Abstract
Human CA150, a transcriptional activator, binds to and is co-deposited with huntingtin during Huntington's disease. The second WW domain of CA150 is a three-stranded β-sheet that folds in vitro in microseconds and forms amyloid fibers under physiological conditions. We found from exhaustive alanine scanning studies that fibrillation of this WW domain begins from its denatured conformations, and we identified a subset of residues critical for fibril formation. We used high-resolution magic-angle-spinning NMR studies on site-specific isotopically labeled fibrils to identify abundant long-range interactions between side chains. The distribution of critical residues identified by the alanine scanning and NMR spectroscopy, along with the electron microscopy data, revealed the protofilament repeat unit: a 26-residue nonnative β-hairpin. The structure we report has similarities to the hairpin formed by the Aβ(1–40) protofilament, yet also contains closely packed side-chains in a “steric zipper” arrangement found in the cross-β spine formed from small peptides from the Sup35 prion protein. Fibrillation of unrelated amyloidogenic sequences shows the common feature of zippered repeat units that act as templates for fiber elongation.
Keywords: FBP28, folding, misfolding, protein
Amyloid fibrils are formed in disease states and also in vitro by many proteins and peptides (1). There are high-resolution crystal structures for fibrillar forms of amyloidogenic peptides derived from the Sup35 prion protein (2). These peptides form self-assembling homotypic β-structure “zippers.” Solid-state NMR and mutagenesis data have been used to construct models of larger repeating units found in the Aβ(1–40) and Aβ(1–42) protofilaments implicated in Alzheimer's disease (3, 4). The repeat unit is a β-hairpin where side-chain–side-chain interactions, rather than hydrogen bonds, define the interface between strands.
Here, we have studied the amyloidogenic properties of the second WW domain of human CA150 (CA150.WW2; ref. 5), which is identical in sequence to the murine FBP28 WW domain, a small three-stranded antiparallel β-sheet protein (ref. 6; Fig. 1A) that folds in μs with apparent two-state kinetics (7, 8). CA150.WW2 forms highly ordered amyloid fibers in vitro over a period of minutes to hours under physiological conditions (37°C and pH 7.0) (9), which makes them especially amenable to structural and kinetic studies. CA150 is codeposited with huntingtin in Huntington's disease and has been proposed to be involved in regulating the onset of neurodegeneration (5, 10–12). We measured the effects of exhaustive alanine scanning on the fibrillation kinetics of a 40-residue CA150.WW2 construct (Fig. 1B) to determine the role of each side chain in fibrillation. Further, we obtained an unprecedented level of structural information by using a comprehensive isotope-labeling strategy for the fibrillar samples that were characterized by using magic angle spinning (MAS) NMR spectroscopy. We calculated a structural model based on 25 long-range NMR distance constraints, consistent with the results from independent alanine scanning and EM experiments.
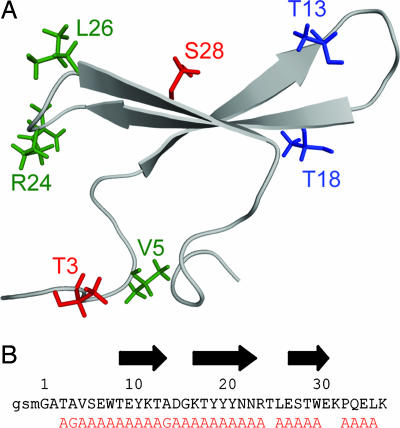
Fig. 1.
Cartoon of the structure of native CA150.WW2 [PDB ID code 1E0L (6)]. (A) Side chains that interact with each other in the fibrillar but not in the native state are marked in identical colors. (B) Sequence of CA150.WW2. The sequence is numbered according to the solution structure with the register of the native β-strands indicated by black arrows. The first three residues (gsm) originate from the expression vector and are numbered −2, −1, and 0, respectively. Flanking residues were also present in synthetic peptides used in this study. The mutations examined in this study are shown in red.
Results
Alanine Scanning of Fibrillation Kinetics.
