Abstract
In haloarchaea, light-driven ion transporters have been modified by evolution to produce sensory receptors that relay light signals to transducer proteins controlling motility behavior. The proton pump bacteriorhodopsin and the phototaxis receptor sensory rhodopsin II (SRII) differ by 74% of their residues, with nearly all conserved residues within the photoreactive retinal-binding pocket in the membrane-embedded center of the proteins. Here, we show that three residues in bacteriorhodopsin replaced by the corresponding residues in SRII enable bacteriorhodopsin to efficiently relay the retinal photoisomerization signal to the SRII integral membrane transducer (HtrII) and induce robust phototaxis responses. A single replacement (Ala-215–Thr), bridging the retinal and the membrane-embedded surface, confers weak phototaxis signaling activity, and the additional two (surface substitutions Pro-200–Thr and Val-210–Tyr), expected to align bacteriorhodopsin and HtrII in similar juxtaposition as SRII and HtrII, greatly enhance the signaling. In SRII, the three residues form a chain of hydrogen bonds from the retinal's photoisomerized C13 C14 double bond to residues in the membrane-embedded α-helices of HtrII. The results suggest a chemical mechanism for signaling that entails initial storage of energy of photoisomerization in SRII's hydrogen bond between Tyr-174, which is in contact with the retinal, and Thr-204, which borders residues on the SRII surface in contact with HtrII, followed by transfer of this chemical energy to drive structural transitions in the transducer helices. The results demonstrate that evolution accomplished an elegant but simple conversion: The essential differences between transport and signaling proteins in the rhodopsin family are far less than previously imagined.
C14 double bond to residues in the membrane-embedded α-helices of HtrII. The results suggest a chemical mechanism for signaling that entails initial storage of energy of photoisomerization in SRII's hydrogen bond between Tyr-174, which is in contact with the retinal, and Thr-204, which borders residues on the SRII surface in contact with HtrII, followed by transfer of this chemical energy to drive structural transitions in the transducer helices. The results demonstrate that evolution accomplished an elegant but simple conversion: The essential differences between transport and signaling proteins in the rhodopsin family are far less than previously imagined.
Keywords: bacteriorhodopsin, phototaxis, retinal, sensory rhodopsin, transport
Microbial rhodopsins are photochemically reactive membrane-embedded proteins with seven transmembrane α-helices that form a pocket for the chromophore retinal. They are widespread in the microbial world in prokaryotes (bacteria and archaea) and in eukaryotes (fungi and algae) (1–3). A striking characteristic of these photoactive proteins is their wide range of seemingly dissimilar functions. Some are light-driven transporters, such as the proton pumps bacteriorhodopsin (BR) in haloarchaea (4) and proteorhodopsin in marine bacteria (5). Others are light sensors, such as the phototaxis receptors sensory rhodopsins (SR) I and II in haloarchaea (6–8). These sensory rhodopsins relay signals by protein–protein interaction to SR integral membrane transducer proteins (Htr) I and II, respectively. The signal relay mechanism from SR receptors to their cognate Htr transducers has become a focus of interest in part because of its importance to the general understanding of communication between integral membrane proteins, about which little is known.
A close relationship between transport and sensory signaling activities was revealed when SRI was separated from its tight complex with HtrI and was found to exhibit light-driven electrogenic proton transport across the membrane (9). The transport activity of SRI is blocked by HtrI binding. Such latent proton transport activity was shown also in SRII and similarly was blocked by HtrII binding (10, 11). These findings demonstrated that the essential features of the proton transport mechanism are conserved in the signal transduction mechanisms of SRI and SRII (12). Phylogenetic analysis strongly supports that rhodopsin photosensors like SRI and SRII evolved from light-driven proton pumps (3). The pumps are individual functional units without the need for interaction with other proteins and, therefore, readily undergo lateral gene transfer, duplication, and modification to develop a functional interaction with signal transduction machinery of the new host. What changes would be required to convert a proton pump into a sensory receptor; in other words, how extensive do the modifications need to be to evolve the new sensory function? We set out to answer this question by site-specific mutagenesis of BR to confer on the protein the ability to transmit signals through HtrII, which normally exists as a subunit of the SRII–HtrII heterotetrameric complex.
The photochemical reaction cycles (6–8) and atomic structures of BR from Halobacterium salinarum (13) and SRII from the related haloarchaeon Natronomonas pharaonis (14, 15) are well characterized. The crystal structure of N. pharaonis SRII bound to an N-terminal fragment of HtrII has provided atomic details of the two proteins' interaction surface in the periplasm and within the membrane (16), and interaction of the HtrII membrane-proximal domain with the cytoplasmic domain of the receptor has been demonstrated by fluorescent probe accessibility and FRET measurements, EPR of spin-labels, and in vitro binding of HtrII peptides to SRII (17–19). The results from several different methods show that light-induced structural changes occur all along the SRII–HtrII interface (refs. 17, 20–22; V. Bergo, E. Spudich, K. Rothschild, and J.L.S., unpublished data). However, which, if any, of these changes are essential and whether they are involved in the signal relay process within the complex or in later steps in signal propagation are unknown (23).
