through a sucrose cushion, immunoprecipitation was performed as described previously (55). Ten micrograms of affinity-purified polyclonal Orc2 antibody or an equivalent amount of preimmune serum was added to 500 μg chromatin-DNA. Coprecipitated DNA was analyzed by real-time PCR (LightCycler) according to the manufacturer's instructions by use of the parameters and primer pairs described previously (55, 61).
Replication assay.
Plasmids (2 μg) were transfected into human embryonic kidney cell line HEK293/EBNA1 or human osteosarcoma cell line 143 cells stably expressing EBNA1, by using Polyfect (QIAGEN) according to the manufacturer's instructions. One day posttransfection, the cells were transferred to 15-cm plates. The transfection efficiency was determined by counting GFP-positive cells. Only transfections with comparable efficiencies were taken in account. For long-term replication assays, cells were subjected to puromycin selection (180 ng/ml) for 2 to 3 weeks. The plasmids were extracted by the method of Hirt (22) as follows. Cells were washed on the plate with TEN (10 mM Tris [pH 8.0], 2 mM EDTA, 100 mM NaCl), covered with 1.5 ml TEN, and lysed with an equal volume of 200 mM NaOH, 1% sodium dodecyl sulfate. After neutralization with 750 μl 5 M NaCl, the lysate was kept on ice for 4 to 5 h or overnight and centrifuged at 15,300 rpm for 30 min at 4°C. The supernatant was extracted twice with phenol-chloroform, and the DNA was precipitated with isopropanol. After dissolving in TE, 500 ng of each DNA sample extracted from a single transfection was digested for plasmid rescue with DpnI and electroporated into DH10B bacteria (Invitrogen). To linearize plasmids for Southern blot analysis, 2 μg of Hirt DNA was incubated with DpnI and BamHI. DNA was subjected to electrophoresis on 0.8% Tris-acetate-EDTA gels and transferred to a nitrocellulose membrane. The membrane was hybridized to 32P-labeled p2832. For short-term replication experiments, transfected cells were grown for 4 days without selection. Plasmid DNA was enriched as described above, and 500 ng of each transfected sample was digested with DpnI. To monitor the DpnI digestion, one nontransfected sample was spiked with 500 ng pUC. The samples were transfected into DH10B bacteria by electroporation. To measure the total DNA recovered as reference, 5 ng DNA not digested with DpnI was electroporated in parallel.
Nascent strand analysis.
Nascent strand analysis was performed as described previously (28). HEK293/EBNA1 or A39 cells (2 × 107) were washed with PBS and resuspended in PBS with 10% glycerol. Cells were lysed for 10 min in slots of a 1.2% alkaline agarose gel (50 mM NaOH, 1 mM EDTA). Afterwards, DNA was separated by electrophoresis overnight (low-melting-temperature agarose; Biozym). Fragments of from 0.8 to 1.3 kb were extracted from the gel by use of a QIAquick gel extraction kit (QIAGEN). Nascent strand abundance was determined by quantitative real-time PCR.
Separation of BrdU-substituted DNA.
HEK293/EBNA1 cells transfected with the different replicator plasmids were drug selected for 2 weeks and then transferred for 18 h to a cell culture medium containing 5-bromodeoxyuridine (BrdU; 10 μM) to increase the buoyant density of replicated DNA for CsCl equilibrium gradient centrifugation. Total DNA was isolated from 1 ×107 cells and digested with the restriction enzymes BamHI and DpnI. CsCl gradients were adjusted to a refractive index of 1.403 and spun in a Beckman SW41 rotor at 40,000 rpm for 48 h. Fractions of 250 μl were collected, and their refractive indexes were determined. The content of bulk genomic DNA was determined with a fluorometer (Beckmann). Samples were diluted, precipitated, and further analyzed by quantitative PCR.
Primers.
All primers used in this study are summarized in Table S3 in the supplemental material and are also available on http://haema143.gsf.de/.
RESULTS
Establishment of a plasmid replication assay.
The goal of the origin-trapping assay was to design a functional tool capable of effectively identifying active origins. We made use of the latent origin of replication of Epstein-Barr virus (oriP), a replicator that supports extrachromosomal replication of plasmids once per cell cycle in an ORC-dependent manner. We reasoned that since ORC-binding sites colocalize with chromosomal replication start sites, human ORC-binding sequences might confer plasmid DNA replication activity in human cell lines on oriP plasmids lacking the viral replicator element DS. If so, a plasmid replication assay might be suitable as a functional screen for origin trapping.
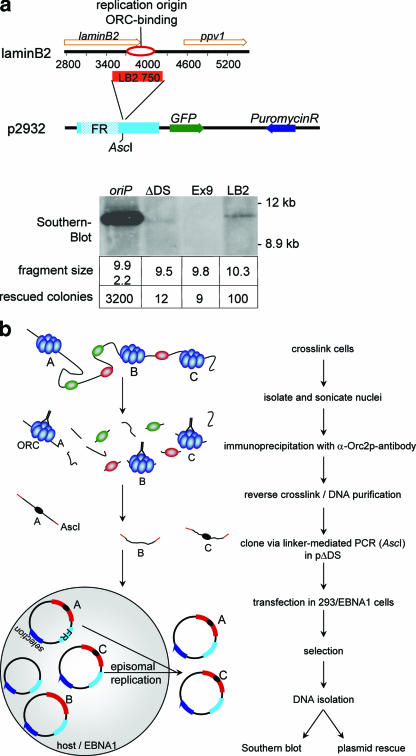
To assess the feasibility of this idea, a 750-bp fragment of the human lamin B2 origin (oriLB2) that binds to ORC in vivo (18) and the control fragment (Ex9) located in exon 9 of the Mcm4 gene, 6.5 kb upstream of the Mcm4 origin (oriMcm4) (35). The exon9 fragment does not bind ORC and was cloned into an oriP plasmid lacking the DS origin but retaining the viral maintenance element FR (Fig. 1a). These plasmids were separately transfected into HEK293 cells expressing the viral protein EBNA1, which binds to FR and DS (36, 69). After 2 weeks of antibiotic selection, the amount of plasmid maintained in each transfected culture was determined by two methods, Southern blot hybridization and plasmid rescue (Fig. 1a). The high signal intensity in Southern blots and the large number of colonies obtained by plasmid rescue indicated efficient replication of the positive control oriP plasmid, while the negative control ΔDS plasmid yielded only a faint signal in Southern blots and few colonies by plasmid rescue. The plasmid bearing the 750-bp oriLB2 fragment reproducibly yielded colony numbers up to eightfold greater than the ΔDS plasmid did. In contrast, only residual replication activity similar to that of ΔDS was observed with the plasmid containing the nonorigin fragment exon9 (Fig. 1a). These results indicate that an ORC-binding human DNA fragment conferred transient replication activity on the ΔDS plasmid, albeit with an activity lower than that of the wild-type oriP.
FIG. 1.
Establishment of the origin-trapping assay. (a) Schematic representation of test replicators and organization of the lamin B2 locus. The lamin B2 origin is located in the 3′ region of the lamin B2 gene (top). The basic vector ΔDS contains the maintenance element FR of EBV, the GFP and puromycin genes, and an AscI site used for cloning of Orc2p-binding fragments. The lamin B2 origin (oriLB2; positions 3495 to 4267 of the gene under accession no. M94363) was cloned in place of the DS element by use of the AscI site. Similarly, the exon9 fragment (Ex9) located near the Mcm4 origin (positions 6342 to 6729, accession no. U63630) was inserted. These plasmids were transfected into HEK293/EBNA1 cells and analyzed for transient replication competence by Southern blotting and plasmid rescuing. For Southern blotting, plasmids were digested with BamHI and DpnI. The sizes of the BamHI fragments are indicated below each lane of the blot. The number of E. coli colonies obtained after transfection of 500 ng Hirt-extracted DNA from an exemplary experiment with each plasmid is also indicated. At least three experiments were performed for each analyzed origin, and results are summarized in Table 1. (b) Schematic diagram of the origin-trapping assay (see Results for details).