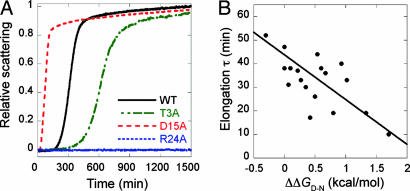
Twenty-nine residues of CA150.WW2 were mutated to Ala (Fig. 1B) and A4 and A14 to Gly (8). W8A, N22A, and P33A did not express in sufficient yield. Most mutants showed typical nucleation-like kinetics of fibrillation, as monitored by light scattering at pH 7.0 and 37°C (Fig. 2A), with characteristic lag, growth, and stationary phases, typically complete within hours. The time courses were analyzed with a model-free approach (13) (see Materials and Methods) (Table 1). Some mutants had a second slower kinetic phase that did not appear to involve further incorporation of monomers.
Fig. 2.
Fibrillation kinetics of CA150.WW2 variants. (A) Representative light-scattering transients for variants with fibrillation kinetics significantly different from wild-type CA150.WW2. The data shown for R24A are representative of mutants of abrogated fiber formation with no changes in light scattering observed, even after 6,000 min. (B) Denatured state population correlates with fibrillation rate. The apparent elongation time constant τ (where τ = 1/k, and k is the elongate rate) significantly correlated with the degree of native state destabilization reported (8). The solid line shows the best linear fit weighted by the standard errors determined from triplicate measurements.
Table 1.
Alanine scanning of CA150.WW2 fibrillation kinetics at 37°C and pH 7.0
| Construct | τ, min | τ1/2, min | τlag, min | ΔΔGD-N, kcal/mol |
|---|---|---|---|---|
| WT | 38 ± 2 | 300 ± 10 | 225 ± 10 | |
| T3A | 67 ± 2 | 620 ± 30 | 490 ± 30 | 0.09 ± 0.18 |
| A4G | 47 ± 10 | 330 ± 60 | 260 ± 60 | 0.16 ± 0.09 |
| V5A | Fiber KO | Fiber KO | Fiber KO | 0.10 ± 0.17 |
| S6A | 37 ± 1 | 320 ± 40 | 260 ± 40 | 0.22 ± 0.12 |
| E7A | 43 ± 15 | 1600 ± 200 | 1500 ± 200 | 0.52 ± 0.16 |
| W8A | ND | ND | ND | ND |
| T9A | 40 ± 10 | 340 ± 40 | 260 ± 40 | 0.93 ± 0.09 |
| E10A | 26 ± 4 | 170 ± 15 | 120 ± 15 | 0.50 ± 0.10 |
| Y11A | Fiber KO | Fiber KO | Fiber KO | 0.63 ± 0.11 |
| K12A | 95 ± 5 | 270 ± 40 | 80 ± 40 | ND |
| T13A | 19 ± 1 | 120 ± 10 | 80 ± 10 | 0.81 ± 0.17 |
| A14G | 8 ± 1 | 40 ± 5 | 25 ± 5 | 0.50 ± 0.22 |
| D15A | 17 ± 3 | 70 ± 2 | 35 ± 7 | 0.42 ± 0.09 |
| G16A | 19 ± 1 | 140 ± 10 | 100 ± 10 | 1.33 ± 0.27 |
| K17A | 30 ± 3 | 40 ± 5 | −20 ± 8 | 0.35 ± 0.08 |
| T18A | 44 ± 1 | 310 ± 40 | 220 ± 40 | 0.54 ± 0.17 |
| Y19A | 36 ± 1 | 255 ± 5 | 185 ± 5 | 0.67 ± 0.13 |
| Y20A | Fiber KO | Fiber KO | Fiber KO | ND |
| Y21A | 10 ± 2 | 30 ± 2 | 10 ± 4 | 1.7 ± 0.10 |
| N22A | ND | ND | ND | ND |
| N23A | 31 ± 3 | 260 ± 8 | 200 ± 10 | 0.07 ± 0.06 |
| R24A | Fiber KO | Fiber KO | Fiber KO | 0.78 ± 0.17 |
| L26A | Fiber KO | Fiber KO | Fiber KO | 0.56 ± 0.12 |
| E27A | 33 ± 3 | 180 ± 10 | 115 ± 10 | 1.02 ± 0.13 |
| S28A | 38 ± 1 | 230 ± 15 | 155 ± 15 | −0.01 ± 0.21 |
| T29A | Fiber KO | Fiber KO | Fiber KO | 0.33 ± 0.11 |
| W30A | Fiber KO | Fiber KO | Fiber KO | 0.76 ± 0.14 |
| P33A | ND | ND | ND | ND |
| Q34A | 52 ± 2 | 630 ± 40 | 525 ± 40 | ND |
| E35A | 33 ± 2 | 190 ± 20 | 125 ± 20 | 0.27 ± 0.10 |
| L36A | 38 ± 12 | 240 ± 80 | 165 ± 80 | 0.91 ± 0.14 |
Errors were determined from triplicate measurements and are repor ted at the 95% confidence level (2× standard error). Fiber KO, no fibers were observed within the time scale of our measurements (typically ≥ 5,000 min). ND, not determined, because these mutants either did not express in sufficient quantities or were too insoluble for subsequent characterisation. τ, t1/2, and tlag were determined from light-scattering kinetics as described in Materials and Methods. ΔΔGD-N values are those previously reported for FBP28 (8), which is identical in sequence to CA150.WW2.