On its distal end, HtrII binds a cytoplasmic histidine kinase, CheA. CheA phosphorylates a phosphoregulator CheY that binds to the flagellar motor switch and controls the swimming reversal probability of the cell. The minimal modifications we need to make to enable BR to transmit photosignals through HtrII to CheA kinase would tell us the least extent of evolutionary changes needed to confer the new sensory function to the pump. Also, successful conversion of the pumping function to signaling would show us the core structure needed for signal relay from SRII to HtrII. BR and SRII share only 26% identity in primary sequence and, thus, a priori the changes required might be extensive. Here, we report that introduction into BR of merely one hydrogen bonding residue that in SRII bridges the retinylidene chromophore and the surface residues confers a small but detectable signaling activity. Moreover, substitutions of two more residues that enable hydrogen bonding of BR to the two HtrII transmembrane helices (Fig. 1) greatly enhances the signaling activity such that H. salinarum cells exhibit robust phototaxis responses.
Fig. 1.
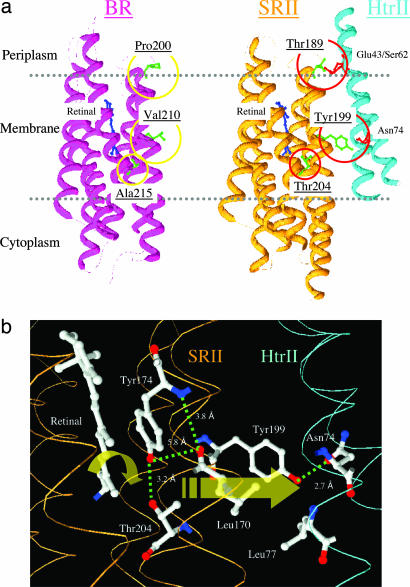
Locations of residues in this study. (a) X-ray crystal structures of BR (PDB ID code 1C3W) and SRII/HtrII (PDB ID code 1H2S) complex in the dark, indicating the three hydrogen-bonding residues in SRII introduced into BR in this study: Pro-200, Val-210, and Ala-215 in BR that correspond to Thr-189, Tyr-199, and Thr-204 in SRII, respectively. Tyr-199 bonds with HtrII Asn-74; Thr-189 bonds with HtrII Glu-43 and Ser-62; and Thr-204 with retinal pocket residue Tyr-174, which is conserved in BR as Tyr-185. (b) Detail of the x-ray SRII/HtrII structure (16), which focuses on the midmembrane SRII–HtrII interface containing the core signal relay structure identified in this work.
Results and Discussion
The first modification was the addition of a flexible linker of 11 residues connecting the cytoplasmic C terminus of BR and cytoplasmic N terminus of HtrII to ensure a 1:1 stoichiometry, as exists in the SRII–HtrII complex (24–26). The fusion of SRII and HtrII by their cytoplasmic ends in this manner permits normal phototaxis responses, indicating the linker does not influence signaling (see Fig. 4a). Analogously, the linker does not inhibit the proton pumping activity of BR measured in right-side-out membrane vesicles derived from H. salinarum transformants (Fig. 2). In addition, the photocycle of BR linked to HtrII is unchanged except for a small component of slow cycling (Fig. 3) addressed below. The cells expressed BR linked to HtrII, as confirmed by SDS/PAGE, and did not exhibit phototaxis responses. These results indicate that there is very little interaction of WT BR with HtrII, consistent with binding studies with the purified proteins (27).
Fig. 4.
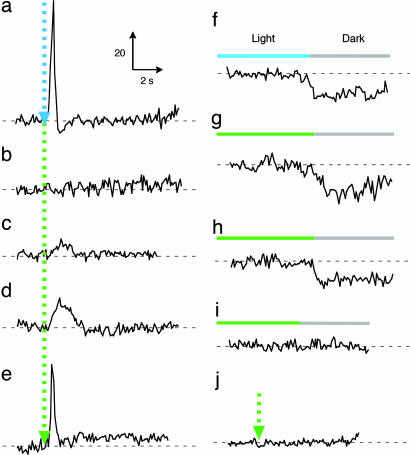
Phototaxis responses of BR–HtrII complexes. Swimming reversal frequency responses of cell populations measured by stimulus (at dotted arrow) effects on the ratio of rate of change of direction (RCD) to speed (SPD) by computer-assisted motion analysis. There were 200–2,500 cells assayed for each trace. “BR” designates BR in the following description: 100-ms 500-nm stimulus for SRII/HtrII (a), 500-ms 580-nm stimulus for P200T/V210Y-BR (b), 500-ms 550-nm stimulus for A215T-BR (c), P200T/V210Y/A215T-BR/NpHtrII (d), and P200T/V210Y/A215T-BR/G83F-NpHtrII (j), and 100-ms 550-nm stimulus for P200T/V210Y/A215T-BR/HsHtrII (e). BR mutants containing the A215T mutation have shorter absorbance maxima (λmax, 550 nm) than WT BR (λmax, 580 nm) (27). Swimming reversal frequency transients to a step-down in continuous light as indicated (f–i%). (f) SRII/HtrII; (g) P200T/V210Y/A215T-BR/NpHtrII; (h) P200T/V210Y/A215T-BR/HsHtrII; and (i) P200T/V210Y/A215T-BR/G83F-NpHtrII.
Fig. 2.