Orc2p-binding elements support replication of ΔDS plasmid DNA.
The origin-trapping assay combines a ChIP protocol and a plasmid replication assay (Fig. 1b). To prepare a library of potential human replicators, we enriched Orc2p-binding fragments from the established human mini-EBV cell line A39 by ChIP cloning (61). Cross-linked chromatin fragments containing DNA of 1 kb or less were generated by sonication and partial digestion with micrococcal nuclease. Chromatin fragments were immunoprecipitated with antibodies directed against the human Orc2p, and the precipitates were pooled after the precipitation step. To assess the specificity of this approach, we determined the oriP-specific enrichment of Orc2p as an internal control. Using primer set 4.2, which binds near the oriP origin, DS, we measured at least a 10-fold enrichment of Orc2-bound DNA relative to what was seen for a distal reference fragment (data not shown) (55). Coprecipitated Orc2p-binding fragments of 200 to 1,000 bp were size selected, amplified by AscI linker-mediated PCR, purified, and cloned into the ΔDS plasmid by use of the AscI site present in the inserted linker. This library was then screened to identify ΔDS plasmids bearing human DNA elements able to support extrachromosomal replication.
Plasmids that preferentially contained 200- to 500-bp inserts were tested in pools of four to six origin candidates for transient replication activity in HEK293 cells stably expressing EBNA1. This cell line was chosen because it is easily transfected with high efficiency and is established as a cell line that supports oriP-dependent replication (36). After 2 weeks of selection, plasmids that had been extrachromosomally maintained were rescued back into Escherichia coli and analyzed by restriction enzyme digestion (see Table S1 in the supplemental material). Two readouts reflect the stringency of this replication assay: (i) the number of colonies obtained after transformation into E. coli compared to that obtained with ΔDS and (ii) the enrichment of a dominant candidate fragment in each pool. In 10 out of 20 pools, up to fourfold more colonies were obtained with plasmids bearing a human DNA fragment than with the negative control ΔDS. Most replication-competent pools contained one predominant clone (see Table S1 in the supplemental material).
Potential origins of replication serve as chromosomal initiation sites.
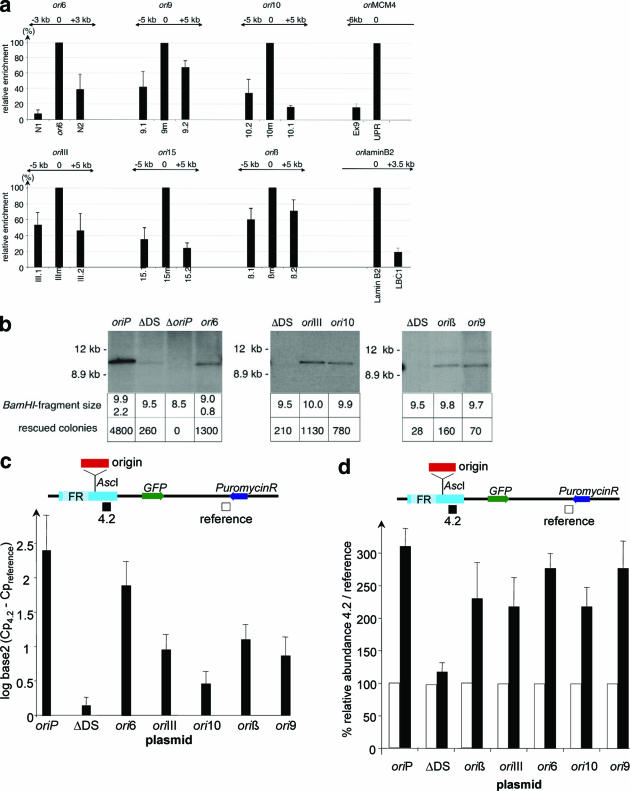
To test whether the isolated sequences were indeed active origins of replication, we measured in the mini-EBV cell line A39 and in HEK293/EBNA1 cells their initiation activities in their native chromosomal context with the nascent strand abundance assay (5, 13, 35). Nascent strand abundance at the hypoxanthine phosphoribosyltransferase, MCM4, and lamin B2 origins was measured as a positive control (Fig. 2a and data not shown) (13, 18, 35). A panel of primer pairs was generated for each potential replicator, encompassing up- and downstream elements of the corresponding chromosomal regions (for chromosomal locations and primers, see Tables S1 and S2 in the supplemental material). The relative amount of nascent strand DNA in the neighboring regions was determined and compared with that at the potential origin, set as 100%. Nascent DNA enrichment ranging from 10 to 75% at the distal regions was determined for the six potential origins compared to the reference locus (Fig. 2a). Measurements of nascent strand abundance at these loci in the HEK293/EBNA1 cell line confirmed that all six trapped sequences reside at active chromosomal replication origins (data not shown).
FIG. 2.
Confirmation of potential human replicators. (a) The abundance of sequences corresponding to the potential human origins was quantified for nascent genomic DNA (800 to 1,300 nucleotide fragments) prepared from 1 × 107 A39 cells. For each potential replicator, we designed primer pairs located 5 kb and 3 kb down- and upstream, respectively, of the origin region. The putative initiation site is located in the middle primer set (m), and this center bar was defined as 100%. The abundance at the flanking regions was calculated relative to that at the potential origin site. The MCM4, lamin B2, and hypoxanthine phosphoribosyltransferase origins (data not shown) served as internal positive controls for each preparation with primer pairs previously described (13, 18, 35). The name of each element is indicated at the top of each histogram, which depicts the average and standard deviation of at least three independent experiments. The middle primer set (m) defines the isolated origins (ori6, ori9, ori10, oriIII, ori15, and oriβ). The primer sets on the left and right define reference fragments. UPR, PCR fragment spanning oriMCM4; N1 and N2, reference fragments of ori6; LBC1, control fragment of oriLB2. (b) Each plasmid bearing a human Orc2-binding sequence was transfected individually into HEK293/EBNA1 cells, and transient replication was evaluated after 2 weeks by plasmid rescue analysis and Southern blotting. The size of the DNA fragment obtained after BamHI digestion is indicated below each lane of the blot. The number of E. coli colonies rescued in the same experiment is indicated below. The result of one exemplary experiment is shown. At least three experiments were performed for each analyzed origin, and results are summarized in Table 1. Similar results were obtained for the human osteosarcoma cell line 143 stably expressing EBNA1 (data not shown). (c) ChIP experiments with HEK293/EBNA1 cells were performed to measure Orc2p association at the potential human replicators inserted at the AscI site relative to that at a distal sequence in the plasmid. Relevant features of the replicator plasmids are diagrammed at the top. The primer set 4.2, near the AscI site, and the reference primer set, located at the puromycin gene, are depicted below. The site-specific enrichment (ΔCp = Cp at 4.2 − Cp at reference) of Orc2p was calculated on a logarithmic scale (log base 2) as the difference between the primer locus 4.2 and the reference. The averages and standard deviations of triplicates are shown. (d) Nascent strand abundance at primer set 4.2 and at the reference primer set was analyzed 4 days after plasmid transfection into HEK293/EBNA1 cells. The histogram shows the abundance of fragment 4.2 relative to that of the reference fragment. The number of nascent strand molecules was first determined by a standard curve for the amplified regions. All plasmids except the negative control, ΔDS, had similar amplification rates. The number of molecules determined for the reference was set at 100% (white bars). The black bars show the ratios between the abundance of the reference and those of the 4.2 sequences in nascent DNA. The graph shows the mean values and standard deviations, indicated by error bars, from three independent experiments.
