Abstract
The role of each histone tail in regulating chromatin structure is elucidated by using a coarse-grained model of an oligonucleosome incorporating flexible histone tails that reproduces the conformational and dynamical properties of chromatin. Specifically, a tailored configurational-bias Monte Carlo method that efficiently samples the possible conformational states of oligonucleosomes yields positional distributions of histone tails around nucleosomes and illuminates the nature of tail/core/DNA interactions at various salt milieus. Analyses indicate that the H4 histone tails are most important in terms of mediating internucleosomal interactions, especially in highly compact chromatin with linker histones, followed by H3, H2A, and H2B tails in decreasing order of importance. In addition to mediating internucleosomal interactions, the H3 histone tails crucially screen the electrostatic repulsion between the entering/exiting DNA linkers. The H2A and H2B tails distribute themselves along the periphery of chromatin fibers and are important for mediating fiber/fiber interactions. A delicate balance between tail-mediated internucleosomal attraction and repulsion among linker DNAs allows the entering/exiting linker DNAs to align perpendicular to each other in linker-histone deficient chromatin, leading to the formation of an irregular zigzag-folded fiber with dominant pair-wise interactions between nucleosomes i and i ± 4.
Keywords: Monte Carlo simulations, nucleosome, DNA/protein complexes, chromatin structure regulation, irregular zigzag
Eukaryotic double-stranded DNA achieves cellular compaction through several hierarchical levels of organization (1). The most fundamental of these involves wrapping of DNA around protein aggregates known as nucleosomes. The nucleosome, whose structure has been determined at high resolution (2), comprises two copies each of the positively charged histones H2A, H2B, H3, and H4. A large portion of each histone chain forms the nucleosome core around which DNA makes ≈1.75 turns, whereas the terminal portion, the histone tail, extends outwards from the core and is much floppier than the rest of the nucleosome. The resulting “beads-on-a-string” nucleosome/DNA complex compacts further at physiological salt conditions and, in the presence of highly charged linker histone proteins (H1 or H5), forms the compact 30-nm chromatin fiber.
The histone tails critically regulate chromatin compaction and function. Electrostatic arguments alone suggest that the compact state of chromatin can be achieved only if the strong DNA/DNA repulsion as well as the entropic penalty loss associated with folding are alleviated. The positively charged histone tails provide the necessary driving force for folding by mediating favorable internucleosomal interactions and screening DNA repulsion. At the same time, gene activation requires that related regions of chromatin become partially unfolded to allow access to transcription machinery. Indeed, acetylation of histone tail residues through the action of histone acetyl transferases results in unfolding of chromatin, possibly through a partial neutralization of charge on the tails (3–5). Histone tail acetylation, along with other forms of functional modifications, provides further evidence for the dominant role of histone tails in regulating chromatin. In addition, the histone tails likely play important roles in the subsequent compaction of chromatin into heterochromatin through fiber/fiber interactions.
Experimental studies dissecting the role of histone tails (6–11) are typically conducted in vitro and rely on self-assembling short fragments of chromatin (oligonucleosomes) from the components: core and linker histones and short “designer” DNA templates. The histone proteins are modified chemically before their assembly, and the impact of each modification is observed directly at the level of the assembled oligonucleosome. These studies have indicated the greater importance of the H3/H4 dimer over the H2A/H2B dimer for producing compact chromatin (7, 8, 9) and the roles of specific lysine residues in the H4 tail in chromatin compaction (10, 11). However, because the tails are very dynamic and their locations cannot be precisely determined, these experiments cannot resolve exactly how and why certain histone tails are more important than others for chromatin folding. Advanced techniques employing ligation and chemical cross-linking can address internucleosomal versus intranucleosomal tail/DNA interactions in chromatin but lack detailed information on how histone tails regulate chromatin folding both in the absence and presence of linker histones (12, 13). Theoretical approaches are thus well poised to address some fundamental questions regarding the nature of interactions between the histone tail and the rest of chromatin; however, the complexity of chromatin and the large spatial and temporal scales involved pose special challenges to modelers.