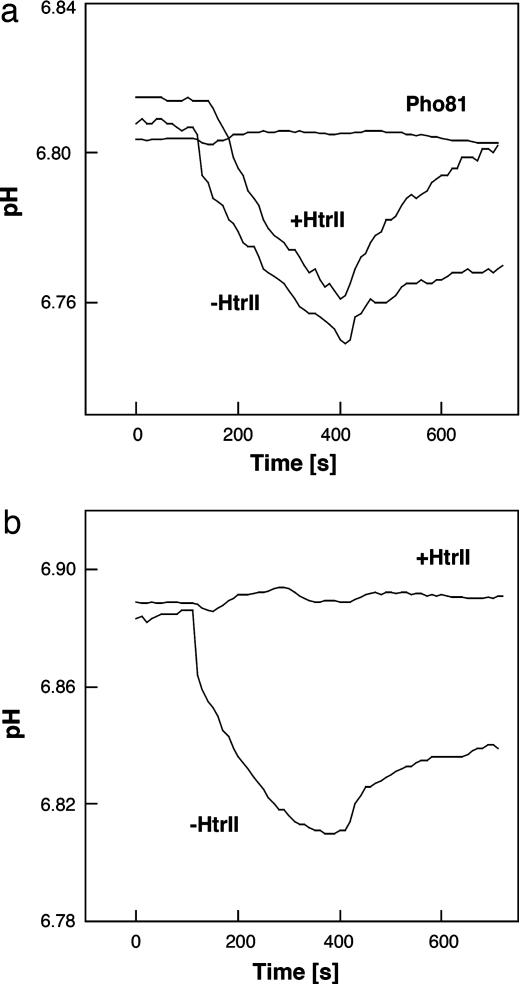
Photoinduced proton transport of WT BR (a) and BR mutant P200T/V210Y (b) with and without HtrII. (a) Pho81Wr− vesicles not expressing BR are included as a negative control. We measured proton pumping activity of BR (WT and P200T/V210Y mutant) with and without HtrII. Light-driven proton transport by BR mutant P200T/V210Y is ceased by association with the transducer protein, HtrII. We infer that association with HtrII closes a cytoplasmic channel of BR as it does in SRI (12).
Fig. 3.
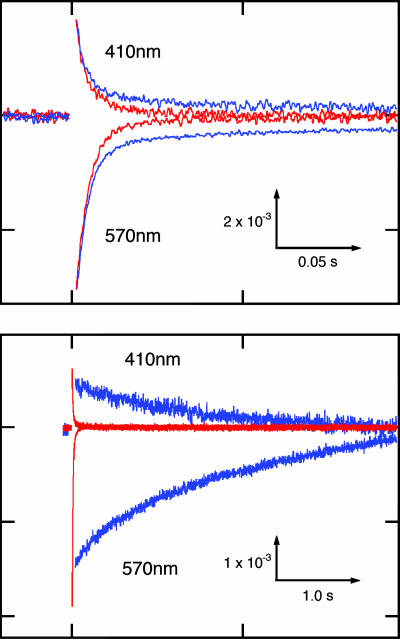
Flash-induced absorption changes of WT BR with (blue) and without (red) HtrII (Upper) and BR mutant P200T/V210Y with (blue) and without (red) HtrII (Lower). Scale in absorption units (ordinate) and time (abscissa). Note the 20-fold difference in the time scales. The M intermediate is monitored by 410 nm and the unphotolysed state is monitored by 570 nm. Flash photolysis data were acquired from membrane samples in 4 M NaCl at pH 7.0 in transformants of H. salinarum strain Pho81Wr−, which lacks rhodopsins and the transducer proteins HtrI and HtrII.
The second modification was to introduce two surface residues into BR that enable hydrogen bonding to HtrII to align BR with the transducer in the same manner as SRII to HtrII. The x-ray crystallographic structure of the SRII/HtrII complex shows two hydrogen-bonded regions at Tyr-199–SRII/Asn-74–HtrII and Thr-189–SRII/Glu-43–HtrII/Ser-62–HtrII (16) (Fig. 1). Previously, it was shown that introducing the corresponding hydrogen-bonding residues into BR enable the purified protein in detergent to exhibit weak but measurable affinity for HtrII (27). Pro-200 and Val-210 in BR correspond to Thr-189 and Tyr-199 in SRII, respectively. The P200T/V210Y double mutant of BR carries out light-driven proton transport, but unlike WT BR linked to HtrII, HtrII presence inhibits transport in the double mutant (Fig. 2). In addition, the hydrogen bond-induced association of BR with HtrII reduces its photochemical reaction cycle rate ≈400-fold (Fig. 3). This reduction in rate is particularly notable because a key difference between transport and sensory rhodopsins is the much slower kinetics of the photochemical reaction cycle of the sensors. Ion-pumping rhodopsins, BR, halorhodopsin, and proteorhodopsin, have been optimized by nature to have relatively fast photocycling rates (≈10 ms), making them efficient pumps, whereas the sensory rhodopsins SRI and SRII have slow photocycles persisting several seconds, which allow the transient accumulation of long-lived signaling states of the receptors to catalyze a sustained phosphorylation cascade (2). The slow photocycle and lack of proton transport of the BR_P200T/V210Y/HtrII complex therefore are similar properties as those of SRI/HtrI and SRII/HtrII complexes, but the artificial BR complex does not mediate phototaxis responses (Fig. 4), demonstrating that further modification is necessary to convert BR into a sensory signaling receptor.
The third and key residue change is the mutation A215T. In SRII, Thr-204 and its hydrogen-bonded partner Tyr-174 are crucial residues for photosignal relay (28). These residues are the first in SRII shown to be essential for phototaxis and provide biological significance to the previous observation that the hydrogen bond between them is greatly strengthened upon the formation of the earliest SRII photointermediate (SRIIk) only when SRII is complexed with HtrII (29). Tyr-174 forms part of the retinal-binding site and is conserved as Tyr-185 in BR. Thr-204 corresponds in position to Ala-215 and is the principal component of the π-bulge on helix G present in both SRII and BR (30). Therefore, we introduced A215T into the P200T/V210Y double mutant of BR. The triple mutant, like the double mutant, exhibits a slow photocycle and a lack of light-driven proton transport (data not shown). In addition, the triple mutant mediates phototaxis responses assessed as transient changes in swimming reversal frequency to pulse and step-down 550-nm light stimuli (Fig. 4 d and g). The triple mutant, like SRII, mediates a transient increase in reversal frequency in response to an increase in extent of photocycling and, conversely, a transient decrease in reversal frequency in response to a decrease in photocycling. These two behavioral responses are the elemental responses biasing the random walk motility pattern to cause migration of cells down a repellent light gradient in prokaryotic phototaxis (31).