V5A, Y11A, Y20A, R24A, L26A, T29A, and W30A did not give detectable changes in light scattering, positive thioflavin T assays, or any detectable fibers by EM after 5 days' incubation (Table 1 and data not shown). These variants varied widely in stability, from wild-type-like (V5A, T29A) to fully unfolded (Y20A) (8).
Many fiber-forming mutants (e.g., A4G, S6A, T9A, T18A, Y19A, N23A, E27A, S28A, E35A, and L36A) had wild-type-like time constants for fiber growth but varying lag times (Table 1). The fibrillation kinetics of some mutants deviated more markedly from wild-type behavior (Fig. 2A). For example, T13A, A14G, D15A, G16A, K17A, Y21A, and E27A had significantly faster fiber growth rates or shorter lag-times than wild type (Table 1). By contrast, T3A, E7A, K12A, and Q34A had slower fibrillation than wild type, due to their longer lag times or slower fiber growth. The time constant for fiber growth correlated significantly with the destabilization of the native state (Fig. 2B), consistent with a link between fibrillation rate and the concentration of unfolded protein, but not the rate constant for unfolding. The lag time, however, had little correlation with the change in native stability (data not shown).
Solution Backbone Dynamics.
The backbone dynamics of wild-type protein was measured at 285 K by using 15N relaxation NMR experiments (14), under low aggregation conditions. The backbone of the first six residues (GATAVS) was significantly more mobile (lower S2 values) than the backbone of the core residues, as expected for a linker region (Fig. 6, which is published as supporting information on the PNAS web site). The higher mobility of the N-terminal residues correlates with an absence of long-range interactions, consistent with the disorder of this region evident in the solution structure [PDB ID code 1E0L (6)].
Peptide Fragment Studies.
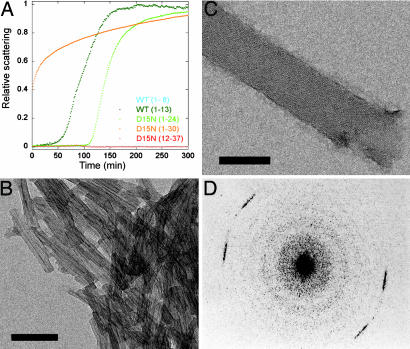
We tested synthetic peptides corresponding to CA150.WW2 fragments for fiber formation. WT[1–8], corresponding to the N-terminal region, was soluble at millimolar concentrations and did not form fibers detectable by light scattering (Fig. 3A), thioflavin T assays, or EM analysis (data not shown). In contrast, the longer peptides that contained residues from the N terminus and β-strand 1 of native protein (WT[1–13], D15N[1–24], D15N[1–30]) did form amyloid-like fibers, implying that residues in the N terminus and first β-strand are necessary and sufficient to form β-sheet aggregates under these conditions. Consistent with this hypothesis, the peptide D15N[12–37], lacking the first 11 residues of CA150.WW2, did not form fibers.
Fig. 3.