Most computational/theoretical models of chromatin (14–21) either have included the effects of histone tails crudely or neglected them. This approach is reasonable to a first approximation because of the separation of time scales that exists between the dynamics of the flexible tails and those of the nucleosomes/linker DNA segments. Recently, we have developed a mesoscopic model of oligonucleosomes (Fig. 1) that comprehensively models the conformational freedom and electrostatics of each histone tail and is amenable to long-time, large-scale sampling by Brownian dynamics (BD) and Monte Carlo (MC) (22). Our model represents each histone tail as a chain of charged coarse-grained beads with customized charges and forcefield parameters for each tail; the nucleosome core and linker DNA are treated by using the Discrete Surface Charge Optimization (DiSCO) model (14, 23, 24) and the discrete elastic chain model (25), respectively. Thorough testing has already demonstrated how the incorporation of flexibility into histone tails leads to better agreement with experimental measurements (diffusion constants, interaction energies, salt-dependent extension of histone tails, and folding of nucleosomal arrays) (22).
Fig. 1.
Modeling the basic motif c1 (nucleosome core, linker DNA, and histone tails) to yield the final model c2: the atomistic nucleosome core n1 is modeled as a rigid body with a uniformly distributed set of charges (n2); the linker DNA d1 is treated by using the discrete elastic chain model d2; and the histone tails [(t1), H3 tail is shown] are represented by using the subunit model t2 and then coarse-grained further to the protein bead chain t3. The tails in c2 are colored blue (H3), green (H4), yellow (H2A), and red (H2B).
Here, we employ the flexible-tail model of oligonucleosomes to elucidate the role of each histone tail in chromatin folding. Our MC methods, including an end-transfer configurational bias method, efficiently sample different configurations of oligonucleosomes and prevent local configurational traps, as observed for Brownian dynamics. The extensive configurational ensembles allow us to extract both the pattern of internucleosomal interactions within chromatin and the positional distribution of each histone tail. These patterns reveal that each histone tail has a special function in regulating chromatin folding and that the cumulative effect is an irregular zigzag arrangement of nucleosomes with regular interaction patterns between each nucleosome and its fourth neighbor along the chain.
Results and Discussion
We next analyze various properties related to the distribution and interaction of the histone tails and the pattern and energetics of tail-mediated internucleosomal interactions. We describe the role of each histone tail in chromatin folding and in the nucleosomal arrangement and evaluate these predictions in light of available experimental data. The broader biological implications are discussed.
Global Histone Tail Pattern.
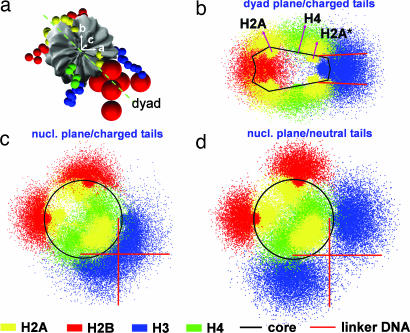
The histone tails differ from each other not only in lengths and charges but also in their location on the surface of the nucleosome core. The location-dependent functions of the tails can be deduced directly from analyses of positional distributions, obtained here by projecting the position vectors of tail beads (relative to the parent nucleosome's center of mass) onto the reference frame of the parent nucleosome described by the orthogonal set of unit vectors {a, b, c}. Tail distributions along the “nucleosomal” plane {a, b} and “dyad” plane {(a − b)/, c} at 0.2 M salt are plotted in Fig. 2. Tail distributions at 0.2 M salt for arrays in which all tail charges are set to zero also help assess the impact of electrostatics on their distributions.
Fig. 2.
Reference frame of a nucleosome core (a) and positional distribution of fully charged histone tails along its dyad (b) and nucleosomal planes (c) and of neutralized tails along a nucleosome plane (d), all at 0.2 M salt. Red arrows in b indicate the mean position/orientation of H4 and H2A tails. H2A*, C termini of H2A histones.
Fig. 2 shows that all histone tails exhibit fairly broad distributions compatible with their highly dynamic/flexible nature. The H2A and H4 tails spread mostly in the direction normal to the nucleosomal plane because of their origin from the flat faces of the nucleosome core (Fig. 2b). The longer H4 tails extend further outwards than the H2A tails. On the other hand, the H2B and H3 tails, which originate from the curved side of the nucleosome core in between the wound DNA supercoil, spread predominantly along the nucleosomal plane (Fig. 2c). The H3 tails, in particular, tend to remain close to the entering/exiting linker DNA. Their point of origin from the nucleosome core, proximal to the linker DNA origin, allows entire tails rather than the ends to participate in strong electrostatic interactions with the linker DNA. Note how the positional distribution of H3 tails “hugs” the mean position of linker DNAs (Fig. 2c) and how their bias toward the linker DNAs disappears when they are neutralized (Fig. 2d). We show later how this unique property of H3 tails impacts chromatin folding. Also note the propensity of H2A (N termini) and H2B tails to distribute along the nucleosome edge farthest from the linker DNAs.
