Abstract
Intermediate filaments (IFs), along with microtubules, microfilaments, and associated cross-bridging proteins, constitute the cytoskeleton of metazoan cells. While crystallographic data on the dimer representing the elementary IF “building block” have recently become available, little structural detail is known about both the mature IF architecture and its assembly pathway. Here, we have applied solution small-angle x-ray scattering to investigate the in vitro assembly of a 53-kDa human IF protein vimentin at pH 8.4 by systematically varying the ionic strength conditions, and complemented these experiments by electron microscopy and analytical ultracentrifugation. While a vimentin solution in 5 mM Tris·HCl (pH 8.4) contains predominantly tetramers, addition of 20 mM NaCl induces further lateral assembly evidenced by the shift of the sedimentation coeficient and yields a distinct octameric intermediate. Four octamers eventually associate into unit-length filaments (ULFs) that anneal longitudinally. Based on the small-angle x-ray scattering experiments supplemented by crystallographic data and additional structural constraints, 3D molecular models of the vimentin tetramer, octamer, and ULF were constructed. Within each of the three oligomers, the adjacent dimers are aligned exclusively in an approximately half-staggered antiparallel A11 mode with a distance of 3.2–3.4 nm between their axes. The ULF appears to be a dynamic and a relatively loosely packed structure with a roughly even mass distribution over its cross-section.
Keywords: 3D structure, vimentin
The cytoskeleton of metazoan animal cells is formed by three major filament systems: microtubules, microfilaments, and intermediate filaments (IFs). They are integrated into a complex network by associated cross-linking proteins (1). In humans, ≈75 different IF proteins are expressed. For instance, keratins are synthesized in epithelia, desmin in muscles, and vimentin in mesenchymal cells, whereas nuclear lamins are present in all cell types (2–5). The main proposed function of IFs is to provide cells with resistance to mechanical stress caused by external forces or internal processes, e.g., during cell division or migration (6).
At present, the molecular architecture of IFs is fundamentally less well understood than that of microtubules and microfilaments (5, 7, 8). Unlike the latter, IFs self-assemble from fibrous, elongated coiled-coil dimers (7, 9). The elementary IF dimer includes a central coiled-coil “rod” domain containing close to 300 residues and non-α-helical N- and C-terminal end domains (10). Recently, atomic structures of several fragments derived from human vimentin and lamin A have been determined by x-ray crystallography (10–12). The elementary dimers appear to be roughly aligned and oriented in both directions along the filament axis; hence the filaments have no net polarity. As yet, the exact arrangement of the dimers within IFs can not be experimentally determined, but some theoretical models have been suggested (5, 7).
IF assembly appears to be driven by a specific hierarchical association of the elementary dimers in two directions: the lateral and the longitudinal. The exact course of events, especially the prevalence of either lateral or longitudinal association at the early stages of assembly, varies across different IF types (5, 13). However, chemical cross-linking experiments (14, 15) suggested that in all mature IFs there are three distinct types of lateral contacts between dimers: the A11 type corresponding to an antiparallel, approximately half-staggered alignment of two dimers with the first halves of their rod domains being in register, the A22 type with an antiparallel, approximately half-staggered alignment of the second halves of their rod domains, and the A12 type in which rod domains are aligned antiparallel and unstaggered (for a schematic diagram, see figure 8A of ref. 5).
Recombinant vimentin in 5 mM Tris·HCl buffer (pH 8.4) yields predominantly A11-type tetramers (16). The filament assembly can be readily initiated by a simultaneous increase of the ionic strength to 50–160 mM and lowering the pH to 7.5 by adding a concentrated buffer/salt solution (17, 18). Within the first second of assembly, formation of so-called unit-length filaments (ULFs) is observed, which result from the lateral association of dimers. Vimentin ULFs exhibit some variability of their composition and apparent width but the mean mass value determined by scanning transmission electron microscopy (EM) is close to 1.7 MDa, i.e., 16 dimers or four octamers (19). Cross-linking studies indicated that the dimers within the ULFs are interacting exclusively via the antiparallel, approximately half-staggered A11 mode (16). Next, the ULFs anneal longitudinally to yield progressively longer filaments. These yet-immature filaments undergo a slow radial compaction from ≈16 nm to ≈11 nm in diameter, a process that evidently involves rearrangements of the dimers relative to each other. After 1 h, long and smooth mature filaments having a uniform width of 10–12 nm are observed.
The aim of the work presented here was to elucidate the detailed assembly pathway of human vimentin in vitro and to structurally characterize the corresponding assembly intermediates. Taking into account their dynamic nature, such intermediates are likely to be very difficult to crystallize and hence cannot be effectively accessed by x-ray crystallography. In contrast, small-angle x-ray scattering (SAXS) can provide nanometer-scale structural information on macromolecules in the solution environment of interest (20, 21). Here, we used SAXS to monitor vimentin assembly as a function of ionic strength at pH 8.4. As the most significant result, we could generate molecular models of distinct assembly intermediates including tetramers, octamers, and ULFs.