The A215T mutation in BR, even without the two mutations that introduce residues that form hydrogen bonds to HtrII, was sufficient to mediate a very small but detectable phototaxis response (Fig. 4c). Our interpretation is that the Tyr-185/Thr-215 hydrogen bond, mimicking the crucial Tyr-174/Thr-204 hydrogen bond in SRII, is sufficient to confer on BR the essential bond for signal relay to HtrII. This effect may be due to HtrII, unattached to BR, attaining the proper positioning with respect to the A215T BR a small fraction of the time through its thermal fluctuations in position in the membrane. In this interpretation, the role of the introduced residues Thr-200 and Tyr-210 would be simply to position HtrII to more efficiently receive the signal from the Tyr-185/Thr-215 pair, thereby increasing the signal relay efficiency in the artificial complex. This hypothesis is supported by the presence of a small component of slow photocycling in WT BR when attached to HtrII (Fig. 3), indicating a slight contribution of the same interaction with HtrII that is greatly enhanced by the double mutation P200T/V210Y.
To confirm that the photosignals from the BR triple mutant indeed are relayed through HtrII and not influencing the flagellar motor in some indirect manner, we linked it to G83F–HtrII. Replacement of the cytoplasmic residue Gly-83 of HtrII by phenylalanine abolishes the phototransducing function of HtrII in the SRII–HtrII complex (32). The BR triple mutant linked to G83F–HtrII showed no phototaxis responses (Fig. 4 i and j), confirming that the BR triple mutant transmits light signals to the motility apparatus through the HtrII protein.
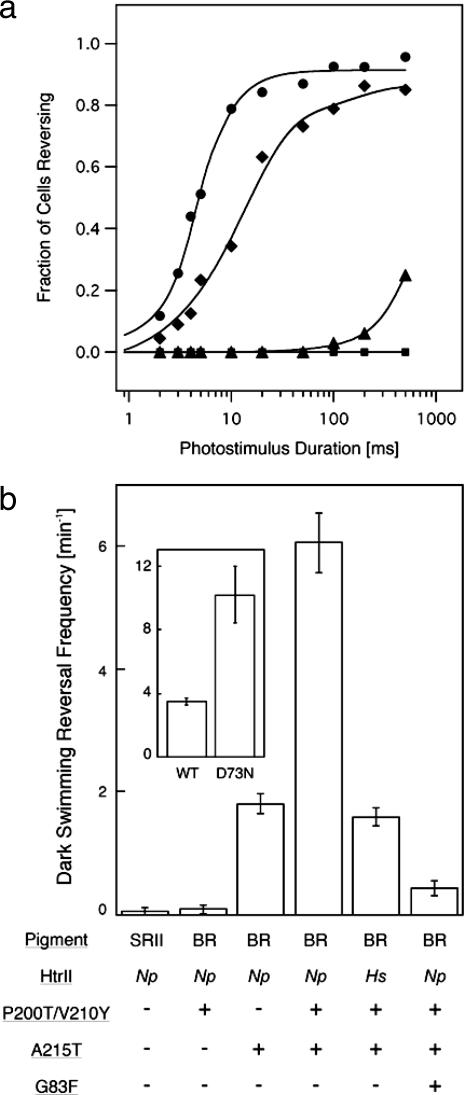
A homolog of N. pharaonis HtrII used above is present in H. salinarum, the same species that contains the BR gene we used in the constructs (6). A crystal structure of HtrII from H. salinarum (HsHtrII) is not available, but HsHtrII is 26% identical to HtrII from N. pharaonis (NpHtrII), and the HsSRII and NpSRII receptors each mediate phototaxis through the other species' HtrII (Toshifumi Nara and J.L.S., unpublished data). Only one (Ser-62) of the three interface hydrogen-bonding residues in NpHtrII is strictly conserved in HsHtrII, but HsSRII conserves Tyr-199, Tyr-174, and Thr-204 (Fig. 1) and is expected to form a structurally similar receptor–transducer interface. Substitution of the NpHtrII in the triple BR mutant-HtrII-linked construct with the HsHtrII dramatically increases nearly 100-fold the efficiency of phototaxis signaling by the triple mutant (Fig. 4e). This enhancement does not appear to be due to an inherently greater signaling activity of HsHtrII than NpHtrII because both transducer proteins show similar signaling efficiencies when bound to their native SRII receptors (Toshifumi Nara and J.L.S., unpublished data). Therefore, the enhancement indicates more efficient coupling of the BR triple mutant with HsHtrII than NpHtrII. Fluence response curves show that the sensitivities of cells containing BR triple mutant/NpHtrII and BR triple mutant/HsHtrII are 0.4% and 35%, respectively, of that of SRII/HtrII-containing cells (see Fig. 6a).
Fig. 6.