Properties of CA150.WW2 aggregates formed under physiological conditions. (A) Light-scattering kinetics for CA150.WW2 peptide fragments. Peptides WT[1–8] and D15N[12–37] gave no appreciable scattering at 37°C, even after 1,600 min. In contrast, WT[1–13], D15N[1–24], and D15N[1–30] all had strong changes in light scattering, consistent with aggregation. The aggregates tested positive for amyloid fibers in thioflavin T assays (data not shown) and by EM. There was no lag phase in the aggregation kinetics for D15N[1–30], which had very low solubility. Residue 15 was changed from Asp to Asn in D15N[1–24], D15N[1–30], and D15N[12–37], because it was exceptionally difficult to synthesize these peptides with the wild-type residue. This mutation did not affect the amyloid-forming tendencies of full-length FBP28 peptides (unpublished data). (B) Negatively stained electron micrograph of the microcrystals formed by WT[1–13] peptide fragments. (C) Low-dose uranyl acetate electron micrograph of a tubular association of amyloid fibrils formed by the Y19F mutant of CA150.WW2. The frayed end of one of these tubes, comprised of ≈100 protofilaments, is evident in the lower right-hand corner of the image, as are the upper and lower face of a tube. (D) Optical diffraction pattern of a Y19F amyloid tube oriented as shown in C. The width of individual protofilaments was determined from this to be 30 Å, with a cross-β spacing of 4.73 Å. (Scale bars, 100 nm.)
The aggregates formed by the shortened fragments (Fig. 3B) had, on examination by EM, a well defined ultrastructure that consisted of clusters of homogenous crystalline blocks. The gross morphology and fibrillar properties of the short peptide fragments of CA150.WW2 were quite similar to those formed in vitro by fragments of other proteins (2, 15, 16, 17–19) and amino acid homopolymers (20). In contrast, the fibrillar deposits formed by full-length CA150.WW2 were significantly larger than those formed by the shorter peptide fragments, had more complex ultrastructures, and the individual protofilaments (30 Å width with a cross-β spacing of 4.73 Å) were clearly visible [Fig. 3 C and D and previously reported data (9)].
Solid-State NMR and Structure Calculations.
Five different isotopically labeled samples of the Y19F mutant of CA150.WW2 were prepared for solid-state NMR spectroscopy by using the scheme of Castellani et al. (21): uniformly labeled with 13C15N (u-CA150.WW2); uniformly 15N-labeled with 13C labeling using either 2-13C-labeled glycerol or 1,3-13C-labeled glycerol (2- CA150.WW2 and 1,3-CA150.WW2, respectively); and uniformly labeled with 2H,13C,15N, except for the labile deuterons that were exchanged with 1H (d-CA150.WW2). The Y19F mutant was used, because it reproducibly formed highly homogeneous fibers (Fig. 3C), with very similar cross-β spacing to wild-type CA150.WW2 amyloid fibers.
MAS NMR spectra of CA150.WW2 Y19F showed signals of intermediate line width for residues M0–E35. In most spectra, the signals of the first and last two residues of our construct (G-2, S-1, L36, and K37) were absent. Two sets of signals with similar intensity were observed for S28 and at least two of the tyrosines, suggesting that these residues have multiple conformations. The Cα, Cβ, and most side-chain carbon resonances of residues G1, A2, T3, A4, V5, S6, T9, E10, K12, T13, A14, D15, G16, K17, T18, F19, R24, T25, L26, S28, T29, K32, P33, and Q34 were unambiguously assigned. However, we could not unambiguously assign the signals for W8, W30, N22, N23, E7, E27, E31, and E35 because of spectral overlap.
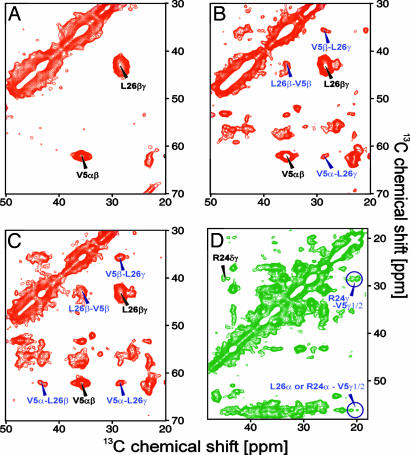
The cross-peaks linking residues V5 and L26 were the predominant features in the spectra of 2-CA150.WW2 (Fig. 4A–C). The correlations V5α-L26γ, V5β-L26β, and V5β-L26γ appeared at a proton-driven spin-diffusion (PDSD) mixing time of 200 ms. A correlation between V5α and L26β was not observed at a mixing time of 200 ms but became evident at a mixing time of 400 ms. This demonstrates that V5α is further from L26β than L26γ, which suggests that these side chains are arranged in a parallel head-to-tail fashion. The correlations V5γ1/γ2-R24γ observed in the spectra of 1,3-CA150.WW2 supported further that the side chain of V5 was situated between the side chains of L26 and R24 in a zipper-like arrangement. Similar interactions were observed between T3 and S28. A complete set of cross-peaks, involving all aliphatic carbon signals, defined the interaction of T13 and T18. These data suggested the presence of a β-hairpin defined by interdigitating side chains, separated by a turn involving A14, D15, G16, and K17. This agrees well with TALOS predictions of the φ and ψ angles using chemical-shift values (22), consistent with two regions of β-structure (residues T3-T13 and T18-S28) connected by a loop between A14 and K17. Residues A14–G16 also formed a flexible dynamic loop in native protein (Fig. 6). However, the interactions between V5-R24, V5-L26, T3-S28, and T13-T18 found in the fibrillar form cannot exist in the native structure of CA150.WW2 (Fig. 1).