To assess the effectiveness of the origin-trapping assay in enriching for putative origin fragments, we compared the chromosomal replication initiation activity of the six replicator candidates shown in Fig. 2a with that of one of the DNA sequences in groups 3 and 7, which contained no obvious origin candidates in the transient replication screen (see Table S1 in the supplemental material). None of these sequences showed any chromosomal initiation activity (data not shown). These findings indicate that the origin-trapping assay selected quite effectively for active chromosomal replication origins.
Replicators support extrachromosomal replication but not long-term establishment.
To further characterize the novel origins as replicators, we reexamined them individually in plasmid replication assays after 2 weeks of drug selection. Five of the six plasmids with potential origin sequences generated colony numbers similar to those obtained with a plasmid containing oriLB2; these colony numbers were 2.3- to 4.4-fold higher than those obtained with ΔDS (Fig. 2b; Table 1). The remaining candidate, ori15, did not support significant plasmid replication activity and was therefore not further analyzed (data not shown). We confirmed the results of the replication assay for these replicators with another EBNA1-expressing cell line, 143/EBNA1 (69), clearly verifying these replicators as active replication origins in two different cell lines.
TABLE 1.
Summary of results for human replicators in plasmid contexta
| Origin plasmid | Value after 2-wk selection
|
Avg test plasmid/ΔDS plasmid ratio after 4 days (no selection) | Extrachromosomal maintenance for 6 wk (under selection) | Relative site-specific enrichment ratio forc:
|
||
|---|---|---|---|---|---|---|
| Avg no. of colonies rescued (no. of exptsb) | Avg test plasmid/ΔDS plasmid ratio | Orc2 binding | Nascent strand | |||
| oriP | 3,828 ± 445 (7) | 21.6 | 20.4 | Yes | 5.28 | 3.08 ± 0.3 |
| ΔDS | 177 ± 68 (7) | 1 | 1 | No | 1.14 | 1.18 ± 0.15 |
| oriLB2 | 712 ± 246 (3) | 4.0 | ND | No | ND | ND |
| ori6 | 577 ± 20 (3) | 3.3 | 6 | No | 3.81 | 2.27 ± 0.21 |
| ori10 | 600 ± 224 (3) | 3.4 | 2.2 | No | 1.38 | 2.11 ± 0.31 |
| oriβ | 457 ± 161 (3) | 2.6 | 4.5 | No | 2.2 | 2.25 ± 0.5 |
| oriIII | 773 ± 248 (3) | 4.4 | 5.4 | No | 1.91 | 2.11 ± 0.47 |
| ori9 | 289 ± 75 (3) | 2.3 | 2.4 | No | 1.81 | 2.27 ± 0.4 |
The replication and maintenance competences of the individual plasmids were tested in transient replication assays. Exemplary results thereof are shown in Fig. 1a and 2b. Colony numbers were normalized to the absolute amount of extracted DNA by calculating the ratio between 500 ng of DpnI-treated and 5 ng of untreated DNA sample. ND, not determined.
At least three independent experiments were performed with each plasmid.
The Orc2 binding and the start site mapping data indicate the enrichment (n-fold) of the origin-proximal PCR fragment 4.2 versus the reference. Each experiment was performed in triplicate.
To assess whether the origin-containing plasmids could also be maintained in human cells without antibiotic selection, equal numbers of origin and negative control (ΔDS) plasmid molecules were separately transfected into HEK293/EBNA1 cells, cultivated for 4 days, and recovered from the cells. If the recovered plasmid DNA had undergone replication in human cells, it should be resistant to DpnI digestion and would generate colonies after electroporation into E. coli. Thus, the replication competence of each plasmid was calculated as the ratio of the colony number generated by DpnI-treated relative to untreated DNA. The replication competences of the five origin plasmids in human cells without drug selection were two- to sixfold greater than that of the negative control ΔDS (Table 1). These results confirm that the trapped genomic fragments support plasmid replication in human cells.
Next, we investigated plasmid maintenance in HEK293/EBNA1 human cells after 1 month. The cells were transfected with wild-type oriP, the negative control plasmid ΔDS, and ΔDS derivatives containing the isolated replicator ori6 or oriLB2. The cells were placed under selection 2 days after transfection, and puromycin-resistant populations were grown for 2 weeks. At this time point, cultures were split in half and cultured in the presence or absence of selection. Plasmid loss in the absence of puromycin was monitored every third day. At different times following transfection, plasmid DNA was isolated by Hirt extraction (22) and digested with DpnI, and 500 ng of DNA was transfected into E. coli. We found that ΔDS was lost from HEK293/EBNA1 cells 6 days after withdrawal of selection, whereas the ori6- and oriLB2-containing plasmids were maintained for 9 days under nonselective conditions. HEK293/EBNA1 cells transfected with the origin plasmids (ori6, ori9, and oriLB2) and control plasmids (oriP and ΔDS) were also kept under selection and analyzed 2, 4, and 6 weeks posttransfection for the extrachromosomal status of plasmids. Plasmid DNA was isolated from the different cell lines and transfected back into E. coli. None of the isolated small DNA fragments maintained long-term episomal replication of their ΔDS derivatives for at least 6 weeks (Table 1; also data not shown).
The trapped human replicators serve as ORC binding and initiation sites in the ΔDS plasmid.
The new human replicator elements described here were selected as Orc2p-binding fragments able to confer replication competence to a defective oriP plasmid. To assess whether Orc2p is indeed specifically associated with the human replicator elements, ChIP experiments were performed. The replicator plasmids were individually transfected into HEK293/EBNA1 cells and drug selected for 2 weeks prior to the ChIP. As positive and negative controls, we used the oriP and ΔDS plasmids. The association of Orc2p with the origin sequences was compared to that of Orc2p at a distal reference site (see map in Fig. 2c). As expected, oriP showed reproducible enrichment of Orc2p at the wild-type viral origin relative to what was seen for a distal reference fragment, whereas ΔDS exhibited no Orc2p enrichment at primer set 4.2 near the AscI site (Fig. 2c). The replicator ori6 demonstrated enrichment of Orc2p at primer set 4.2 that was 3.8-fold greater than that for the reference site (Fig. 2c; Table 1). The sequences of the oriIII, oriβ, ori9, and ori10 elements were significantly enriched in Orc2p-chromatin immunoprecipitates compared to the corresponding sequences in the negative control ΔDS but to a lower extent than the sequences for ori6 and oriP were.
To investigate whether the newly identified human replicators serve as the preferred initiation sites for plasmid replication, we performed nascent strand abundance analysis. As shown in Fig. 2d, the 4.2 sequence was greatly enriched relative to the reference in nascent DNA from cells transfected with each of the human replicator plasmids and the oriP plasmid. As expected, no preferred initiation site was observed in the ΔDS plasmid. In addition, the copy number of nascent strand DNA obtained from ΔDS-transfected cells was several hundredfold lower with that obtained from the same number of transfected cells containing the human origin plasmids, indicating a severe reduction in replication competence. From these results, we conclude that the human replicator elements isolated by the origin-trapping assay function in their plasmid context as site-specific origins of DNA replication.
Once-per-cell-cycle plasmid replication is EBNA1 independent.