Role of Each Histone Tail.
Next, we quantify the extent to which each histone tail interacts with the rest of chromatin by computing the frequency of attachment to a specific component of chromatin (i.e., number of occurrences divided by the total sampled tail configurations). A tail is considered “attached” if the shortest distance between its beads and that of the component is smaller than 80% of the excluded volume size parameter (σ). Specifically, we compute in Fig. 3 the fraction of time each tail interacts with: parent nucleosomes (intranucleosomal), fTCintra; other nucleosomes (internucleosomal), fTCinter; entering/exiting linker DNA of parent nucleosomes, fTLintra; and other linker DNAs, fTLinter; the fraction of time that tails remain unattached/free is then given by (1 − fTCintra − fTCinter − fTLintra − fTLinter). We also compute the extension of each histone tail, lext from the average distance between the last tail bead and the nearest nucleosomal charge. Tail interactions/extensions are compared for three folded states of the oligonucleosome: an extended oligonucleosome at 0.01 M salt, a moderately folded oligonucleosome at 0.2 M salt, and a highly compact fiber at 0.2 M salt where repulsion among linker DNAs is set to zero.
Fig. 3.
Representative 48-unit oligonucleosome at 0.2 M salt (a) and analyses and snapshots of boxed regions in a to highlight different tail interactions (b–g%). Analyzed systems involve oligonucleosomes without linker histones at 0.2 M (□) and 0.01 M (▵), and “compact” oligonucleosomes at 0.2 M salt (×). In each box (b–g%), a cartoon image depicts the interaction plotted as a frequency for the time that tails mediate: internucleosomal interactions (b), attach to parent nucleosomes (c), remain unattached (d), attach to linker DNA not associated with parent nucleosome (e), and attach to linker DNA associated with parent nucleosome (f). Plot (g) provides tail extension lengths. Results are averaged over the two copies of each tail. H2A*, C termini of H2A histones.
Overall, Fig. 3 shows that, at low salt (0.01 M; red triangles), the histone tails either remain free (Fig. 3d) or interact exclusively with parent nucleosomal DNA (Fig. 3c) and parental linker DNA (Fig. 3f); internucleosomal interactions almost are nonexistent (Fig. 3 b and e). At higher salt (0.2 M; blue squares), electrostatic screening of linker DNA causes the tails to mediate internucleosomal interactions (Fig. 3 b and e) with wound and linker DNA of adjacent nucleosomes (mostly through their ends) at the expense of intranucleosomal interactions (Fig. 3 c and f) (except H3; see below). With increasing salt, the tails extend outward from the nucleosome as a result of greater screening of the electrostatic attraction between tails and wound DNA of parent nucleosomes (22, 26), further encouraging internucleosomal interactions (Fig. 3g). Still, the fraction of tail-mediated internucleosomal interactions is quite small (<5%) in linker-histone deficient oligonucleosomes (Fig. 3b). By setting linker DNA repulsions to zero to crudely mimic the effect of linker histones, we observe a large number of internucleosomal interactions upon further compaction of chromatin (Fig. 3b).
Significantly, we can identify a specific role for each histone tail in regulating chromatin structure and higher-level folding as follows.
The H3 tails have a unique tendency to attach to the linker DNA entering/exiting the parent nucleosome cores; in fact, the tails remain attached 60–65% of their time to the two linker DNAs (Fig. 2d). The proximity of the H3 tails to the linker DNAs implies that they may be strongly involved in screening electrostatic repulsion between the linker DNAs in addition to mediating internucleosomal interactions.
The H4 histone tails mediate the largest number of internucleosomal interactions for moderately folded arrays at 0.2 M salt, and even more so in compact chromatin where they spend as much as 11% of their time bound to neighboring nucleosomes (Fig. 3b). We speculate that the H4 tails play equally dominant roles in mediating internucleosomal interactions within compact chromatin that contains linker histones. This dominance of the H4 tails may be related to their optimal position on the flat portion of the nucleosome core close to the linker DNAs from where they can bind to neighboring nucleosomes (Fig. 2b); this is especially relevant to compact chromatin where nucleosomes tend to stack themselves parallel to each other. Even though the N termini of H2A histones also originate from the flat portion of the nucleosome core, they cannot mediate internucleosomal interactions to the same extent as the H4 tails because of their slightly shorter length and distant location from the linker DNAs (Fig. 2b).