Results
Vimentin Assembly in Different Solution Environments as Revealed by SAXS.
Initial SAXS measurements were done with solutions of recombinant temperature-sensitive mutant K139C of human vimentin. This mutant variant yields a lower viscosity increase than the WT after assembly initiation at room temperature, because of strongly attenuated longitudinal annealing of its ULFs (16). Therefore we originally used this mutant to avoid possible macroscopic aggregation at relatively high concentrations (up to ≈15 mg/ml) needed for SAXS measurements. Eventually, we could repeat the SAXS experiments with recombinant WT human vimentin with no serious aggregation problems. Moreover, we demonstrated that the difference between the WT and the mutant variant was only noticeable at later assembly stages, i.e., during longitudinal growth of the filaments (see below).
The measurements were carried out in a 5 mM Tris·HCl buffer (pH 8.4), first without additional salt and thereafter gradually increasing the NaCl concentration to 100 mM (Table 1). Salt addition had a prominent effect on the scattering profile of the vimentin solution: the innermost portion of the scattering curve became progressively steeper, pointing to an increased particle size (Fig. 1A). A qualitatively similar effect was observed when pH was lowered to 7.5 or 7.0 without NaCl addition (data not shown).
Table 1.
Samples used for SAXS measurements and their composition
| NaCl, mM | Sample (concentration, mg/ml) | Volume fractions, % |
χ2 | R | ||
|---|---|---|---|---|---|---|
| Tetramer | Octamer | 32-mer | ||||
| 0 | K139C (4.2/7.5) | 100 ± 1 | 0 ± 0 | 0 ± 0 | 2.6 | 0.03 |
| WT (2/3) | 100 ± 3 | 0 ± 0 | 2 ± 2 | 0.8 | 0.08 | |
| WT (6.6/7.7) | 79 ± 5 | 0 ± 0 | 22 ± 1 | 1.1 | 0.04 | |
| 6 | K139C (4.5/6.6) | 74 ± 1 | 26 ± 1 | 0 ± 0 | 2.9 | 0.04 |
| 12 | K139C (4.5/6.6) | 34 ± 1 | 66 ± 1 | 0 ± 0 | 4.2 | 0.04 |
| 20 | K139C (1.6) | 3 ± 0 | 97 ± 1 | 0 ± 0 | 0.5 | 0.05 |
| K139C (2.5) | 1 ± 1 | 71 ± 5 | 29 ± 2 | 0.7 | 0.07 | |
| K139C (6.5) | 0 ± 0 | 46 ± 2 | 54 ± 1 | 1.4 | 0.05 | |
| K139C (18) | 0 ± 0 | 20 ± 1 | 80 ± 1 | 4.6 | 0.07 | |
| 50 | K139C (3.9/5.1)/WT (3.9/5.7) | 9 ± 1 | 2 ± 1 | 90 ± 1 | 3.6 | 0.05 |
| 75 | K139C (2.1/4/8/17) | 0 ± 0 | 0 ± 0 | 100 ± 1 | 1.2 | 0.06 |
| 100 | K139C (3.7/4.9)/WT (6.9) | 0 ± 0 | 0 ± 0 | 100 ± 3 | 1.5 | 0.08 |
The buffer used was 5 mM Tris·HCl, pH 8.4. Quasimonodisperse solutions used to build the tetramer, octamer, and the ULF models are shown in bold. In nearly all conditions, the SAXS data collected for the WT and the K139C mutant were not significantly different and are shown together. In most cases, no pronounced dependence of the SAXS curve on the sample concentration was present. For definitions of the goodness-of-fit χ2 and the R factor between the experimental data and calculated scattering see Supporting Text.
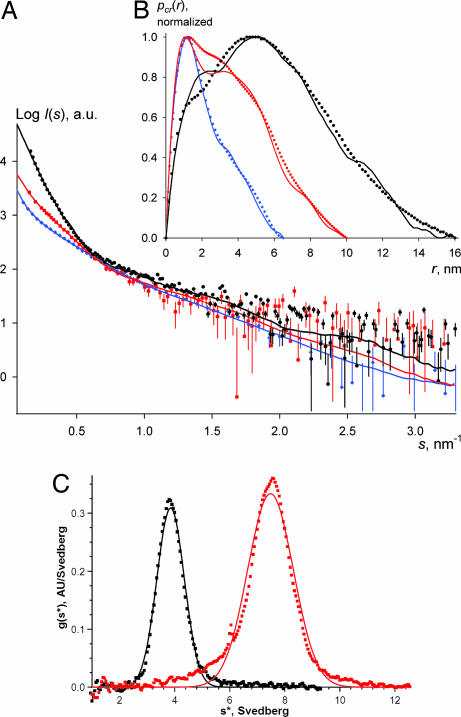
Fig. 1.