Fig. 4.
Solid-state NMR spectra of CA150.WW2 Y19F amyloid fibers recorded at 900 MHz, 10.5 kHz MAS, and 284 K. Side-chain correlations within the same amino acid are annotated in black, and cross-peaks due to interactions between side chains of different amino acids are labeled in blue. (A) 2-CA150.WW2, PDSD, 20 ms mixing. (B) 2-CA150.WW2, PDSD, 200 ms mixing. (C) 2-CA150.WW2, PDSD, 400 ms mixing. (D) 1,3-CA150.WW2, PDSD, 200 ms mixing. The appearance of extra cross-peaks in A–C with longer mixing times identifies the interactions between atoms, which are more distant than those observed using shorter mixing times.
We extracted 25 long-range distance constraints from the MAS NMR spectra and calculated a structure of six repeat units of the protofilament (of which the inner four are shown in Fig. 5A), by using hydrogen-bond constraints in the manner of Riek and coworkers (3). Simulated annealing gave a β-hairpin structure with tight side-chain packing stapling the ends together (Fig. 5 B and C). These residues form a heterotypic steric zipper (2). The hairpin becomes somewhat expanded between residues T9-K12 and Y20-N22, because the Y11 and Y20 side chains are longer than the others in the zipper. In this region, the different structure calculations show variations in the hydrogen bonding and cross-hairpin interactions. Calculations including two ambiguous constraints assigned to an interaction between T9 and Y20 did not change the overall structure in this region, especially the packing density. The smaller spacing of the backbone is reestablished before the turn region due to the tight packing of residues T13 and T18, as found in β-arch structures (23). In summary, the protofilament structure of CA150.WW2 is a β-hairpin with variable separation of the opposing backbone fragments, including both tight (hydrophobic) and loose (hydrophilic) zipper-like subsections. Residues 13–16 form a disordered loop in both the native and fibrillar form, and the structural promiscuity of this region allows native and nonnative β-hairpins to be formed by competing reactions.
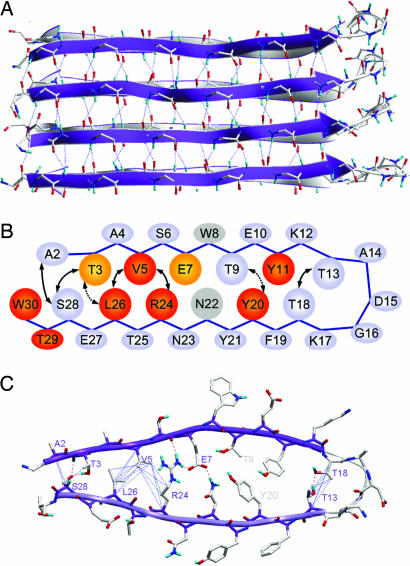
Fig. 5.