The H2A/H2B tails are less important than the H3/H4 tails in terms of mediating internucleosomal interactions (Fig. 3b). The C termini of H2A tails, especially, are too short to mediate any internucleosomal interactions for moderately folded chromatin at 0.2 M but mediate more internucleosomal interactions in compact chromatin (Fig. 3b). The positional distribution of the N termini H2A and H2B tails indicates that they are mostly distributed along the edge of the nucleosome farthest from the linker DNAs (Fig. 2c; tail snapshots in Fig. 3d) and thereby along the periphery of chromatin fibers, making them ideal for mediating fiber/fiber interactions during oligomerization of chromatin. In fact, the fiber/fiber interactions we observe in longer oligonucleosomes indeed are mediated through the H2A/H2B tails (Fig. 4).
Fig. 4.
Representative configurations of oligonucleosomes with 6 (a), 12 (b), 24 (c), and 48 (d) nucleosomes at 0.2 M salt. The nucleosome cores are shown as white cylinders, and the DNAs are shown as red cylindrical tubes. The histone tails are omitted for clarity.
To assess the relative impact of each histone tail in maintaining folded chromatin, we compute the sedimentation coefficients (S20,w) of 12-unit oligonucleosomes [for comparison with experiments (27)] with selectively neutralized tails at 0.01 M and 0.2 M salt concentrations by using the Kirkwood–Bloomfield formula (14, 28, 29) (Table 1). We note a large reduction in the computed S20,w from 39 S to 33 S between the regular arrays and those with neutralized H3 tails at 0.2 M salt, suggesting a drastic unfolding of arrays. Moreover, the impact of H3 neutralization is more dramatic than that attributable to neutralization of other tails. In fact, oligonucleosomes in which all tails are neutralized yield S20,w of 31 S, which is only marginally lower than that corresponding to H3 neutralized arrays. This finding suggests that the H3 tails are more important than the H4 tails for moderately folded oligonucleosomes, possibly because of their dual role in linker repulsion screening and mediating internucleosomal interactions, even though H4 tails mediate more internucleosomal interactions. This additional screening role of H3 tails uniquely posits them to prevent complete chromatin unfolding at dilute salt (0.01 M). Indeed, the neutralization effect of H2A and H2B tails, a moderate unfolding of nucleosomal arrays (Table 1), suggests certain roles in chromatin folding other than mediation of fiber/fiber interactions.
Table 1.
Sedimentation coefficients (S20,w) and tail-mediated internucleosomal (ETC) and linker/linker electrostatic energies (ELL) for 12-unit nucleosomal arrays at different salt concentrations (Cs)
| Array type | Cs, M | S20,w, S | ETC, kcal/mol | ELL, kcal/mol |
|---|---|---|---|---|
| Regular | 0.01 | 30.0 ± 0.1 | −1.71 ± 0.04 | 6.61 ± 0.05 |
| 0.2 | 39.0 ± 1.2 | −0.75 ± 0.16 | 0.72 ± 0.09 | |
| H3† | 0.01 | 25.8 ± 0.1 | −0.31 ± 0.02 | 3.50 ± 0.01 |
| 0.2 | 33.2 ± 0.4 | −0.13 ± 0.01 | 0.13 ± 0.01 | |
| H4† | 0.01 | 29.8 ± 0.1 | −0.58 ± 0.01 | 6.55 ± 0.04 |
| 0.2 | 35.6 ± 0.4 | −0.14 ± 0.02 | 0.62 ± 0.03 | |
| H2A† | 0.01 | 29.6 ± 0.1 | −1.00 ± 0.02 | 6.48 ± 0.08 |
| 0.2 | 35.0 ± 0.3 | −0.19 ± 0.03 | 0.56 ± 0.02 | |
| H2B† | 0.01 | 29.6 ± 0.1 | −0.89 ± 0.02 | 6.45 ± 0.06 |
| 0.2 | 36.1 ± 0.8 | −0.24 ± 0.08 | 0.62 ± 0.02 | |
| H2A*† | 0.01 | 30.0 ± 0.1 | −1.21 ± 0.02 | 6.63 ± 0.05 |
| 0.2 | 38.5 ± 1.0 | −0.47 ± 0.06 | 0.68 ± 0.03 | |
| All† | 0.01 | 25.8 ± 0.2 | 0.0 | 3.23 ± 0.03 |
| 0.2 | 31.2 ± 0.2 | 0.0 | 0.14 ± 0.01 | |
| Compact | 0.2 | 49.7 ± 2.3 | −2.50 ± 0.67 | 0.0 |
Tabulated energies are given “per core.” Data are presented as ±SD.