Photostimulus-induced and dark swimming reversal frequencies. (a) Fluence/response curves of cells containing SRII/HtrII (circles), BR triple mutant/HsHtrII (diamonds), BR triple mutant/NpHtrII (triangles), and WT BR/NpHtrII (squares). Km values from Michaelis–Menton fits to the fluence/response curves were used to estimate sensitivities of cells expressing BR triple mutant/HsHtrII and BR triple mutant/NpHtrII (35% and 0.4%, respectively, of that conferred by SRII-HtrII). (b) Dark swimming reversal frequency of cells expressing WT and mutated BR and SRII. Increase in frequency corresponds to constitutive activity in the dark (37). The values were determined from duplicate measurements of ≈30 cells imaged by infrared illumination and tracked for 2 min. (b Inset) Dark reversal frequencies of cells containing the WT and D73N mutant of H. salinarum SRII–HtrII complexes studied in ref. 31.
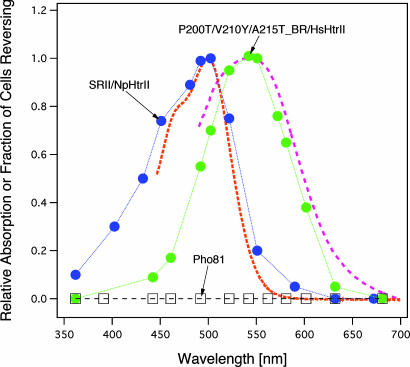
The BR triple mutant/HsHtrII construct produced a sufficiently robust response to permit action spectroscopy. The action spectrum of the BR signaling complex is red-shifted 50 nm from that of the SRII signaling complex and matches closely the shift in absorption of BR A215T mutants from that of SRII (Fig. 5). The good agreement of both BR and SRII signaling complex action spectra with their absorption spectra confirms the fidelity of our quantification of motility behavioral responses.
Fig. 5.
Action spectra for the repellent response in transformants containing SRII/NpHtrII (blue circles), BR triple mutant/HsHtrII (green circles), and Pho-81 devoid of rhodopsins (squares). The absorption spectra (dotted lines) were measured by using the membrane fraction in the presence of 0.1% n-dodecyl-β-d-maltoside (26). We measured phototaxis responses as swimming reversal frequency changes to a 5-ms photostimulus for SRII/NpHtrII and 10-ms photostimuli for the BR triple mutant/HsHtrII and Pho-81.
Measurements of the spontaneous reversal frequencies of the mutants in the dark provide further confirmation and insight into the function of the mutant BR proteins. The dark reversal frequency is a measure of the CheA kinase-activating activity of the cell's taxis transducers (e.g., HtrII), as shown by numerous studies in bacterial chemotaxis (33–36). SRII, the natural partner of HtrII, does not alter this activity in the dark (Fig. 6b), but mutated forms, such as the partially constitutively activated D73N-SRII, increase the dark reversal frequency (ref. 37; Fig. 6b Inset). The P200T/V210Y double mutant of BR has no effect, indicating no dark signal is transmitted. In contrast, the A215T mutation in BR linked to HtrII increases the dark reversal rate. This result means that the introduction of this single mutation changes the structure of HtrII to increase its kinase-activating activity. Therefore, the A215T mutation must alter the structure of BR at the BR–HtrII interface to partially activate HtrII in the dark state of the complex. This dark effect of A215T is greatly enhanced, just as its photosignaling efficiency is enhanced, by the P200T/V210Y mutations (Fig. 6b). Our interpretation is that the hydrogen bonds that pin the receptor to the transducer are important for aligning the two proteins but do not necessarily participate in a significant way in the signal relay process. Therefore, remarkably, it appears that a single hydrogen bond (i.e., that is between Thr-204 and Tyr-174 in SRII) spells the essential difference between a sensory receptor and a proton pump in the microbial rhodopsin family.
Concluding Remarks.
These results prove that the distinct functions of active transport and signaling can be produced by a small number of modifications of the same protein structure. The nature of the changes needed to convert the proton pump BR into a signaling receptor also provides insight into the essential core of the signal relay mechanism. In SRII, the Tyr-174–Thr-204 pair form a hydrogen bond that lies between the C13 C14 double bond of the retinal and residues Leu-170 and Tyr-199, which form part of the SRII–HtrII interface by van der Waals contact and hydrogen bonding to HtrII (Fig. 1b). Photoisomerization results in an unusually large storage of energy in the Tyr-174–Thr-204 hydrogen bond in the earliest SRII intermediate K only when SRII is bound to HtrII (29). In BR, Tyr-185 (the residue corresponding to Tyr-174 in SRII) is structurally active as early as the K intermediate (38), supporting our interpretation that an early event in signal transduction is the structural alteration of Tyr-174 by retinal isomerization. We calculate strengthening of the Tyr-174–Thr-204 hydrogen bond by 3.3 kcal·mol−1 from the downshift of the Thr-204 O-H stretching frequency based on a reported relationship between O-H band shifts and bond enthalpy (39), which is a large portion of the total energy stored in the K intermediate (12 kcal·mol−1 in BR; ref. 40). This destabilizing bond energization likely results from a local steric constraint between the retinal C14-H and Thr-204, caused by C13
C14 double bond of the retinal and residues Leu-170 and Tyr-199, which form part of the SRII–HtrII interface by van der Waals contact and hydrogen bonding to HtrII (Fig. 1b). Photoisomerization results in an unusually large storage of energy in the Tyr-174–Thr-204 hydrogen bond in the earliest SRII intermediate K only when SRII is bound to HtrII (29). In BR, Tyr-185 (the residue corresponding to Tyr-174 in SRII) is structurally active as early as the K intermediate (38), supporting our interpretation that an early event in signal transduction is the structural alteration of Tyr-174 by retinal isomerization. We calculate strengthening of the Tyr-174–Thr-204 hydrogen bond by 3.3 kcal·mol−1 from the downshift of the Thr-204 O-H stretching frequency based on a reported relationship between O-H band shifts and bond enthalpy (39), which is a large portion of the total energy stored in the K intermediate (12 kcal·mol−1 in BR; ref. 40). This destabilizing bond energization likely results from a local steric constraint between the retinal C14-H and Thr-204, caused by C13 C14 double-bond rotation (41). The HtrII dependence of this reaction indicates that the bonded Tyr-Thr pair is coupled to the receptor–transducer interface, where chemical signal relay occurs. Therefore, relaxation of the steric hindrance, shown to occur in later photocycle steps in which intermediates responsible for signaling are formed (28), is expected to be communicated as a structural change to the interface. Evidence for such coupling is that the nearby Tyr-199–Asn-74 hydrogen bond (Fig. 1b) is greatly perturbed, probably disrupted, in the signaling state (20). Our results argue for crucial signal relay chemistry occurring within the membrane locally between the isomerizing double bond of retinal and the adjacent residues in the receptor–transducer interface.