Structural model of the CA150.WW2 protofilament. (A) View of the long axis of the protofilament. The strands of the parallel β-sheet in the background (gray) were formed by residues 20–28, with residues 3–11 forming the β-sheet in the foreground (violet). Residues N-terminal to A2 and C-terminal to T29 are not shown, because they had no detectable regular structure. Each hairpin is linked to others by backbone hydrogen bonds used as constraints (dashed lines) and buried side-chain interactions (not shown). (B) Cartoon representation of the nonnative β-hairpin. The long-range interactions detected by MAS NMR and used for the structure calculations are shown by black arrows and define the interface between the β-strands. Dotted arrows indicate ambiguous distance constraints that could be fitted into the structure after the calculation. Residues that eliminated or significantly decelerated fibrillation when mutated to Ala are marked in red and orange, respectively (Table 1). We could not determine the effects of mutating W8 and N22 (light gray), because the corresponding Ala variants could not be expressed in sufficient quantities for characterization. (C) One repeat unit, viewed down the fiber axis, with colors encoding atom type. It is a hairpin formed by two β-strands of nonnative register linked by a flexible loop region between residues T13 and T18. The interface between strands is well packed and includes a salt bridge between E7 and R24. This interaction was not an intrinsic restraint in the model but a consequence of the periodicity dictated by the V5-R24, V5-L26, T13-T18, and T3-S28 interactions identified using solid-state MAS NMR. The width of the ordered region of the hairpin (T3-T13 = 31 Å) is consistent with the dimensions determined using EM (Fig. 3). Blue dotted lines represent the long-range interactions identified by MAS NMR, which were used as restraints in structure calculations; hydrogen bonds are indicated by dashed red lines.
Discussion
Folding vs. Fibrillation.
Kinetic, mutagenic, and structural data conspired to show that the unfolded state of CA150.WW2 either folds productively or forms fibrils with a completely different set of internal interactions. There was a strong correlation between the fibrillation rate and increased occupancy of the unfolded state (Fig. 2B). Wild-type CA150.WW2 is only marginally stable, even under native conditions, and an appreciable population of unfolded protein always exists [ΔGD-N of wild-type CA150.WW2 is ≈2.4 kcal/mol at 283 K (7, 8), so that ≈1.5% is unfolded]. There is always a pool of rapidly formed and relatively unstructured molecules that can form fibrils. This situation is similar to that for disease-causing amyloidogenic proteins, such as Aβ peptides, amylin, α-synuclein, and prion proteins, where fibrillation initiates from unstructured or natively unfolded conformations (reviewed in ref. 24).
The rate-limiting step in the productive folding of CA150.WW2 involves formation of a hairpin between β-strands 1 and 2 (residues 9–13 and 17–23, respectively) (8, 25), a mechanism found in other WW domain homologues (26). Further, the N-terminal residues (G1-S6) are dynamic in the native state (Fig. 6), make little or no long-range interactions (6), and contribute little to the folding and stability (8). By contrast, mutation of T3, V5, and E7, located at the N-terminal region, inhibited fibrillation. Solid-state NMR measurements also revealed long-range contacts from V5 to R24 and L26 and between T3 and S28, as well as between T13 and T18 (Fig. 4). None of these interactions are present, or indeed possible, in the correctly folded native structure. Partitioning of residues important to protein folding and aggregation also happens in human acylphosphatase (18).
Molecular Model of the CA150.WW2 Protofilament Structure.
Seven alanine mutations eliminated (V5A, Y11A, Y20A, R24A, L26A, T29A, and W30A) and three significantly decelerated (T3A, E7A, and K12A) fiber growth during our experiments (Fig. 1 and Table 1). These key residues were located between residues 3–11 and 20–30, regions that are connected by a relatively unstructured dynamic loop in the native protein (Fig. 6). The interactions between T13 and T18, identified by MAS NMR, define a tight nonnative turn in the protofilament, consistent with a β-hairpin conformation, as proposed for Aβ protofilaments (3, 4). A network of long-range nonnative interactions was present in the CA150.WW2 fibers, between V5-R24, V5-L26, and T3-S28, and this was identified by MAS NMR measurements. These side chains appear to interdigitate tightly into a steric zipper, as found for short peptide fragments of the Sup35 prion protein (2).
Twenty-five long-range distance constraints were used to generate a structural model of the CA150.WW2 protofilament. The protofilament contains vertically stacked intramolecular, nonnative β-hairpins (Fig. 5A) hydrogen-bonded together in parallel. The dimensions of the protofilament structure and cross-β spacing matched those independently determined by EM. Intriguingly, most of the residues that inhibited fibrillation when mutated to alanine mapped to those in the buried core of the β-hairpin (Fig. 5 B and C), providing a structural rationale for the observed mutational effects. Our model contained the stacked aromatic side chains and asparagine ladders proposed to be a general property of amyloid fibers (27), although these were not used as constraint features in the model building.