†Array segments that have been neutralized.
H2A*, C termini of H2A histones.
Links to Experiments.
The tail roles dissected above are in good agreement with experiments. Many studies have attributed regulation of moderately folded chromatin in the absence of linker histones to the H3/H4 tails (7–9). Specifically, a greater unfolding of arrays with intact H2A/H2B and trypsinized H3/H4 histones has been observed in 12-unit nucleosomal arrays than vice versa. Our argument that the H4 tails may be essential for maintaining highly compact chromatin also agrees well with recent experimental observations (10, 11, 30). For example, Dorigo et al. (10) implicate amino acids 14–19 of the H4 N terminus in mediating strong stacking-type of interactions with neighboring nucleosomes, and Shogren-Knaak et al. (11) pinpoint lysine 16 (K16) of the H4 tail as the key residue for maintaining highly compact chromatin in the presence of linker histones. This importance of K16 suggests that specific interactions (such as hydrogen bonding and steric interactions) other than electrostatic attraction may play a key role in mediating internucleosomal interactions here.
Clearly our oligonucleosome model only considers physical nonspecific effects of the histone tails but neglects the effect of such specific interactions. Nonetheless, our study correctly predicts the general tendency of H4 histone tails to electrostatically attach to the core regions and the wound DNA of neighboring nucleosomes. Given the crucial role of the H3/H4 histone tails, it is not surprising that these tails are excellent candidates for various forms of modifications constituting the so-called “histone code.” Indeed, ≈70% of all modifications, of which acetylation and methylation are the most common, occur on the H3 and H4 tails (1, 5, 31). Our prediction that the H2A/H2B tails mediate fiber/fiber interactions also agrees well with the experiments of Hansen and coworkers, who demonstrate that these tails are crucial for oligomerization of nucleosomal arrays at high salt concentrations, whereas the H3 and H4 tails are dispensable (7, 13).
Energetics.
The interplay between various energetic interactions within an oligonucleosome determines the folded state. The dominant interactions are the repulsion between linker DNA (ELL) and attraction between the tails and nonparent nucleosome cores (ETC) (Table 1). At low salt (0.01 M), strong electrostatic repulsion among linker DNAs (≈6.6 kcal/mol per nucleosome) dominates the entire electrostatic energy, triggering oligonucleosome unfolding. At high salt (0.2 M), the repulsion is reduced by an order of magnitude, and the tail/core attraction maintains oligonucleosomes in moderately folded state. Our Brownian dynamics simulations reveal that the histone tails rapidly bind to and detach from nucleosomal DNA with time scales on the order of a hundred nanoseconds (data not shown). Interestingly, the linker DNA/linker DNA repulsion of 0.72 kcal/mol per nucleosome here is balanced by the tail-mediated internucleosomal attraction of −0.75 kcal/mol per nucleosome. Considering that compact chromatin with linker histones has been measured to have an internucleosomal energy of −2.0 kcal/mol per nucleosome (32), our computed value for the internucleosomal energy without linker histones is reasonable and is considerably better than that obtained from our fixed-tail model (14) (−5.8 kcal/mol per nucleosome). The energy for compact chromatin without the linker DNA repulsion terms is −2.5 kcal/mol per nucleosome, in excellent agreement with experiments (32). That the histone tails interactions with nucleosomes are infrequent, short-lived and relatively weak on a time-averaged sense (on the order of kBT) agrees well with the view of nucleosomal arrays as being highly dynamic and locally interconverting between compact and extended states. The weak internucleosomal interactions also may be insufficient to maintain highly bent linker DNAs as in the solenoid model (33).
Internucleosomal Interaction Pattern.
Globally, our longer nucleosomal arrays display fiber-like morphologies with an irregular zigzag arrangement of nucleosomes connected by linker DNAs that crisscross the fiber axis (Fig. 4). Most linker DNAs are straight or slightly bent, consistent with the irregular zigzag model of chromatin (30, 34). The fibers have a radius of gyration (thickness) of 31 nm and packing of 0.4 nucleosomes per nm at physiological salt concentrations (0.2 M).