C14 double-bond rotation (41). The HtrII dependence of this reaction indicates that the bonded Tyr-Thr pair is coupled to the receptor–transducer interface, where chemical signal relay occurs. Therefore, relaxation of the steric hindrance, shown to occur in later photocycle steps in which intermediates responsible for signaling are formed (28), is expected to be communicated as a structural change to the interface. Evidence for such coupling is that the nearby Tyr-199–Asn-74 hydrogen bond (Fig. 1b) is greatly perturbed, probably disrupted, in the signaling state (20). Our results argue for crucial signal relay chemistry occurring within the membrane locally between the isomerizing double bond of retinal and the adjacent residues in the receptor–transducer interface.
Our findings do not exclude that photoinduced conformational changes shared between BR and SRII play a role in signal relay or signal enhancement (42). The maximum signaling efficiency we observed with the mutated BR is 35% of that of SRII, indicating that other contributions to signaling in SRII remain to be identified. In particular, tilting of helix F, which opens the cytoplasmic channel of BR during its pumping cycle (4), also occurs in SRII, at least when not bound to HtrII (2, 7, 8), and may contribute. However, Tyr-174 and Thr-204 are the only residues so far shown in SRII to be essential for signaling. Thus, the causative link between chromophore photoisomerization and later events that have been detected may well be the early structural change that resolves the steric hindrance between Thr-204 and the retinal C14-H.
Materials and Methods
Strain for Transformation and Expression.
H. salinarum strain Pho81Wr− (43), which lacks the four native rhodopsins (BR, halorhodopsin, SRI, and SRII) and the two transducer proteins (HtrI and HtrII) and is carotenoid pigment-deficient and restriction-deficient, was used for transformation by following the protocol described in ref. 44. To obtain high expression levels, the strong bop promoter was used for expression of all genetic constructs instead of the native promoters (44). The expression levels calculated from amplitudes of the pigments' main absorption bands in the visible spectrum for SRII/HtrII, BR(P200T/V210Y)/NpHtrII, BR(A215T)/NpHtrII, BR(P200T/V210Y/A215T)/NpHtrII, and BR(P200T/V210Y/A215T)/HsHtrII complexes in transformants used in this study were all similar, namely 2.0 × 104, 1.5 × 104, 3.1 × 104, 1.3 × 104, and 1.0 × 104 molecules per cell, respectively.
Plasmid Construction.
The pYS001 plasmid was modified from plasmid pJS010 that encodes the WT SRII/HtrII fusion gene (45). The NcoI and NsiI fragment from the pJS010 plasmid was ligated to the NcoI and NsiI site of the pGEM-T vector. The 3′ end of sopII was mutated by PCR to create a SpeI restriction site, after which the NcoI and NsiI fragment was ligated into NcoI and NsiI sites of pJS010. This plasmid encodes six histidines in the C terminus, and it was named pYS001 (28). For the preparation of BR/NpHtrII and BR/HsHtrII expression plasmids, the 5′ and 3′ end of the bop gene was mutated by PCR to introduce a NcoI and SpeI restriction site, respectively. The resulting NcoI-SpeI fragments were ligated with the large fragment of NcoI/SpeI-treated pYS001 and pTN603 vector, respectively. PTN603 encodes the WT SRII/HsHtrII fusion gene (Toshifumi Nara and J.L.S., unpublished data). The stop codon was deleted during amplification, generating a linker region between SRII and HtrII that contains 11 residues (Thr-Ser-Ala-Ser-Ala-Ser-Asn-Gly-Ala-Ser-Ala; 5′-CTAGTGCGTCGGCGTCGAACGGCGCGTCGGCG-3′). The underline indicates the added restriction site for SpeI. The mutant genes were constructed by PCR with the two-step mutagenesis method (46). For the preparation of WT BR and mutant genes, the 5′ and 3′ end of the bop genes were mutated by PCR to introduce NcoI and ApaI restriction sites, respectively. The resulting NcoI-ApaI fragments were ligated with the large fragment of NcoI/ApaI-treated pYS001 vector. Theses plasmids encode six histidines in the C terminus. All constructed plasmids were analyzed by using an automated sequencer to confirm the expected nucleotide sequence.
Membrane Vesicle Preparation.