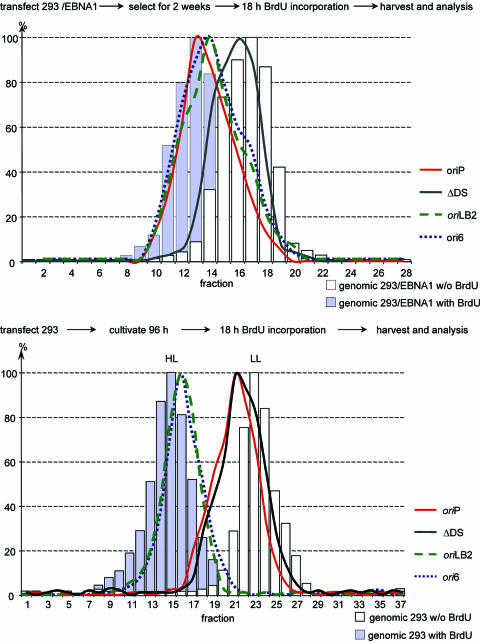
The results shown in Fig. 2, which are summarized in Table 1, indicate that the newly identified human DNA origins have several properties predicted for a human chromosomal replicator. Another property predicted for a human replicator is that it should direct initiation of DNA replication only once per cell cycle. The ability of the newly identified origins to initiate replication once per cell cycle can be measured in their plasmid context with the classic Meselson-Stahl density approach (2, 45). To test this prediction, we analyzed ori6 and oriLB2, as well as wild-type oriP and ΔDS control plasmids, with this approach. HEK293/EBNA1 cells transfected with the different plasmids were drug selected for 2 weeks and then transferred to medium containing 10 μM BrdU for 18 h. Total DNA was isolated, digested with BamHI, and fractionated by equilibrium density gradient centrifugation. The amount of genomic DNA in each fraction was determined by measuring the absorbance at 260 nm and confirmed by quantitative PCR using a primer pair located at oriMCM4 (35). The distribution of the plasmid sequences within the fractions was determined by real-time PCR, using the plasmid-specific primer set 4.2. We found that ΔDS, which lacks an efficient replication origin, incorporated BrdU only very inefficiently (Fig. 3, upper panel), consistent with a previous report stating that an EBV plasmid containing only the FR element of oriP supported replication poorly (36). In contrast, the oriP control plasmid and the oriLB2 and ori6 plasmids shifted from a being light fraction to having a density near that of a heavy-light hybrid (Fig. 3, upper panel). These data clearly demonstrate that plasmids containing these two human chromosomal origins replicated efficiently once in the HEK293/EBNA1 cell cycle.
FIG. 3.
Human replicator plasmids are duplicated once per cell cycle. Equimolar amounts of wild-type oriP, ΔDS, ori6, and lamin B2 plasmids were introduced independently into HEK293/EBNA1 (upper panel) and HEK293 (lower panel) cells. Ninety-six h after transfection, the cells were labeled with bromodeoxyuridine for 18 h. Total DNA was isolated from 1 × 107 cells, digested with BamHI, and separated by equilibrium centrifugation in a CsCl density gradient. The refractive index and the amount of total genomic DNA (absorbance at 260 nm) were determined for every second fraction and confirmed by PCR amplification at the lamin B2 origin (primer pair lamin B2 [Fig. 2]). Plasmid DNA in the light-light (LL) and heavy-light (HL) hybrid fractions was digested with DpnI and quantified using real-time PCR and a plasmid-specific primer pair that amplifies a sequence containing DpnI sites. For each of four independent experiments, the level of replicated test plasmid was set to 100%, and the relative DNA concentration was plotted versus fraction number. No DNA was detected at the density at which heavy-heavy hybrid fraction DNA should migrate.
We have previously shown that EBNA1 is in a complex with ORC, thus recruiting ORC to the latent EBV origin oriP (61). Therefore, it is formally possible that EBNA1, by binding to the FR, recruits ORC to the different plasmids. To test this possibility, we asked whether the plasmids containing human replicators are duplicated independent of EBNA1. Some residual EBNA1-independent replication of oriP plasmids might occur in the first 48 h after transfection, but continued replication at 96 h after transfection is dependent on EBNA1 (3). The density transfer experiments were repeated in HEK293 cells transiently transfected for 96 h with the same plasmids as before. After being labeled with BrdU for 18 h, total DNA was isolated, digested with BamHI and DpnI, and fractionated by CsCl gradient centrifugation as before. The fractions were collected and analyzed as before. At 18 h after the addition of BrdU, when most cells had progressed through S phase, the ΔDS plasmid and the oriP plasmid remained near the light density, indicating that they did not replicate in the absence of EBNA1 (Fig. 3, lower panel). In contrast, the ori6- and oriLB2-containing plasmids had shifted to the heavy-light hybrid fraction (Fig. 3, lower panel). At 96 h after transfection, the transfected cells still contain a large number of nonreplicated input plasmids. To test the completeness of the DpnI digestion, we used a plasmid-specific primer pair that contains two DpnI recognition sites and amplifies only DNA fragments generated by cellular replication. These control reactions gave an amplification level the same as that with primer pair 4.2. These data indicate that the plasmids containing human chromosomal origins replicated independently of EBNA1.
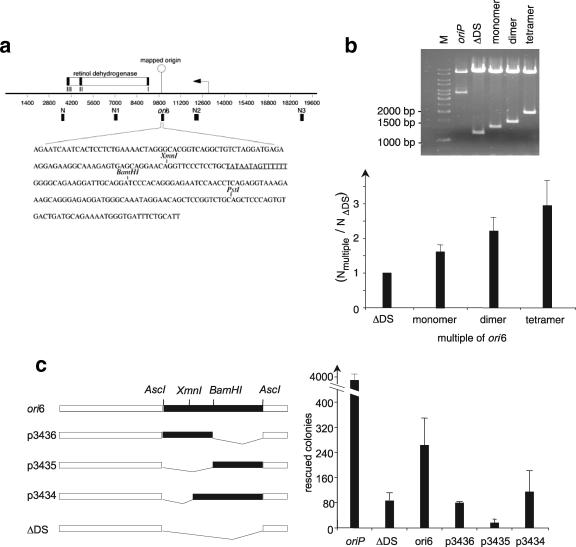
Characterization of the replication origin ori6.
The replication efficiency of the isolated replicator elements is moderate in comparison to that of oriP. To analyze whether the efficiency can be improved, we multimerized ori6 (Fig. 4a) and tested the different plasmids in transient replication assays (Fig. 4b). As a positive control we used oriP (data not shown), and as a negative control we used ΔDS (Fig. 2b). The multimerization of the ori6 sequence resulted in a linearly increased replication efficiency, indicating that additional elements might be required for an improved origin activity.
FIG. 4.
Genetic analysis of ori6. (a) The Orc2-precipitated ori6 sequence is located in the 5′ untranslated region of the retinol dehydrogenase gene. The genomic organization of this region is shown on top. The arrowhead depicts the transcription start site, and the putative exons are shown as black boxes (I to III). The location of the cloned Orc2-binding fragment is shown below the line (ori6), as are the locations of the control regions amplified by quantitative PCR (N to N3). The 249-bp-long fragment, including the restriction enzymes used for the deletion mutants as well as an AT-rich region (underlined), is depicted below. (b) ori6 fragments were multimerized in a head-to-tail fashion. The ori6 sequence was PCR amplified from the ori6 plasmid with one primer containing an AscI restriction site and one containing an AflIII restriction site. After the PCR, ori6 fragments were ligated to each other by a head-to-tail cloning strategy and ligated into plasmid p2932 (ΔDS). Replication competence was determined as described above in three independent transient replication assays. (c) Deletion analysis of ori6 located at the retinol dehydrogenase gene. Different deletion mutants of ori6 are depicted on the left. Using the indicated internal restriction sites, XmnI and BamHI, generated the deletion mutants. The replication competences of the different mutants were determined in transient replication assays with HEK293/EBNA1 cells and quantified by counting the rescued colonies. The histogram shows the average values and standard deviations for triplicates.