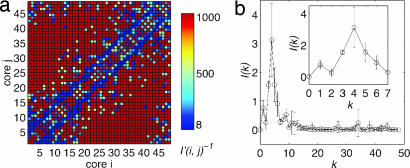
To examine interactions between nucleosomes within a chromatin fiber, we present in Fig. 5a a two-dimensional map of the intensity of histone tail-mediated interactions between nucleosomes i and j, I′(i, j), by computing the fraction of time either nucleosome's histone tail is “attached” to the other nucleosome, as defined earlier.
Fig. 5.
Internucleosomal interactions within a 48-unit oligonucleosome at 0.2 M salt. Color-coded map showing intensity of interaction between nucleosome i and j (a) and interaction intensity I(k) versus linker DNA separation k (b).
A striking pattern emerges: the strongest interactions occur along the two parallel lines offset by four nucleosomes from the diagonal, i.e., nucleosome i interacts most extensively with nucleosomes i ± 4. The same dominant pattern is evident from Fig. 5b, which shows a one-dimensional projection of map by using the transformation I(k) = Σi I′(i, i ± k), where I(k) represents interaction intensity between nucleosomes separated by k linkers. Long-range interactions among nucleosomes (away from the block diagonal) are caused by sharp bending of chromatin (e.g., see representative oligonucleosomes, Fig. 4).
This pattern of interactions likely emerges because of the mean angle of ≈92° between consecutive nucleosome triplets that we observe in the oligonucleosome configurations. This nucleosome triplet geometry is close to the natural linker DNA entry/exit angle of 90° built into our model. We can show (see Appendix 1, which is published as supporting information on the PNAS web site) that this triplet angle naturally promotes (i, i ± 4) interactions; in contrast, triplet angles of 60° promote interactions between nucleosomes i and i ± 3. Altering this balance of interactions (linker DNA repulsion vs. internucleosomal attraction), through the addition of linker histones, for example, will alter the interaction pattern markedly.
The results obtained here along with the new model and sampling approaches open avenues for studying other fascinating problems dealing with chromatin. Certainly, delineating the mechanism by which linker histones compact chromatin is still unresolved. Our analyses suggest that linker histones reduce the effective angle between the entering/exiting linker DNA and thus alter the global chromatin morphologies substantially. The details of this effect present challenges for future studies. Of course, a reasonable description of the linker histones and their precise location on the nucleosome is required; setting repulsions between linker DNAs to zero to mimic the effect of linker histone is a gross simplification. Our studies to date also suggest the presence of sharp kinks/bends within chromatin fibers, mostly mediated through H2A/H2B tails. A thorough analysis of such fiber/fiber interactions may lead to a better understanding of chromatin oligomerization and its folding into heterochromatin.
Conclusion
We have elucidated the physical role of each histone tail in chromatin folding by using our mesoscopic model of oligonucleosomes in conjunction with tailored MC sampling methods. Analysis of the positional distributions of histone tails relative to parent nucleosomes and the nature of their interaction with other chromatin components reveals: (i) the dominance of the H4 tails in mediating internucleosomal interactions, (ii) the important role of the H3 tails in screening electrostatic repulsion between entering/exiting linker DNAs and mediating internucleosomal interactions, and (iii) the tailored role of the H2A/H2B tails in mediating fiber/fiber interactions and oligomerization. These collective interactions lead to a global helical zigzag arrangement where nucleosomes interact most strongly with their fourth neighbor nucleosomes. This result immediately suggests that linker histones compact chromatin fibers further by altering this interaction pattern.
Computational Methods
Flexible-Tail Model.
Fig. 1 sketches our procedure for constructing the flexible-tail model of oligonucleosomes discussed in more detail in ref. 22. The nucleosome core minus histone tails is treated as a rigid body whose surface is uniformly spanned by 300 discrete charges. The charges are optimized by using the Discrete Surface Charge Optimization (DiSCO) algorithm to reproduce the electric field around the nucleosome core (24). Each charge is also assigned an effective excluded volume to prevent overlap of nucleosome core with the other components. The linker DNA is represented as a chain of charged beads with appropriate excluded volume, stretching, bending, and twisting terms in its forcefield (14, 18, 25). The salt-dependent charge on each bead is parameterized by using the procedure of Stigter (35). Our simulations address linker-histone deficient chromatin with 21-nm-long linker DNA represented by using a chain of six linker beads. The angle enclosed by entering/exiting linker DNAs at each nucleosome core is set to 90° (corresponding to 1.75 DNA turns).