Vimentin assembly monitored by SAXS and AUC. (A) Experimental SAXS curves for the K139C vimentin mutant at: 4.2 mg/ml in 5 mM Tris·HCl (pH 8.4) (blue diamonds) overlaid with the calculated scattering from the final tetramer model (blue line), 1.6 mg/ml in the same buffer in the presence of 20 mM NaCl (red squares) overlaid with the calculated scattering from the final octamer model (red line), and 4 mg/ml in the same buffer in the presence of 75 mM NaCl (black circles) overlaid with the calculated scattering from the final ULF model (black line). (B) Corresponding cross-sectional distance distributions pcr(r). (C) Distribution of sedimentation coefficients as determined from AUC velocity runs at 17°C with a K139C vimentin solution at 2 mg/ml in 5 mM Tris·HCl (pH 8.4) (black squares) and 4 mg/ml in 5 mM Tris·HCl (pH 8.4) in the presence of 12 mM NaCl (red squares).
Because the vimentin rod domain is ≈46 nm in length and 2–3 nm in diameter (16), lateral vimentin oligomers were expected to have highly elongated shapes. For such particles, the experimental SAXS curve can be used to calculate the distribution pcr(r) of interatomic distances within the particle cross-section (see Supporting Text, which is published as supporting information on the PNAS web site). The value of r at which this distribution vanishes to zero corresponds to the maximal dimension of the particle cross-section. The obtained pcr(r) functions were also indicative of a gradual increase of particle thickness, suggesting progressive lateral association of the tetramers (Fig. 1B). For each pH and ionic strength condition, measurements with samples at ≈2–15 mg/ml indicated that the dependence of oligomerization on protein concentration was only pronounced with <50 mM NaCl at pH 8.4 (Table 1 and Fig. 5, which is published as supporting information on the PNAS web site). Moreover, in all conditions studied the SAXS data for the WT and the K139C mutant were not significantly different (Table 1), pointing to nearly identical lateral assembly properties.
Importantly, by a systematic screening it was possible to find conditions where a single oligomeric species (tetramer, octamer, or ULF) was predominant. The corresponding SAXS data were used to build and refine the atomic models of each oligomer, as will be explained below. Thereafter it became possible to determine the volume fractions of each oligomer in the remaining samples (Table 1).
Probing Early Stages of Assembly Using Analytical Ultracentrifugation (AUC).
Recently, we used AUC sedimentation velocity runs to show that a 0.1 mg/ml WT vimentin solution in 5 mM Tris·HCl (pH 8.4) without NaCl contains a homogenous population of tetramers with an apparent sedimentation coefficient s20,w of 4.7S (16). Here we have studied a K139C mutant vimentin solution at a concentration of 2 mg/ml comparable to that used for SAXS measurements. A single species with s20,w = 4.2S (Fig. 1C) was found yielding a corrected value for a diluted solution s20,wo of 4.6S, indicating that even at 2 mg/ml the K139C mutant features “normal” vimentin tetramers. Furthermore, the K139C solution at 4 mg/ml in 5 mM Tris·HCl (pH 8.4) with 12 mM NaCl revealed a predominant species with s20,w of 8.0S and a minor fraction of species with smaller s (Fig. 1C). We suggest that the 8S species correspond to an octamer. Indeed, the lateral association of two tetramers should yield a rod-like particle with about the same length but twice the mass of a tetramer (2 × 214 kDa), which would roughly double the sedimentation coefficient compared with a tetramer.
EM of Partially Annealing ULFs.
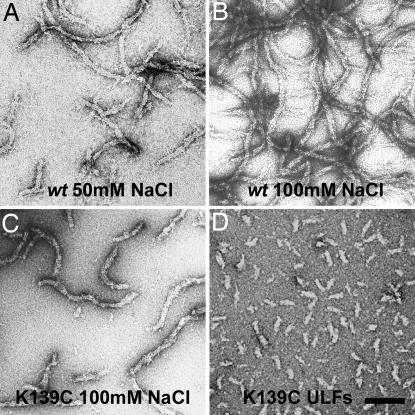
Electron micrographs of negatively stained WT samples at ≈5 mg/ml in 5 mM Tris·HCl (pH 8.4) with 50 mM NaCl revealed short filaments comprised of several ULFs annealed in the longitudinal direction (Fig. 2A), as evident from the comparison with the ULFs assembled by the “standard” protocol used in earlier studies (18, 19) (Fig. 2D). The tendency for the longitudinal annealing further increased in 100 mM NaCl (Fig. 2B). The K139C mutant assembled at ≈5 mg/ml also revealed considerable annealing of the ULFs but to a lesser extent than the WT (Fig. 2C). Importantly, the longitudinal assembly did not result in any considerable reduction of the apparent filament width, which remained at the same value (16–20 nm) as for isolated ULFs (19) (Fig. 2D).
Fig. 2.
Electron micrographs of negatively stained vimentin samples. (A and B) WT assembled at ≈5 mg/ml in 5 mM Tris·HCl buffer (pH 8.4) in the presence of 50 and 100 mM NaCl, respectively. (C) K139C mutant at ≈5 mg/ml in 5 mM Tris·HCl (pH 8.4) with 100 mM NaCl. (D) K139C mutant assembled for 2 s under standard conditions (0.1 mg/ml protein, 25 mM Tris·HCl, pH 7.5, and 50 mM NaCl). (Scale bar: 100 nm.)