We also performed a deletion analysis of ori6 in order to determine whether or not one particular DNA motif of ori6 was required for ORC binding and replication efficiency. The different ori6 mutants (Fig. 4c, left) were tested in a transient replication assay. For comparison, we used oriP, ori6, and ΔDS and observed that not one particular element is crucial for extrachromosomal replication but that the deletion of any element results in a loss of replication function (Fig. 4c right). These results indicated that the whole ori6 sequence is needed for initiation of replication.
Binding of ORC and MCM subunits at origins in different phases of the cell cycle.
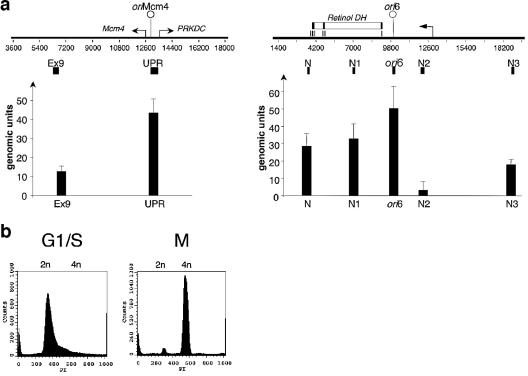
The colocalization of the newly identified human replicators with chromosomal initiation sites (Fig. 2) and their abilities to bind ORC and direct once-per-cell-cycle plasmid replication in an EBNA1-independent fashion (Fig. 2 and 3) suggested that in their native chromosomal context, these replicators are likely licensed during G1 phase to initiate replication. This prediction was tested by measuring Orc2p enrichment at the potential origins ori6 and ori9 in their native chromosomal contexts in ChIP assays relative to preimmune control precipitates and the input DNA (35, 60). In initial experiments with asynchronously proliferating HeLa cells, we found that the origin sequence (UPR) (Fig. 5a) located between the Mcm4 and PRKDC genes was enriched threefold in Orc2p immunoprecipitates relative to the origin-distal exon9 control sequence (Fig. 5a). Chromatin from the same ChIP experiments was moderately enriched in sequences at ori6 relative to the reference sequences in the flanking retinol dehydrogenase gene (Fig. 5a, N and N1) and more highly enriched relative to reference sites N2 and N3. Similarly, the ori9 replicator sequence displayed moderate enrichment in the Orc2p precipitate relative to reference sequence 9.2 but significant enrichment relative to control sequence 9.1 (data not shown).
FIG. 5.
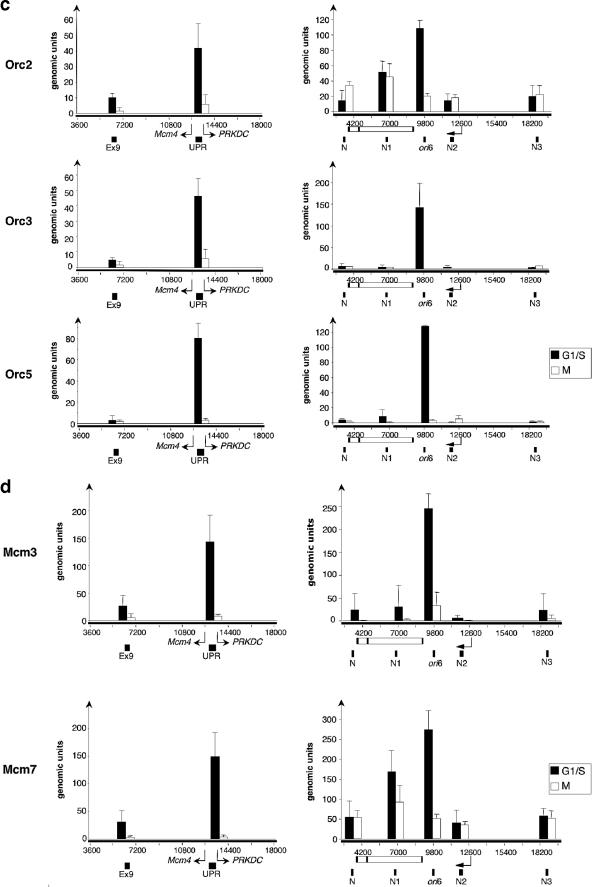
Orc2p localizes near newly identified origins in chromatin from human cells. (a) Cross-linked chromatin fragments prepared from asynchronously growing HeLa cells were enriched by ChIP using antibodies against Orc2p. DNA sequences that coprecipitated with Orc2p were quantified using primer sets specific for the Mcm4 origin (left) and ori6 (right). The genomic units were calculated relative to values for serially diluted genomic DNA by quantitative real-time PCR as described previously (60). Diagrams of the genomic regions flanking the Mcm4 and ori6 origins show the locations of primer sets (small black boxes) and the start sites and direction of gene transcription (arrow). UPR is the PCR fragment spanning oriMCM4, whereas exon9 (Ex9) is a reference site located 6.5 kb apart from oriMCM4 (60). DH, dehydrogenase. (b) Flow cytometry of HeLa cells synchronized at G1/S with thymidine and at G2/M with nocodazole. PI, propidium iodide. (c and d) ChIP analysis with antibodies directed against Orc2, Orc3, Mcm3, and Mcm7 was performed using HeLa cells synchronized in G1/S and G2/M. Coprecipitated genomic sequences surrounding the MCM4 (c) and ori6 (d) replicators were quantified by real-time PCR using the primer sets used for the experiment shown in panel a (27). The results of at least three independent experiments are shown.
To monitor the cell cycle dependence of ORC localization, ChIP assays were performed using chromatin isolated from HeLa cells synchronized at G1/S with a double thymidine block and in mitosis with nocodazole (Fig. 5b). The specific enrichment of Orc2p, Orc3p, and Orc5p in G1/S at the ori6 and ori9 origins was similar to that at oriMcm4 (Fig. 5c) (see Fig. S1 in the supplemental material) (60). This enrichment was much greater than that observed with chromatin from asynchronous cultures (Fig. 5a). Mcm3p and Mcm7p immunoprecipitates of chromatin fragments from G1/S cells were also efficiently enriched for the origin-specific sequences relative to the more distal sequences (Fig. 5d). As expected, the licensing factors Mcm3p and Mcm7p were released from the origins in mitosis, and the amplification at the sites was as low as that at the neighboring regions (Fig. 5d). Similarly, no enrichment of Mcm4, ori6, or ori9 origin sequences was detected in all ORC immunoprecipitates prepared from mitotic chromatin (Fig. 5c) (see Fig. S1 in the supplemental material). These observations confirm and extend a previous report of cell cycle-regulated association of Orc2p at the oriLB2 origin (1).
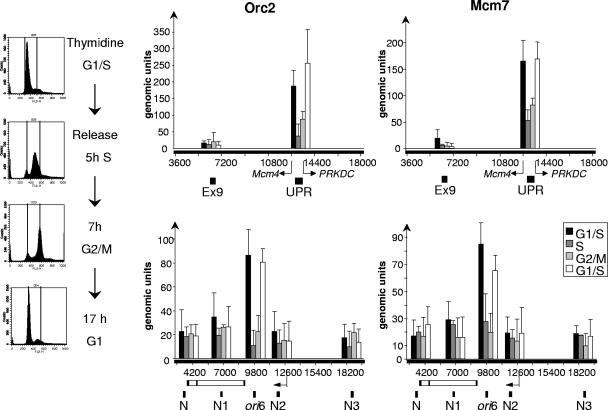
To rule out the possibility that excess nocodazole caused the loss of ORC-binding at origins in mitotic chromatin, we repeated the ChIP experiments using cells that had been synchronized at G1/S as described above and then released into medium without any inhibitor. The site-specific association of Orc2p and Mcm7p was measured at ori6, ori9, and oriMcm4 in chromatin immunoprecipitated from blocked cells and from cells harvested at 5 h (S phase), 7 h (G2/M), and 17 h (G1) after release from the G1/S block (Fig. 6). Both Orc2p and Mcm7p were specifically enriched at origins during early cell cycle phases (G1 and G1/S), while little Orc2p or Mcm7p was detected at origins in later stages of the cell cycle. These results are in accord with those shown in Fig. 5c and d and demonstrate that ORC and MCM associate with these human origins in G1 HeLa cells and are largely absent from these sites in mitotic chromatin.