There are 10 histone tails per nucleosome core: the N termini of H2A, H2B, H3, and H4 histones, and the C termini of H2A histones (denoted by H2A*). Each histone tail is modeled as a chain of coarse-grained beads (each bead represents five amino acid residues) with a forcefield comprising of stretching, bending, and excluded volume terms (22). The stretching and bending potentials for interbead lengths and bond angles (defined by three consecutive beads) are represented by harmonic potentials with parameters that reproduce configurational properties of the atomistic histone tails. A Lennard–Jones potential models excluded volume for each protein bead. Appropriate charges assigned to each protein bead mimic the electrostatics of the atomistic tails. Each histone chain is attached to the nucleosome core by using a stiff spring. All parameters follow ref. 22 except for σtl (tail/linker DNA excluded volume parameter), which is 2.9 nm.
MC Methods.
Five different MC moves are used to efficiently sample from the ensemble of oligonucleosome conformations under constant temperature conditions. Global pivot moves are implemented by randomly choosing one of the linker beads or nucleosome cores, picking a random axis passing through the chosen component, and then rotating the shorter part of the oligonucleosome about this axis by an angle chosen from a uniform distribution within [0, 20°]. Local translation and rotation moves also begin first by choosing a randomly oriented axis passing through randomly picked linker bead/nucleosome core. In a translation move, the chosen component is shifted along the axis by a distance sampled from a uniform distribution in the range [0, 0.6 nm], whereas in a rotation move, it is rotated about the axis by an angle uniformly sampled from the range [0, 36°]. All three MC moves are accepted/rejected based on the standard Metropolis criterion.
For efficient sampling of histone-tail conformations, we employ the configurational bias MC method (36, 37). The idea is to randomly select a histone chain and regrow it bead-by-bead by using the Rosenbluth scheme (38), beginning with the bead attached to the nucleosome core. We also employ an extension of the configurational bias approach that we have recently developed, called end-transfer configurational bias MC, to enhance the sampling of oligonucleosome conformations. The main concept of this method is to transfer randomly picked terminal portions of an oligonucleosome from one end to the other and regrow them by using a Rosenbluth scheme (38). As this method gives extremely low acceptance ratios in low-salt conditions, we employ it only at high-salt conditions. Details on the two methods are provided in Appendixes 2 and 3, which are published as supporting information on the PNAS web site. In addition, to prevent histone tail beads from penetrating the nucleosome core during tail regrowth and end-transfer moves, the volume enclosed within the nucleosomal surface is discretized, and any insertion attempts that place the tail beads within this volume are rejected automatically.
Simulation Details.
All simulations are conducted at T = 293 K and two monovalent salt concentrations: 0.01 M and 0.2 M. For 0.2 M salt simulations, the five MC moves (pivot, translation, rotation, tail regrowth, and end transfer) are attempted with frequencies 0.2:0.1:0.1:0.5:0.1, respectively. For the 0.01 M salt simulations, the first four MC moves are attempted with frequencies 0.2:0.1:0.1:0.6. Typical ensemble sizes vary from 10 to 50 million MC steps. We employ five different oligonucleosome configurations as starting conditions in our MC simulations (Fig. 6, which is published as supporting information on the PNAS web site). All five configurations yield a similar ensemble of oligonucleosome conformations as confirmed by the convergence of sedimentation coefficients obtained for 12-unit oligonucleosomes at 0.2 M salt conditions (Fig. 7, which is published as supporting information on the PNAS web site). In addition to regular oligonucleosome simulations at varying array sizes, two special types of simulations are conducted: (i) neutral tail simulations, where one or more of the histone tails is neutralized to dissect their role in chromatin compaction, and (ii) compact chromatin simulations, where the charge on the linker DNA beads has been set to zero to yield highly compact chromatin so as to assess the impact of compaction on histone tail interactions.
Inspection of representative oligonucleosomes at 0.2 M salt (Fig. 4) reveals that fiber-like morphologies representative of chromatin fibers emerge only for longer arrays (24 and 48 nucleosomes). See the convergence of frequency of internucleosomal interactions in Fig. 8, which is published as supporting information on the PNAS web site. We thus employ 24-unit oligonucleosomes for all analyses unless stated otherwise.
Supplementary Material
Acknowledgments
We thank Dr. Qing Zhang for many useful discussions. This work was supported by National Science Foundation Grant MCB-0316771, National Institutes of Health Grant R01 GM55164, and the donors of the American Chemical Society Petroleum Research (Award PRF39225-AC4 to T.S.).