These EM observations were complementary to the SAXS measurements, which were not sensitive to the longitudinal annealing of the ULFs. Indeed, the lowest resolution of the SAXS data measured corresponded to a maximal particle dimension of ≈42 nm, i.e., somewhat less than the 46-nm-long vimentin dimer. In other words, nearly the same SAXS signal should be expected for solutions containing either isolated ULFs or longitudinal assemblies of the latter.
3D Model of the Vimentin Tetramer.
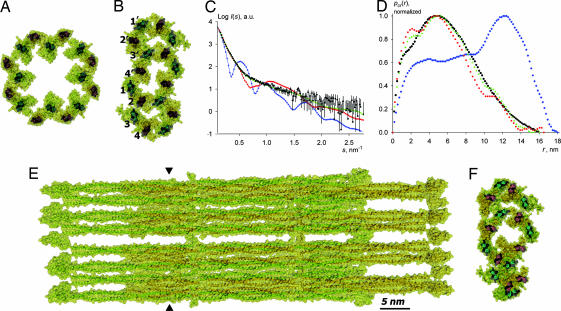
The tetramer model was optimized on the basis of SAXS data collected under the standard conditions, i.e., in 5 mM Tris·HCl (pH 8.4) buffer. The starting model was created by using a pair of atomic models of the vimentin dimer (10), which were placed antiparallel and approximately half-staggered with residues 191 of the two dimers being in register, to form an A11-type lateral contact (2, 22) (Fig. 3A). During subsequent refinement, the two dimers were treated as rigid bodies, whereas a single parameter, the separation of the two dimer axes dtetra (Fig. 3B), was varied so as to optimize the goodness-of-fit χ2 between the calculated and experimental scattering curves. A clear minimum was obtained for dtetra = 3.4 nm (see Fig. 6, which is published as supporting information on the PNAS web site). Rotating one of the dimers by the angle ω (see Fig. 3A) relative to the other dimer did not produce a significant effect on the χ2 value within the ω range of ±10°, so the two dimer axes were kept parallel in the final model. Because the head domain was known to be critically involved in the tetramer formation (13, 18), one of the head domains of each dimer was qualitatively modeled to fold back onto the rod domain of the adjacent dimer (Fig. 3A). The calculated scattering from the final tetramer model (Movie 1, which is published as supporting information on the PNAS web site) produced a good fit to the experimental data (χ2 = 2.2, Fig. 1A, blue diamonds), and the cross-sectional distance distribution functions pcr(r) calculated from the model and the experiment were also in good agreement (Fig. 1B).
Fig. 3.
Vimentin tetramer and octamer. (A) Side view of the tetramer as a ribbon diagram overlapped with a semitransparent surface (yellow). (B) Cross-sectional view at the position indicated by triangles in A. (C) Side view of the octamer. (D) Cross-sectional view of the octamer.
Octamer Model.
As stated above, AUC runs with a K139C mutant solution at 4 mg/ml in 5 mM Tris·HCl (pH 8.4) in the presence of 12 mM NaCl indicated the prevalence of octameric particles. In line with this finding, the SAXS data for the K139C solution at 1.6 mg/ml in the same buffer but with 20 mM NaCl suggested the presence of particles with about twice the diameter of a tetramer, as evident from comparison of the corresponding pcr(r) functions (Fig. 1B, red squares and blue diamonds, respectively). An initial atomic model for the octamer was created by aligning two previously constructed A11 tetramers in such a way that the two closest dimers (one from each tetramer) were engaged in a further A11 contact (Fig. 3C). The best fit to the experimental data were found by using the two tetramers as rigid bodies and systematically varying two parameters: the axis-to-axis distance docta between the nearest dimers of the two tetramers and the puckering angle α (see Fig. 3D; to reduce the number of parameters, the angle α was assumed to be the same for both tetramers: α1 = α2). The best octamer model reveals a “chair-like” conformation when viewed down the dimer axes (Fig. 3D) with α = 107° and docta = 3.2 nm, and it produces good fits to both the SAXS curve (χ2 = 0.57; Fig. 1A, red squares) and the experimental pcr(r) function (Fig. 1B). Introduction of a relative tilt by the ω angle (see Fig. 3A) within each of the A11 contacts worsened the fits, and in the final model all dimer axes were kept parallel. The cross-section of the final octamer model (Fig. 3D and Movie 2, which is published as supporting information on the PNAS web site) fits inside an oval with dimensions 10.5 nm × 5.0 nm if the peripheral head domains are not taken into account.
The ULF Model.
In accordance with the EM observations of the ULFs (Fig. 2), the scattering curve for the K139C mutant in 5 mM Tris·HCl (pH 8.4) with 75 mM NaCl yielded a pcr(r) distribution with a maximum at 4.8 nm (Fig. 1A, black circles). Moreover, the largest cross-sectional dimension was 16 nm, which is ≈1.5 times that of the octamer. The starting model for the ULF was constructed from four octamers. Initially, a structure with exact 4-fold symmetry was assembled, whereby the outermost dimers of adjacent octamers were brought together to form A11 contacts (Fig. 4A). Such a structure had an excessively large cross-section, as evident from its pcr(r) distribution (Fig. 4D, blue diamonds). Moreover, the calculated SAXS curve for this model exhibited distinct peaks typical for a “hollow cylinder” structure, in contrast to the rather smooth experimental curve (Fig. 4C).