FIG. 6.
Cell cycle-dependent localization of Orc2p and Mcm7p at human chromosomal origins. HeLa cells were synchronized at G1/S with a double thymidine block and released into fresh medium for 5, 7, or 17 h. The cell cycle distribution of each population was determined by flow cytometry (left). The histograms (right) show ChIP enrichment of Orc2p and Mcm7p at the MCM4 and ori6 origins during the different stages of the cell cycle. The data shown are the mean values and standard deviations for three independent experiments. Diagrams of the genomic regions flanking the MCM4 and ori6 origins are shown in Fig. 5.
DISCUSSION
We have established a novel two-step origin-trapping assay and validated it as a useful tool to identify active origins of replication in human cells. The trapped origins are recognized by ORC in vivo and serve as replication start sites in a plasmid and in their native chromosomal contexts.
An effective screen for active human replication origins.
This origin-trapping strategy differs from earlier approaches developed in the Calos and Knippers laboratories in several respects. Keller and colleagues at the Knippers laboratory used the ChIP technique to enrich ORC-binding sequences that were then used to identify ORC-binding elements in the corresponding genomic regions, followed by nascent strand analysis to confirm them as active origins (29). Although the efficiency of this approach was not determined quantitatively, 2 out of 20 precipitated and cloned elements were reported to show high ORC enrichment in confirmatory ChIP experiments. These observations highlight the need for a screen, such as that used in our origin-trapping assay, to functionally assess the origin activity of coprecipitated ORC-binding fragments.
Krysan and coworkers at the Calos laboratory used an unbiased cloning approach to identify human DNA sequences that supported autonomous replication of oriP-like plasmids in human cells under drug selection for 2 months (33). Larger fragments supported autonomous replication more efficient than that of smaller fragments, but the possibility that the DNA sequence influenced replication competence of these plasmids was not excluded. It is possible that large human DNA fragments are more likely to contain an origin. Alternatively, large fragments may favor establishment of epigenetic features that allowed plasmid maintenance for several months in human cells. To minimize the contributions of largely uncharacterized epigenetic events, we used a relatively short selection period of 2 weeks. Consistent with this possibility, we found that oriP-like plasmids containing trapped small human fragments (less than 1,000 bp) support plasmid replication but were not maintained for longer than a month.
Newly identified human origins have properties of chromosomal replicators.
The five human replicators identified in our assay replicate once in each cell cycle in the context of a ΔDS plasmid, and replication begins near the human origin fragment in these plasmids and in the native chromosomal context in human cells. Although the newly isolated origins share no sequence homology, all of them are located in a distance of 7 to 10 kb from matrix attachment regions (see Table S2 in the supplemental material). All newly identified origins are located in actively or putatively transcribed regions (see Table S2 in the supplemental material). This strongly supports the hypothesis that transcription may be supportive of origin activity or origin selection. ORC associated with each newly identified human origin in context of the ΔDS plasmids and was localized at the corresponding sites in human chromosomes during the G1 phase of the cell cycle. Unexpectedly, Orc2p was not detected at the analyzed origins in mitosis, which appeared to contradict the biochemical analysis of cell cycle-fractionated chromatin showing that Orc2p and other components of ORC might be stably associated with origins throughout cell division (44, 48, 54). However, Orc2p is known to associate with nonorigin elements in vivo (50, 51). It is important to differentiate between the global chromatin binding of Orc proteins as it is studied with chromatin binding experiments (43) and their specific binding to replication origins as it is studied with ChIP experiments. Chromatin binding experiments measure physical properties of complexes bound to chromatin and allow the identification and characterization of subcomplexes with different affinities. ChIP experiments, however, allow for the characterization of complexes at specific sites of the chromatin.
From the discussion above, it is quite possible to conclude that only a minority of ORC is actually bound to origins in a cell cycle-dependent manner and that the majority is involved in other chromatin-associated processes that are not cell cycle regulated. We confirmed that three different Orc subunits and, by implication, ORC itself as well as Mcm3p and Mcm7p, localize at the chromosomal oriMCM4 and at the five new origins in human cells in G1, dissociate during S phase and G2/M, and relocalize at origins in G1 (Fig. 5c and d and 6). The behavior of the Mcm proteins at oriMCM4 and ori6 is in line with that previously reported (1, 17, 35, 60). The origin association of different Orc subunits along the cell cycle is currently less clear. It is generally accepted that Orc1p is selectively released from origins and, in some cases, degraded (1, 55, 60). The reduced Orc2-binding level at ori6 and oriMCM4 in mitosis is in agreement with reports for oriLB2 (1, 17) but differs from what has been reported for oriP of EBV and the c-myc origin (11, 17, 55). The exact point in time of Orc2p-Orc5p release during S phase is currently unknown. Orc6p has not been analyzed in this respect on chromosomal origins. The laboratories of Falaschi and Knippers reported that Orc2p is associated with origins during S phase (1, 59), but we did not detect ORC subunits at origins during S phase. One possible explanation for the initially contradictory-seeming results is that the laboratories of Falaschi and Knippers analyzed Orc2p binding at oriMCM4 and oriLB2 in early S phase, whereas our samples were examined later in S phase (Fig. 6). Furthermore, it is also possible that Orc2p-Orc6p are not dissociating from origins during S phase but experience a conformational change. Individual experimental setups (cross-linking condition, UV or formaldehyde cross-linking) might lead to differing results. The exact point in time of a conformational change has to be determined in detail. Our findings, which are summarized in a model in Fig. 7, argue against possible epitope masking in mitosis and extend reports of ORC dynamics at human origins. Taken together, the properties of the trapped human origins are those expected for chromosomal replicators.
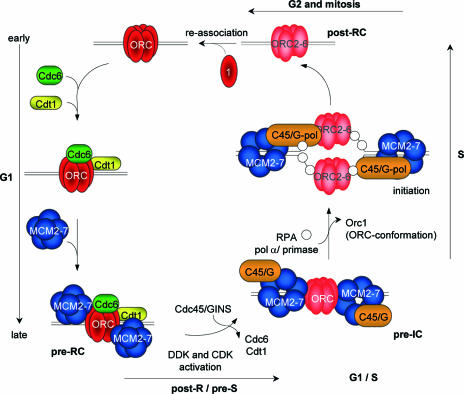
FIG. 7.
Model for the origin cycle at a human origin. The origin recognition complex is assembled at origins in the early stages of G1. Cdc6 and Cdt1 are required for the association of the Mcm2-7 protein family, resulting in the formation of the prereplicative complex (pre-RC). After activation of the DBF4-dependent kinase (DDK) and cyclin-dependent kinase (CDK) complexes, the transition from the pre-RC to the preinitiation complex (pre-IC) occurs at G1/S and is characterized by the formation of the CMG complex (consisting of Cdc45p, Mcm2-7p, and GINS) (47). After origin activation, Orc1p is selectively released from origins. This leads probably to a conformational change of the Orc2p-Orc6p subcomplex, resulting in a decreased sensitivity to formaldehyde and UV cross-linking (this study and reference 1). The exact time point of ORC reassociation has to be determined.
Human replicators as extrachromosomal replication origins.