Membrane vesicles were prepared from H. salinarum Pho81Wr− transformants, expressing each of the genetic constructs as described in ref. 47. Briefly, cells in suspension were disrupted by sonication to produce vesicles, centrifuged at low speed to remove unbroken cells and debris, and the membrane vesicles finally were pelleted by centrifugation at 45,000 rpm (rotor type 70 Ti; Beckman, Fullerton, CA), for 30 min at 4°C, resuspended in 50 mM Tris·Cl (pH 7.0) containing 4 M NaCl, and stored at 4°C.
Proton-Pumping Measurements.
Membrane vesicles were washed with 4 M NaCl three times by centrifugation and finally suspended in 4 M NaCl/10 mM MgCl/0.1 mM Mes buffer at pH values shown in Fig. 2. The samples were irradiated with yellow light with a 520-nm long pass filter (Y52). A hot mirror was placed in front of the projector lamp (tungsten halogen, 150 W) to remove heat radiation. The pH was monitored by a pH electrode (F72 pH meter; Beckman), which was connected to a personal computer through an RS232C cable.
Laser Flash Photolysis.
Flash-induced absorption changes in vesicle suspensions in the millisecond to seconds time window were acquired on a personal computer with a digital oscilloscope program (ClampX 8.2) by following a Nd-YAG laser flash (Continuum, Santa Clara, CA; Surelight I; 532 nm, 6 ns, 40 mJ) in a lab-constructed flash photolysis system as described in ref. 48. Monitoring wavelengths are shown in Fig. 3. Transients (128–1,024) were collected and averaged for each measurement. All measurements were performed at 20°C.
Motility Analysis.
Phototaxis responses were measured as transient swimming reversal frequency changes of cell populations in response to pulse photostimuli and to step-down photostimuli. Swimming reversals were measured by stimulus-induced effects on the ratio of rate of change of direction (RCD) to speed (SPD) by computer-assisted motion analysis of individual cell tracks. The software used for the analysis was CellTrack 1.2 Beta (Motion Analysis, Santa Clara, CA); 200-2,500 cells were assayed for each trace. Maximal stimuli from a 100 W Hg lamp through a heat filter and 40-nm band-pass interference filters delivered through microscope optics were as follows: 100-ms 500-nm stimulus for SRII/HtrII, 500-ms 580-nm stimulus for P200T/V210Y-BR, 500-ms 550-nm stimulus for A215T-BR, P200T/V210Y/A215T-BR/NpHtrII, and P200T/V210Y/A215T-BR/G83F-NpHtrII and 100-ms 550-nm stimulus for P200T/V210Y/A215T-BR/HsHtrII. Note that BR mutants containing the A215T mutation have shorter absorbance maxima (λmax, 550 nm) than WT BR (λmax, 567 nm). We measured phototaxis responses also as swimming reversal frequency transients induced by a step-down from 60-s continuous light to darkness (Fig. 4).
Constitutive activity of the cells was assessed by individual cell tracking as increases in spontaneous reversal frequency in the dark. The values were determined from duplicate measurements of ≈30 cells imaged by infrared illumination and tracked for 2 min as described in ref. 37.
Acknowledgments
We thank Elena Spudich, Kevin Ridge, Oleg Sineshchekov, and Jun Sasaki for valuable discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant R37GM27750 and the Robert A. Welch Foundation.
Abbreviations
- BR
bacteriorhodopsin
- SR
sensory rhodopsins
- Htr
SR integral membrane transducer
- NpHtrII
HtrII from Natronomonas pharaonis
- HsHtrII
HtrII from Halobacterium salinarum.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ruiz-Gonzalez MX, Marin I. J Mol Evol. 2004;58:348–358. doi: 10.1007/s00239-003-2557-8. [DOI] [PubMed] [Google Scholar]
- 2.Spudich JL, Jung K-H. In: Handbook of Photosensory Receptors. Briggs W, Spudich JL, editors. Weinheim, Germany: Wiley; 2005. pp. 1–24. [Google Scholar]
- 3.Sharma AK, Spudich JL, Doolittle WF. Trends Microbiol. 2006 Sep 26; doi: 10.1016/j.tim.2006.09.006. 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Luecke H, Lanyi JK. Adv Protein Chem. 2003;63:111–130. doi: 10.1016/s0065-3233(03)63005-6. [DOI] [PubMed] [Google Scholar]
- 5.Béjà O., Spudich EN, Spudich JL, Leclerc M, DeLong EF. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 6.Hoff WD, Jung KH, Spudich JL. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 7.Klare JP, Bordignon E, Engelhard M, Steinhoff HJ. Photochem Photobiol Sci. 2004;3:543–547. doi: 10.1039/b402656j. [DOI] [PubMed] [Google Scholar]
- 8.Sudo Y, Kandori H, Kamo N. Recent Res Dev Biophys. 2004;3:1–16. [Google Scholar]