Abbreviation
- MC
Monte Carlo.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Felsenfeld G, Groudine M. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Davey CA. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 3.Tse C, Sera T, Wolffe AP, Hansen JC. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wade PA, Pruss D, Wolffe AP. Trends Bio Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 5.Davie JR. In: Chromatin Structure and Dynamics: State-of-the-Art. Zlatanova J, Leuba SH, editors. Amsterdam, The Netherlands: Elsevier; 2004. pp. 205–240. [Google Scholar]
- 6.Krajewski WA, Ausio J. Biochem J. 1996;316:395–400. doi: 10.1042/bj3160395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tse C, Hansen JC. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 8.Moore SC, Ausió J. Biochem Biophys Res Commun. 1997;230:136–139. doi: 10.1006/bbrc.1996.5903. [DOI] [PubMed] [Google Scholar]
- 9.Hansen JC, Tse C, Wolffe AP. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 10.Dorigo B, Schalch T, Bystricky K, Richmond TJ. J Mol Bio. 2003;372:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 11.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Science. 2006;10:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 12.Zheng C, Lu X, Hansen JC, Hayes JJ. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 13.Gordon F, Luger K, Hansen JC. J Biol Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Zhang Q, Schlick T. Proc Natl Acad Sci USA. 2005;102:8180–8185. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katritch V, Bustamante C, Olson WK. J Mol Biol. 2000;295:29–40. doi: 10.1006/jmbi.1999.3021. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Haim E, Lesne A, Victor J-M. Phys Rev E 64. 2001:051921. doi: 10.1103/PhysRevE.64.051921. [DOI] [PubMed] [Google Scholar]
- 17.Scheissel H, Gelbart WM, Bruinsma R. Biophys J. 2001;80:1940–1956. doi: 10.1016/S0006-3495(01)76164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beard DA, Schlick T. Structure (London) 2001;9:105–114. doi: 10.1016/s0969-2126(01)00572-x. [DOI] [PubMed] [Google Scholar]
- 19.Wedemann G, Langowski J. Biophys J. 2002;82:2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mergell B, Everaers R, Schiessel H. Phys Rev E. 2004;70:011915. doi: 10.1103/PhysRevE.70.011915. [DOI] [PubMed] [Google Scholar]
- 21.Langowski J, Schiessel H. In: Chromatin Structure and Dynamics: State-of-the-Art. Zlatanova J, Leuba SH, editors. Amsterdam: Elsevier; 2004. pp. 397–420. [Google Scholar]
- 22.Arya G, Zhang Q, Schlick T. Biophys J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beard DA, Schlick T. Biopolymers. 2001;58:106–115. doi: 10.1002/1097-0282(200101)58:1<106::AID-BIP100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Beard DA, Schlick T. J Comput Chem. 2003;24:2063–2074. doi: 10.1002/jcc.10337. [DOI] [PubMed] [Google Scholar]
- 25.Allison SA, Austin R, Hogan M. J Chem Phys. 1989;90:3843–3854. [Google Scholar]
- 26.Bertin A, Leforestier A, Durand D, Livolant F. Biochemistry. 2004;43:4773–4780. doi: 10.1021/bi036210g. [DOI] [PubMed] [Google Scholar]
- 27.Hansen JC, Ausio J, Stanik VH, van Holde KE. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 28.Kirkwood JG. J Polymer Sci. 1954;12:1–14. [Google Scholar]
- 29.Bloomfield V, Dalton WO, van Holde KE. Biopolymers. 1967;5:135–148. doi: 10.1002/bip.1967.360050202. [DOI] [PubMed] [Google Scholar]
- 30.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 31.Ausió J, Abbott DW. In: Chromatin Structure and Dynamics: State-of-the-Art. Zlatanova J, Leuba SH, editors. Amsterdam: Elsevier; 2004. pp. 241–290. [Google Scholar]
- 32.Cui Y, Bustamante C. Proc Natl Acad Sci USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widom J, Klug A. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 34.Woodcock CL, Grigoryev SA, Horowitz RA, Whitaker N. Proc Natl Acad Sci USA. 1993;90:9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stigter D. Biopolymers. 1977;16:1435–1448. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- 36.Frenkel D, Mooij GCAM, Smit B. J Phys Cond Matter. 1992;4:3053–3076. [Google Scholar]
- 37.de Pablo JJ, Laso M, Suter UW. J Chem Phys. 1992;96:2395–2403. [Google Scholar]
- 38.Rosenbluth MN, Rosenbluth AW. J Chem Phys. 1955;23:356–359. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.