We found that the replication activity of the plasmids selected in the origin-trapping assay is lower than that of wild-type oriP. This observation is perhaps not surprising, since the latent viral plasmid has evolved to maintain the EBV genome in a lifelong manner in its human host. Thus, the viral DNA binding protein EBNA1 promotes cell cycle-independent ORC binding to DS of oriP, likely rendering the viral replicator resistant to host mechanisms that might modulate origin activity (11, 55). DS interacts via EBNA1 with other elements of oriP, such as FR, possibly to improve origin efficiency in a manner that is not mimicked by the human origin fragments. Another reason could be that additional distal chromosomal elements regulate origin activity. The importance of such distal elements has been demonstrated for origins located near the human β-globin and the hamster DHFR loci (12, 27, 31). In embryonic extracts from frogs, flies, or fish, many genomic sites can be used as replication origins. If these conditions change as cells differentiate, most of these sites are suppressed, but others are enhanced. Furthermore, mammalian ORCs do not bind to specific sequences in vitro, and yet ORC appears to be bound preferentially to specific replication origins in vivo. It is therefore not surprising that, as mentioned in the introduction, mammalian DNA fragments that support extrachromosomal replication are not as active as origins that are defined mainly by sequence, such as ARS elements in yeast or oriP of EBV (14, 19, 23, 66). A deletion analysis of ori6 supported the idea that not one particular DNA motif is required for ORC binding and replication efficiency. The multimerization of the same sequence resulted in an increased efficiency, further indicating that additional elements might be required for an improved origin activity (Fig. 4).
The new chromosomal replicator plasmids also differ in several important respects from pEPI, another recently described plasmid that supports autonomous replication in human cells (7, 59). ORC binding to pEPI is not localized to a particular site in vivo but rather is delocalized with a slight preference for AT-rich regions, much like the weak DNA sequence preference of purified human ORC observed in vitro (65). Replication start sites in pEPI were also distributed throughout the plasmid, despite the fact that each plasmid replicated once per cell cycle (59). The differences between pEPI and the newly identified human origin plasmids suggest the existence of at least two different mechanisms to promote plasmid replication in mammalian cells and yet restrict it to once per cell cycle.
The establishment of autonomously replicating plasmids (oriP and pEPI) is an infrequent event in mammalian cells (25, 36). Regardless of their replication competence, plasmids are lost from cells at a rate of >25% per cell generation during the first 2 weeks after transfection (36). In contrast, cells selected to maintain established oriP plasmids have a 2 to 4% rate of plasmid loss after withdrawal of selection (30, 62). Thus, it seems likely that epigenetic events, in addition to replication competence, contribute to the long-term establishment of oriP plasmids. To exclude cell-type-specific features, we used different EBNA1-expressing cell lines (HEK293/EBNA1 and 143/EBNA1). The nature of the epigenetic events that facilitate plasmid maintenance in mammalian cells is currently unclear. It is possible that the DS element of oriP provides additional features that support the long-term establishment of wild-type oriP plasmids. Such events are probably not supported by the chromosomal replicator in the oriP plasmids analyzed in this study.
Our origin-trapping assay is based on the assumption that at least a subset of human origins is also defined by DNA sequence. Some origins, such as the lamin B2 and Mcm4 origins, are localized within a few kilobases and should be identifiable by origin trapping. Other replicators have an initiation zone with dispersed start sites, such as DHFR and myc origin. These replication origins are probably defined by a combination of DNA sequence elements, local chromatin structure, and DNA topology (53) and may be difficult to isolate in a trapping assay. Origins of replication in higher eukaryotes are also influenced by transcriptional activity, as seen for Drosophila melanogaster and Sciara (38, 40), and by differentiation and development (15, 42, 49). A recent analysis of origins located in the X-inactivation center confirmed an association between replication, transcription, and epigenetic regulation, although there is no clear dependency (58). Whether such origins can be identified by modifications of the trapping assay remains unknown.
The high genome complexity in mammalian cells and the associated decrease in assay sensitivity hampered the identification and characterization of origins. Clearly, there is a need for additional models in studying the features of human replication origins. While this work was in review, two new methods for isolating mammalian replication origins were published (46, 64). The two-dimensional gel-mapping method identified DNA sequences containing active but inefficient origins, such as DHFR origin (46, 64). In one of these reports, Mesner et al. showed that all isolated origins derived from active initiation sites. In a different approach, Todorovic et al. generated a library of nascent strand DNA fragments that identified replication origins with higher resolution, but it was dependent on an additional fine-mapping step by quantification of nascent strand DNA by competitive PCR (46, 64). Thus, both recently published origin-mapping methods are based on physical features of the replicated DNA rather than on functional properties. The two-step origin-trapping assay reported here is based not only on the physical property of ORC binding but also on a functional selection for origins. Our assay allows for the first time the identification of active, efficient replication origins which also support replication of extrachromosomal DNA. Taken together, these three new origin-mapping methods should enable the identification and characterization of new mammalian origins, their composition and biochemical properties, and their roles in chromosome structure and function in mammalian cells.
Supplementary Material
Acknowledgments
We thank W. Hammerschmidt for continuous support and discussion and D. Schaarschmidt for help with ChIP experiments and for critically reading the manuscript. We thank Natalie Grober for technical assistance and are grateful to M. Leffak for sharing the nascent strand abundance protocol. We thank A. Thomae and J. Baltin for discussion and comments on the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Sche 470-4 and SFB 646).
Footnotes
Published ahead of print on 5 September 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdurashidova, G., M. B. Danailov, A. Ochem, G. Triolo, V. Djeliova, S. Radulescu, A. Vindigni, S. Riva, and A. Falaschi. 2003. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 22:4294-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiyar, A., C. Tyree, and B. Sugden. 1998. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 17:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aladjem, M. I., and E. Fanning. 2004. The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep. 5:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aladjem, M. I., M. Groudine, L. L. Brody, E. S. Dieken, R. E. Fournier, G. M. Wahl, and E. M. Epner. 1995. Participation of the human beta-globin locus control region in initiation of DNA replication. Science 270:815-819. [DOI] [PubMed] [Google Scholar]
- 6.Austin, R., T. Orr-Weaver, and S. Bell. 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 13:2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baiker, A., C. Maercker, C. Piechaczek, S. B. Schmidt, J. Bode, C. Benham, and H. J. Lipps. 2000. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat. Cell Biol. 2:182-184. [DOI] [PubMed] [Google Scholar]
- 8.Bell, S. P., and B. Stillman. 1992. Nucleotide dependent recognition of chromosomal origins of DNA replication by a multi-protein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 9.Biamonti, G., S. Paixao, A. Montecucco, F. A. Peverali, S. Riva, and A. Falaschi. 2003. Is DNA sequence sufficient to specify DNA replication origins in metazoan cells? Chromosome Res. 11:403-412. [DOI] [PubMed] [Google Scholar]
- 10.Bogan, J. A., D. A. Natale, and M. L. DePamphilis. 2000. Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell. Physiol. 184:139-150. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimbora, D. M., D. Schubeler, A. Reik, J. Hamilton, C. Francastel, E. M. Epner, and M. Groudine. 2000. Long-distance control of origin choice and replication timing in the human β-globin locus are independent of the locus control region. Mol. Cell. Biol. 20:5581-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, S. M., B. P. Brylawski, M. Cordeiro-Stone, and D. G. Kaufman. 2002. Mapping of an origin of DNA replication near the transcriptional promoter of the human HPRT gene. J. Cell. Biochem. 85:346-356. [DOI] [PubMed] [Google Scholar]