Fig. 4.
Vimentin ULF. (A) Cross-section of the initial model featuring a 4-fold symmetrical arrangement of octamers. (B) Refined model with a flattened cross-section. (C) Calculated scattering from the initial symmetrical model (blue diamonds), flattened model (red squares), and the final randomized model (green triangles) overlaid with the experimental data for the K139C mutant vimentin variant at 4 mg/ml in 5 mM Tris·HCl buffer (pH 8.4) with 75 mM NaCl (black circles). (D) Corresponding pcr(r) functions. (E) Side view of the final randomized model. (F) Cross-section of the position indicated by triangles in E in the final randomized model.
The clearly skewed experimental pcr(r) distribution (Fig. 4D, black circles) suggests particles with an oval rather than a round cross-section (20). Correspondingly, we attempted to “flatten” the cross-section of the model while keeping the octamers rigid (Fig. 4B). To keep the proper A11 contacts between the octamers, it was necessary to invert the “handedness” of the chair-like conformation of every other octamer (compare the octamers built of dimers 1–4 and 1′–4′, Fig. 4B). After optimization, the pcr(r) function for the best flattened model (Fig. 4D, red squares) became reasonably close to the experimental one, and there was a good agreement between the corresponding scattering curves up to the resolution of s = 0.5 nm−1 (Fig. 4C), indicating that the overall shape was correct.
By introducing small random rotations of the individual octamers about their centers of mass, it was possible to further improve the fit (Fig. 4 C and D, green triangles). A family of closely related ULF models (one of them shown in Fig. 4 E and F and Movie 3, which is published as supporting information on the PNAS web site) could be constructed, all providing sufficiently good fits to the experiment with χ2 values not exceeding 2.0. Because of the intrinsic limitations of the SAXS method [resolution limits and spherical averaging (21, 22)], it was not possible to obtain a single model with atomic accuracy, all obtained ULF models have oval (see above) cross-sections with dimensions of ≈16 × 9 nm, and they correlate well with the experimental pcr(r) distribution.
Discussion
Application of SAXS Toward Investigating IF Assembly.
During earlier SAXS experiments using tissue-derived keratin samples (23), no sufficiently homogenous solutions of assembly intermediates could be found. Until now, direct structural information on the IF assembly was predominantly obtained by EM observations either by negative staining or rotary metal shadowing that were unable to resolve individual dimers within higher-order assemblies (7). Our SAXS data indicate that the assembly of vimentin occurs via distinct lateral oligomers, including the tetramer, the octamer, and the ULF, the latter comprising four octamers (Movies 1–3). It should be noted that highly elongated vimentin assembly intermediates are challenging objects for a SAXS study. Although methods of particle shape restoration from the SAXS data alone without the use of additional information (21) exist, much more reliable modeling of vimentin oligomers could be achieved by a “reverse” approach, i.e., by first constructing initial atomic models from individual dimers complying with available steric restraints, and then refining them against the measured SAXS data. All vimentin samples measured could be interpreted as mixtures containing only the three above- mentioned types of oligomers in different percentages, yielding excellent fits to the experimental data (Fig. 7, which is published as supporting information on the PNAS web site). The percentages of higher-order oligomers consistently increased upon the addition of salt, lowering the pH, or increasing the protein concentration (Table 1).
Importantly, we conclude that the next substantially populated and long-lived assembly intermediate beyond the tetramer is the octamer. This finding is paralleled by observations of unraveling vimentin IFs by both EM (24–26) and atomic force microscopy (27) that consistently report the presence of up to four individual, probably octameric, subfilaments (“protofibrils”). Moreover, in vitro-assembled vimentin IFs were shown to have some polymorphism, whereby their mass-per-length value exhibited a distribution with distinct peaks separated by increments corresponding to the addition of a single octamer (18, 28).
Both our present SAXS data and the data of Ip et al. (26) consistently indicate that at pH 8.4 the lateral assembly of vimentin tetramers into ULFs is mostly complete once the NaCl concentration is increased to 50 mM (see Table 1). In addition, EM reveals that there is already a pronounced longitudinal annealing of the WT vimentin ULFs under these conditions (Fig. 2). Furthermore, the EM images and, in particular, the SAXS data clearly indicate that annealing of the ULFs at pH 8.4 does not lead to any significant decrease of their diameter. In contrast, the standard in vitro vimentin assembly protocol by increasing the ionic strength and lowering the pH to 7.5 yields mature, radially compacted filaments that appear ≈33% thinner than the ULFs by negative staining EM (19).
Tetramers, Octamers, and ULFs Represent Distinct Intermediates of Vimentin Assembly.