- 9.Bogomolni RA, Stoeckenius W, Szundi I, Perozo E, Olson KD, Spudich JL. Proc Natl Acad Sci USA. 1994;91:10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmies G, Engelhard M, Wood PG, Nagel G, Bamberg E. Proc Natl Acad Sci USA. 2001;98:1555–1559. doi: 10.1073/pnas.031562298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudo Y, Iwamoto M, Shimono K, Sumi M, Kamo N. Biophys J. 2001;80:916–922. doi: 10.1016/S0006-3495(01)76070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spudich JL. Cell. 1994;79:747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 13.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 14.Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Royant A, Nollert P, Edman K, Neutze R, Landau EM, Pebay-Peyroula E, Navarro J. Proc Natl Acad Sci USA. 2001;98:10131–10136. doi: 10.1073/pnas.181203898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 17.Yang CS, Sineshchekov O, Spudich EN, Spudich JL. J Biol Chem. 2004;279:42970–42976. doi: 10.1074/jbc.M406504200. [DOI] [PubMed] [Google Scholar]
- 18.Sudo Y, Okuda H, Yamabi M, Fukuzaki Y, Mishima M, Kamo N, Kojima C. Biochemistry. 2005;44:6144–6152. doi: 10.1021/bi047573z. [DOI] [PubMed] [Google Scholar]
- 19.Bordignon E, Klare JP, Doebber M, Wegener AA, Martell S, Engelhard M, Steinhoff HJ. J Biol Chem. 2005;280:38767–38775. doi: 10.1074/jbc.M509391200. [DOI] [PubMed] [Google Scholar]
- 20.Bergo VB, Spudich EN, Rothschild KJ, Spudich JL. J Biol Chem. 2005;280:28365–28369. doi: 10.1074/jbc.M505555200. [DOI] [PubMed] [Google Scholar]
- 21.Wegener AA, Klare JP, Engelhard M, Steinhoff HJ. EMBO J. 2001;20:5312–5319. doi: 10.1093/emboj/20.19.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moukhametzianov R, Klare JP, Efremov R, Baeken C, Goppner A, Labahn J, Engelhard M, Buldt G, Gordeliy VI. Nature. 2006;440:115–119. doi: 10.1038/nature04520. [DOI] [PubMed] [Google Scholar]
- 23.Spudich JL. Trends Microbiol. 2006 Sep 25; doi: 10.1016/j.tim.2006.09.005. 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Sudo Y, Iwamoto M, Shimono K, Kamo N. Photochem Photobiol. 2001;74:489–494. doi: 10.1562/0031-8655(2001)074<0489:ppbtic>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Klare JP, Bordignon E, Doebber M, Fitter J, Kriegsmann J, Chizhov I, Steinhoff HJ, Engelhard M. J Mol Biol. 2006;356:1207–1221. doi: 10.1016/j.jmb.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Hippler-Mreyen S, Klare JP, Wegener AA, Seidel R, Herrmann C, Schmies G, Nagel G, Bamberg E, Engelhard M. J Mol Biol. 2003;330:1203–1213. doi: 10.1016/s0022-2836(03)00656-9. [DOI] [PubMed] [Google Scholar]
- 27.Sudo Y, Yamabi M, Kato S, Hasegawa C, Iwamoto M, Shimono K, Kamo N. J Mol Biol. 2006;357:1274–1282. doi: 10.1016/j.jmb.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 28.Sudo Y, Furutani Y, Kandori H, Spudich JL. J Biol Chem. 2006 Sep 6; doi: 10.1074/jbc.M605907200. 10.1074/jbc.M605907200. [DOI] [PubMed] [Google Scholar]
- 29.Sudo Y, Furutani Y, Shimono K, Kamo N, Kandori H. Biochemistry. 2003;42:14166–14172. doi: 10.1021/bi035678g. [DOI] [PubMed] [Google Scholar]
- 30.Cartailler JP, Luecke H. Structure (London) 2004;12:133–144. doi: 10.1016/j.str.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Spudich JL, Bogomolni RA. Annu Rev Biophys Biophys Chem. 1988;17:193–215. doi: 10.1146/annurev.bb.17.060188.001205. [DOI] [PubMed] [Google Scholar]
- 32.Yang CS, Spudich JL. Biochemistry. 2001;40:14207–14214. doi: 10.1021/bi010985c. [DOI] [PubMed] [Google Scholar]
- 33.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson JS, Ames P, Studdert CA. Curr Opin Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Baker MD, Wolanin PM, Stock JB. BioEssays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 36.Bourret RB, Hess JF, Borkovich KA, Pakula AA, Simon MI. J Biol Chem. 1989;264:7085–7088. [PubMed] [Google Scholar]
- 37.Spudich EN, Zhang W, Alam M, Spudich JL. Proc Natl Acad Sci USA. 1997;94:4960–4965. doi: 10.1073/pnas.94.10.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braiman MS, Mogi T, Marti T, Stern LJ, Khorana HG, Rothschild KJ. Biochemistry. 1988;27:8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- 39.Garczarek F, Gerwert K. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 40.Birge RR, Cooper TM, Lavrence AF, Masthay MB, Zhang CF, Zidovetzki R. J Am Chem Soc. 1991;113:4327–4328. [Google Scholar]
- 41.Sudo Y, Furutani Y, Wada A, Ito M, Kamo N, Kandori H. J Am Chem Soc. 2005;127:16036–16037. doi: 10.1021/ja056203a. [DOI] [PubMed] [Google Scholar]
- 42.Spudich JL, Lanyi JK. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 43.Perazzona B, Spudich EN, Spudich JL. J Bacteriol. 1996;178:6475–6478. doi: 10.1128/jb.178.22.6475-6478.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krebs MP, Spudich EN, Spudich JL. Protein Expression Purif. 1995;6:780–788. doi: 10.1006/prep.1995.0009. [DOI] [PubMed] [Google Scholar]
- 45.Jung KH, Spudich EN, Trivedi VD, Spudich JL. J Bacteriol. 2001;183:6365–6371. doi: 10.1128/JB.183.21.6365-6371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen B, Przybyla AE. BioTechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 47.Spudich EN, Spudich JL. J Biol Chem. 1993;268:16095–16097. [PubMed] [Google Scholar]
- 48.Chen X, Spudich JL. J Biol Chem. 2004;279:42964–42969. doi: 10.1074/jbc.M406503200. [DOI] [PubMed] [Google Scholar]