- 14.Coverley, D., and R. A. Laskey. 1994. Regulation of eukaryotic DNA replication. Annu. Rev. Biochem. 63:745-776. [DOI] [PubMed] [Google Scholar]
- 15.Cvetic, C., and J. C. Walter. 2005. Eukaryotic origins of DNA replication: could you please be more specific? Semin. Cell Dev. Biol. 16:343-353. [DOI] [PubMed] [Google Scholar]
- 16.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh, M., M. Kemp, G. Liu, M. Ritzi, A. Schepers, and M. Leffak. 2006. Differential binding of replication proteins across the human c-myc replicator. Mol. Cell. Biol. 26:5270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giacca, M., L. Zentilin, P. Norio, S. Diviacco, D. Dimitrova, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, S. Riva, et al. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, D. M. 2004. In search of the holy replicator. Nat. Rev. Mol. Cell Biol. 5:848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzel, S. S., P. J. Krysan, C. T. Tran, and M. P. Calos. 1991. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol. Biol. Cell 11:2263-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 23.Hyrien, O., C. Maric, and M. Mechali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 24.Jacob, F., S. Brenner, and F. Cuzin. 1963. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 28:329-348. [Google Scholar]
- 25.Jenke, A. C., T. Eisenberger, A. Baiker, I. M. Stehle, S. Wirth, and H. J. Lipps. 2005. The nonviral episomal replicating vector pEPI-1 allows long-term inhibition of bcr-abl expression by shRNA. Hum. Gene Ther. 16:533-539. [DOI] [PubMed] [Google Scholar]
- 26.Jeon, Y., S. Bekiranov, N. Karnani, P. Kapranov, S. Ghosh, D. MacAlpine, C. Lee, D. S. Hwang, T. R. Gingeras, and A. Dutta. 2005. Temporal profile of replication of human chromosomes. Proc. Natl. Acad. Sci. USA 102:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2:797-806. [DOI] [PubMed] [Google Scholar]
- 28.Kamath, S., and M. Leffak. 2001. Multiple sites of replication initiation in the human beta-globin gene locus. Nucleic Acids Res. 29:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller, C., E. M. Ladenburger, M. Kremer, and R. Knippers. 2002. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 277:31430-31440. [DOI] [PubMed] [Google Scholar]
- 30.Kirchmaier, A. L., and B. Sugden. 1995. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J. Virol. 69:1280-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitsberg, D., S. Selig, I. Keshet, and H. Cedar. 1993. Replication structure of the human beta-globin gene domain. Nature 366:588-590. [DOI] [PubMed] [Google Scholar]
- 32.Krysan, P. J., and M. P. Calos. 1991. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol. Cell. Biol. 11:1464-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Biol. Cell 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krysan, P. J., J. G. Smith, and M. P. Calos. 1993. Autonomous replication in human cells of multimers of specific human and bacterial DNA sequences. Mol. Cell. Biol. 13:2688-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladenburger, E. M., C. Keller, and R. Knippers. 2002. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22:1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leight, E. R., and B. Sugden. 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell. Biol. 21:4149-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemaitre, J. M., E. Danis, P. Pasero, Y. Vassetzky, and M. Mechali. 2005. Mitotic remodeling of the replicon and chromosome structure. Cell 123:787-801. [DOI] [PubMed] [Google Scholar]
- 38.Lunyak, V. V., M. Ezrokhi, H. S. Smith, and S. A. Gerbi. 2002. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol. Cell. Biol. 22:8426-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacAlpine, D. M., and S. P. Bell. 2005. A genomic view of eukaryotic DNA replication. Chromosome Res. 13:309-326. [DOI] [PubMed] [Google Scholar]
- 40.MacAlpine, D. M., H. K. Rodriguez, and S. P. Bell. 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 18:3094-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machida, Y. J., J. L. Hamlin, and A. Dutta. 2005. Right place, right time, and only once: replication initiation in metazoans. Cell 123:13-24. [DOI] [PubMed] [Google Scholar]
- 42.Machida, Y. J., J. K. Teer, and A. Dutta. 2005. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J. Biol. Chem. 280:27624-27630. [DOI] [PubMed] [Google Scholar]
- 43.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendez, J., X. H. Zou-Yang, S. Y. Kim, M. Hidaka, W. P. Tansey, and B. Stillman. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9:481-491. [DOI] [PubMed] [Google Scholar]
- 45.Meselson, M., F. W. Stahl, and J. Vinograd. 1957. Equilibrium sedimentation of macromolecules in density gradients. Proc. Natl. Acad. Sci. USA 43:581-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mesner, L. D., E. L. Crawford, and J. L. Hamlin. 2006. Isolating apparently pure libraries of replication origins from complex genomes. Mol. Cell 21:719-726. [DOI] [PubMed] [Google Scholar]
- 47.Moyer, S. E., P. W. Lewis, and M. R. Botchan. 2006. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 103:10236-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natale, D. A., C. J. Li, W. H. Sun, and M. L. DePamphilis. 2000. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 19:2728-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norio, P., S. Kosiyatrakul, Q. Yang, Z. Guan, N. M. Brown, S. Thomas, R. Riblet, and C. L. Schildkraut. 2005. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell 20:575-587. [DOI] [PubMed] [Google Scholar]
- 50.Prasanth, S. G., K. V. Prasanth, K. Siddiqui, D. L. Spector, and B. Stillman. 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23:2651-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasanth, S. G., K. V. Prasanth, and B. Stillman. 2002. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297:1026-1031. [DOI] [PubMed] [Google Scholar]
- 52.Rao, H., and B. Stillman. 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. USA 92:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Remus, D., E. L. Beall, and M. R. Botchan. 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23:897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritzi, M., M. Baack, C. Musahl, P. Romanowski, R. A. Laskey, and R. Knippers. 1998. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 273:24543-24549. [DOI] [PubMed] [Google Scholar]
- 55.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 56.Rivella, S., B. Palermo, C. Pelizon, C. Sala, G. Arrigo, and D. Toniolo. 1999. Selection and mapping of replication origins from a 500-kb region of the human X chromosome and their relationship to gene expression. Genomics 62:11-20. [DOI] [PubMed] [Google Scholar]
- 57.Rowley, A., J. H. Cocker, J. Harwood, and J. F. X. Diffley. 1995. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 14:2631-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowntree, R. K., and J. T. Lee. 2006. Mapping of DNA replication origins to noncoding genes of the X-inactivation center. Mol. Cell. Biol. 26:3707-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaarschmidt, D., J. Baltin, I. M. Stehle, H. J. Lipps, and R. Knippers. 2004. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 23:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaarschmidt, D., E. M. Ladenburger, C. Keller, and R. Knippers. 2002. Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 30:4176-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugden, B., and N. Warren. 1988. Plasmid origin of replication of Epstein-Barr virus, oriP, does not limit replication in cis. Mol. Biol. Med. 5:85-94. [PubMed] [Google Scholar]
- 63.Todorovic, V., A. Falaschi, and M. Giacca. 1999. Replication origins of mammalian chromosomes: the happy few. Front. Biosci. 4:D859-D868. [DOI] [PubMed] [Google Scholar]
- 64.Todorovic, V., S. Giadrossi, C. Pelizon, R. Mendoza-Maldonado, H. Masai, and M. Giacca. 2005. Human origins of DNA replication selected from a library of nascent DNA. Mol. Cell 19:567-575. [DOI] [PubMed] [Google Scholar]
- 65.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 67.Walter, J., and J. W. Newport. 1997. Regulation of replicon size in Xenopus egg extracts. Science 275:993-995. [DOI] [PubMed] [Google Scholar]
- 68.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 70.Zhou, J., C. Chau, Z. Deng, W. Stedman, and P. M. Lieberman. 2005. Epigenetic control of replication origins. Cell Cycle 4:889-892. [DOI] [PubMed] [Google Scholar]
- 71.Zhou, J., C. M. Chau, Z. Deng, R. Shiekhattar, M. P. Spindler, A. Schepers, and P. M. Lieberman. 2005. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 24:1406-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.