The tetramer model refined against the SAXS data (Fig. 3A) is a ≈65-nm-long rodlet, which is consistent with EM observations of glycerol-sprayed/rotary metal-shadowed samples (18) and AUC data (16). Furthermore, SAXS measurements indicate that there is a considerable lateral separation of the two dimers within the tetramer: the distance between the two dimers' axes is 3.4 nm, while the coiled coils are clearly separated by ≈1.5 nm between their van der Waals surfaces (Fig. 3A). Such a separation is not unreasonable, as the immediate contact of the acidic rod domains should be electrostatically unfavorable (29). The axial stagger of the two dimers is known from cross-linking and spin labeling/electron paramagnetic resonance studies (2, 22) and could not be further refined because of the low-resolution cut-off of the SAXS data. In addition, the constructed vimentin tetramer reflects the notion that the A11 contact is mediated by the second half (residues ≈35–70) of the head domain (ref. 5 and T.W. and H.H., unpublished work), whereby the electrostatic attraction between the positively charged heads and the acidic rod domains appears as the likely driving force (5, 13). Indeed, a vimentin fragment including only the rod domain does not assemble beyond the dimer in 10 mM Tris·HCl (pH 8.4) (11).
The occurrence of an octameric species in vimentin solutions was suggested (26), but, unlike the tetramer, the octamer was never characterized structurally to any considerable extent. Both our AUC and SAXS data indicate that octamers predominate in concentrated (≈5 mg/ml) vimentin solutions in 5 mM Tris·HCl (pH 8.4) with 12 or 20 mM NaCl (Fig. 1C and Table 1). The SAXS-based octamer model reveals that the association of two tetramers into an octamer is compatible with a chair-like arrangement of the four dimers in cross-section (Fig. 3B). In this arrangement, each pair of dimers nearest to each other (i.e., dimers labeled 1 and 2, 2 and 3, and 3 and 4, respectively, in Fig. 3B) are aligned in the A11 mode. The axial separation between antiparallel dimers was found to be essentially the same (3.2–3.4 nm) regardless of whether the A11 contact occurs within a given tetramer (i.e., between dimers 1 and 2 or 3 and 4) or across the two tetramers (i.e., between dimers 2 and 3). In contrast, the axial separation between parallel dimers (e.g., dimers 1 and 3) is considerably larger (5.1 nm). This finding agrees well with cross-linking studies that indicated that the neighboring dimers within a filament are always antiparallel (14).
In line with the observed skewed cross-sectional distance distribution, the final ULF model built of four octamers has an oval, rather than a round, cross-section with dimensions of ≈16 × 9 nm. In line with this result, the average thickness of glutaraldehyde-fixed vimentin ULFs measured by atomic force microscopy in aqueous solutions after adsorption on graphite was ≈10 nm (27). However, it should be considered that both the pressure applied by the atomic force microscopy tip and the spreading of the relatively loosely packed ULFs on the support may result in some underestimation of their height. Moreover, the negatively stained vimentin ULFs have an apparent width of 16 ± 3 nm (18), with this significant variation being caused by their flattening on the EM grid. Finally, scanning transmission EM measurements of individual ULFs revealed that they vary in mass considerably, suggesting that the number of constituent vimentin chains may also vary (19).
The final SAXS-based ULF model features a fairly even mass distribution over the cross-section (Fig. 4F). There is a relatively small void on the axis of the structure, in contrast to some “hollow tube” models proposed previously (for a discussion, see refs. 5 and 7). While the use of octamers as rigid bodies allowed us to build a reasonable ULF model with a small number of optimization parameters, it is conceivable that ULF formation may also be accompanied by structural rearrangements within the octamers.
Last, but not least, the radial compaction step (not covered in the present study) toward the mature 10- to 12-nm-diameter IFs must involve further significant rearrangements in the filament architecture. On one hand, the A22 and A12 dimer–dimer modes become detectable only in mature IFs (18, 26). On the other hand, based on the cross-sectional dimensions of the ULF obtained in the present investigation, its typical length of 65 nm, and the mass of 32 vimentin chains, one can estimate the average density of the ULF at 0.19 kDa/nm3, as compared with a typical density of 0.81 kDa/nm3 for a protein globule (30). Hence the packing of 16 individual IF dimers into the ULF is clearly far from being dense (Fig. 4E) and leaves plenty of “room” for further structural rearrangements toward considerably more compact, mature IFs.
Concluding Remarks.
It is worth noting that the SAXS signal corresponds to an average over the conformational ensemble in solution. As opposed to the rigid structure of a single molecular model, in reality the rather loosely packed vimentin ULFs should have a considerable degree of conformational dynamics, which also correlates with their “open” nature (15). Moreover, EM observations indicate certain variability of the apparent ULF shape, whereas scanning transmission EM data further suggest some variability in the number of dimers per cross-section (19). Although this polymorphism certainly should be kept in mind, our SAXS measurements point to an ULF comprised of four tetramers as a major species.
In vivo, the presence of a small pool of soluble vimentin tetramers has been demonstrated (31), but the occurrence of other assembly intermediates such as octamers or ULFs needs further investigation. Live cell imaging has indicated the existence of dynein-associated IF precursors that move along microtubules and ultimately fuse to form filaments (32). If the precursor is indeed an ULF, then the capping by dynein should prevent it from annealing with other ULFs moving on the same track.
Finally, there has been a rapid growth of characterized mutations in IF proteins, including keratins, desmin, neurofilament triplet proteins, lamins, and others, that have been found to cause currently incurable human diseases (3, 33, 34). The next major challenge is to establish the molecular mechanism of these diseases and, ultimately, to develop efficient treatment approaches. Here, the in vitro-derived structural information on the IF architecture and assembly is of utmost importance, as seen, e.g., in the recent studies on desminopathies (35–37).
Materials and Methods
Sample Preparation.
WT human vimentin and its K139C mutant were recombinantly expressed in Escherichia coli and purified as described (17). The proteins were stored at −80°C in 8 M urea, 5 mM Tris·HCl (pH 7.5), 1 mM DTT, 1 mM EDTA, 0.1 mM EGTA, and 10 mM methyl ammonium chloride. Before use, the proteins were dialyzed into a 5 Tris·HCl buffer (pH 8.4) with 0.5 mM DTT at room temperature by lowering the urea concentration in a stepwise fashion (6, 4, 2, and 0 M). Dialysis was continued overnight at 4°C against a second volume of the buffer without urea. After addition of the required amount of NaCl as 5 M stock, protein solutions were concentrated to typically 1.6–18 mg/ml by using a Millipore (Billerica, MA) centrifuge concentrator.
SAXS Data Collection and Processing.
The SAXS experiments were performed at the European Molecular Biology Laboratory beamline X33 (Deutsches Elektronen Synchrotron, Hamburg, Germany). A linear gas detector was placed at a distance of 4 m from the 120-μl sample cell. The momentum transfer range was 0.15 < s < 3.4 nm−1 (s = 4πsinθ/λ, where 2θ is the scattering angle and λ of 0.15 nm is the wavelength). Scattering curves were collected in 15 1-min frames to check for radiation damage. The data were processed by using standard procedures with the program PRIMUS (38) and further analyzed with the package ATSAS (39). As a starting point for molecular modeling, a model of human vimentin dimer constructed from available crystal structures of its fragments was used (for details, see ref. 10). The scattering curves from the models were calculated by using CRYDAM, a modified version of CRYSOL (40), whereby the protein solvation shell was represented by a 0.3-nm-thick layer of small beads.
AUC.
Protein samples (4–6 mg/ml) to be used for SAXS measurements were subjected to sedimentation velocity runs at 17°C and 30,000 or 40,000 rpm by using an Optima XLA Beckman (Fullerton, CA) analytical ultracentrifuge. Optical density scans at 284 nm were analyzed to determine the sedimentation coefficient essentially as described (16).
EM.
A small aliquot of a protein sample (≈5 mg/ml) to be used for SAXS measurements was chemically fixed with 0.1% glutaraldehyde for several minutes and then diluted to 0.1 mg/ml. Thereafter a 5-μl aliquot was applied for 1 min onto a glow-discharged carbon-coated copper grid, washed with double-distilled water, and stained with 2% uranyl acetate (pH 4). Electron micrographs were recorded on a H-7000 transmission electron microscope (Hitachi, Tokyo, Japan) operated at 100 kV.
Supplementary Material
Acknowledgments
We thank Drs. M. H. J. Koch and V. V. Volkov for advice on processing SAXS data. This work was supported by grants from the Swiss National Science Foundation (to U.A.), the Swiss Society for Research on Muscular Diseases (to U.A. and S.V.S.), the Molecular and Cell Biology Program of the Russian Academy of Sciences (to S.V.S.), the Deutsche Forschungsgemeinschaft (HE1853 to H.H.), the Canton Basel-Stadt, and the M.E. Müller Foundation. Diffraction data collection was made possible through the European Molecular Biology Laboratory, Hamburg, Germany.
Abbreviations
- IF
intermediate filament
- SAXS
small-angle x-ray scattering
- ULF
unit-length filament
- AUC
analytical ultracentrifugation
- EM
electron microscopy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Herrmann H, Aebi U. Curr Opin Cell Biol. 2000;12:79–90. doi: 10.1016/s0955-0674(99)00060-5. [DOI] [PubMed] [Google Scholar]
- 2.Parry DAD, Steinert PM. Intermediate Filament Structure. Heidelberg: Springer; 1995. [Google Scholar]
- 3.Fuchs E, Weber K. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 4.Herrmann H, Hesse M, Reichenzeller M, Aebi U, Magin TM. Int Rev Cytol. 2003;223:83–175. doi: 10.1016/s0074-7696(05)23003-6. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann H, Aebi U. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 6.Goldman RD, Khuon S, Chou YH, Opal P, Steinert PM. J Cell Biol. 1996;134:971–983. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parry DAD, Steinert PM. Q Rev Biophys. 1999;32:99–187. doi: 10.1017/s0033583500003516. [DOI] [PubMed] [Google Scholar]
- 8.Strelkov SV, Herrmann H, Aebi U. BioEssays. 2003;25:243–251. doi: 10.1002/bies.10246. [DOI] [PubMed] [Google Scholar]
- 9.Aebi U, Haner M, Troncoso J, Eichner R, Engel A. Protoplasma. 1988;145:73–81. [Google Scholar]
- 10.Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P. EMBO J. 2002;21:1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, Burkhard P, Aebi U. J Mol Biol. 2001;306:773–781. doi: 10.1006/jmbi.2001.4442. [DOI] [PubMed] [Google Scholar]
- 12.Strelkov SV, Kreplak L, Herrmann H, Aebi U. Methods Cell Biol. 2004;78:25–43. doi: 10.1016/s0091-679x(04)78002-4. [DOI] [PubMed] [Google Scholar]
- 13.Kreplak L, Aebi U, Herrmann H. Exp Cell Res. 2004;301:77–83. doi: 10.1016/j.yexcr.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Steinert PM, Marekov LN, Parry DA. J Biol Chem. 1993;268:24916–24925. [PubMed] [Google Scholar]
- 15.Herrmann H, Aebi U. Curr Opin Struct Biol. 1998;8:177–185. doi: 10.1016/s0959-440x(98)80035-3. [DOI] [PubMed] [Google Scholar]
- 16.Mucke N, Wedig T, Burer A, Marekov LN, Steinert PM, Langowski J, Aebi U, Herrmann H. J Mol Biol. 2004;340:97–114. doi: 10.1016/j.jmb.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann H, Kreplak L, Aebi U. Methods Cell Biol. 2004;78:3–24. doi: 10.1016/s0091-679x(04)78001-2. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann H, Haner M, Brettel M, Muller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U. J Mol Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann H, Haner M, Brettel M, Ku NO, Aebi U. J Mol Biol. 1999;286:1403–1420. doi: 10.1006/jmbi.1999.2528. [DOI] [PubMed] [Google Scholar]
- 20.Koch MH, Vachette P, Svergun DI. Q Rev Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 21.Svergun DI, Koch MH. Curr Opin Struct Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- 22.Hess JF, Budamagunta MS, Voss JC, FitzGerald PG. J Biol Chem. 2004;279:44841–44846. doi: 10.1074/jbc.M406257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayers Z, Michon AM, Sicre P, Koch MH. J Struct Biol. 1990;103:212–224. doi: 10.1016/1047-8477(90)90039-f. [DOI] [PubMed] [Google Scholar]
- 24.Aebi U, Fowler WE, Rew P, Sun TT. J Cell Biol. 1983;97:1131–1143. doi: 10.1083/jcb.97.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann H, Wedig T, Porter RM, Lane EB, Aebi U. J Struct Biol. 2002;137:82–96. doi: 10.1006/jsbi.2002.4466. [DOI] [PubMed] [Google Scholar]
- 26.Ip W, Hartzer MK, Pang YY, Robson RM. J Mol Biol. 1985;183:365–375. doi: 10.1016/0022-2836(85)90007-5. [DOI] [PubMed] [Google Scholar]
- 27.Mucke N, Kirmse R, Wedig T, Leterrier JF, Kreplak L. J Struct Biol. 2005;150:268–276. doi: 10.1016/j.jsb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann H, Aebi U. Cell Mol Life Sci. 1999;55:1416–1431. doi: 10.1007/s000180050382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. J Mol Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- 30.Matthews BW. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 31.Soellner P, Quinlan RA, Franke WW. Proc Natl Acad Sci USA. 1985;82:7929–7933. doi: 10.1073/pnas.82.23.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon KH, Yoon M, Moir RD, Khuon S, Flitney FW, Goldman RD. J Cell Biol. 2001;153:503–516. doi: 10.1083/jcb.153.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldfarb LG, Vicart P, Goebel HH, Dalakas MC. Brain. 2004;127:723–734. doi: 10.1093/brain/awh033. [DOI] [PubMed] [Google Scholar]
- 34.Omary MB, Coulombe PA, McLean WH. N Engl J Med. 2004;351:2087–2100. doi: 10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- 35.Kaminska A, Strelkov SV, Goudeau B, Olive M, Dagvadorj A, Fidzianska A, Simon-Casteras M, Shatunov A, Dalakas MC, Ferrer I, et al. Hum Genet. 2004;114:306–313. doi: 10.1007/s00439-003-1057-7. [DOI] [PubMed] [Google Scholar]
- 36.Bar H, Strelkov SV, Sjoberg G, Aebi U, Herrmann H. J Struct Biol. 2004;148:137–152. doi: 10.1016/j.jsb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Bar H, Mucke N, Kostareva A, Sjoberg G, Aebi U, Herrmann H. Proc Natl Acad Sci USA. 2005;102:15099–15104. doi: 10.1073/pnas.0504568102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 39.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. J Appl Crystallogr. 2006;39:277–286. [Google Scholar]
- 40.Svergun DI, Barberato C, Koch MHJ. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.