Our model has a buried electrostatic interaction between R24 and E7 (Fig. 5C), similar to the buried electrostatic interactions proposed for the Aβ protofilament (4). As expected, mutation of E7 or R24 seriously perturbed fiber formation (Table 1 and Fig. 2), because a stabilizing interaction would be lost and an unsatisfied charge introduced into the core of the hairpin. Interestingly, mutation of residues that form the flexible turn region in native CA150.WW2 (Fig. 6) significantly increased the pool of unfolded molecules and so accelerated fibrillation. Further experiments are required to understand the roles of T29 and W30 in fibrillation.
Recurrent Structural Motifs in Protofilaments.
The crystal structure of the cross-β spine formed by heptapeptides from Sup35, a yeast prion protein, contains intermolecular β-sheets stabilized by a “steric zipper” formed by the interdigitations of side chains from peptides rotated by a two-fold screw axis (2). Each strand of the zipper is perpendicular to the fiber axis and hydrogen-bonded to those above and below, giving rise to parallel β-sheets with the hydrogen-bonding pattern characteristic of amyloid fibers. The zipper is said to be homotypic where a given region of sequence binds to the same region in another molecule.
Tycko and coworkers (4) used restraints from MAS NMR, fiber diffraction, and EM experiments to generate a structural model of the protofilament formed by the human Aβ(1–40) peptide that contains multiple copies of an intramolecular β-hairpin repeat unit (4). Each β-hairpin has interdigitating hydrophobic side chains from residues 12–24 and 30–40 (i.e., a heterotypic interface), with one buried salt bridge between D23 and K28. As with the cross-β spine of Sup35, each β-strand of the Aβ hairpin forms a continuous parallel β-sheet. Riek and coworkers (3) refined the β-hairpin model for the Aβ(1–42) protofilament using additional (non-NMR) experimental constraints (3). Their protofilament model contains an intermolecular β-hairpin repeat unit that has somewhat different side-chain packing from the earlier model (4).
The structural model of the CA150.WW2 protofilament has similarities to those reported for aggregates of the Aβ(1–40) (4) and Aβ(1–42) peptides (3). All of the models contain many copies of extended β-hairpins arranged into vertically stacked hydrogen-bonded arrays, and there is evidence of buried electrostatic interactions in the “core.” Interestingly, the interactions found in the fibrillar β-hairpins formed by CA150.WW2 and Aβ peptides closely resemble those found in the β-arches of β-solenoid proteins (23). Amyloid fibers, therefore, appear to be further examples of structural interactions already exploited during the evolution of proteins (28).
We found unequivocal evidence for a steric zipper (2) that is heterotypic and staples together the ends of the nonnative β-hairpin by long-range interactions (Fig. 5C). Short fragments of CA150.WW2 (i.e., WT[1–13]), like those of Sup35 (2), also form exceptionally well ordered aggregates, but the repeating unit must be different from that of the full-length domain, because they cannot form the same sequence-distant interactions. Indeed, the fibrils formed from the WW short peptides have a radically different morphology. It seems likely that protofilaments may be stabilized by local and long-range interactions depending on sequence composition, length, and environmental conditions, but that heterotypic or homotypic zippers will be a recurring feature of these aggregates.
Materials and Methods
Reagents.
All reagents were AnalR grade, except thioflavin T (65% dye content), and were purchased from Sigma (St. Louis, MO). Isotopically labeled proteins were expressed in modified K-Mops minimal medium (29), with 0.1% (wt/vol) [15N] ammonium chloride and 0.4% (wt/vol) appropriately labeled carbon sources (CIL, Andover, MA). Peptides were synthesized by using standard fluorenylmethoxycarbonyl chemistry. Protein purity (≥95%) and mass were verified by analytical gel filtration, reverse-phase chromatography, and mass spectrometry. Protein concentrations were determined from molar extinction coefficients (30).
Measurements of Fibrillation Kinetics.
Changes in turbidity were measured with a Varian Cary 500 Scan Spectrometer (Varian, Cary, NC) (9). Lyophilized proteins were diluted in ice-cold 10 mM sodium phosphate, pH 7.0, with sodium azide 0.02% (v/v) to inhibit bacterial growth and stored on ice between manipulations. Before use, each sample was fully dissolved by using two 10-s pulses in a sonicating water bath and filtered through a 0.22-μm disposable filter. Experiments were performed by using disposable plastic cuvettes to minimize cross-seeding. Peptides were added to cuvettes preequilibrated to 37°C to give a final concentration of 100 μM. Changes in light scattering at 350 and 800 nm were recorded every 90 sec for at least 5,000 min. Protein solutions were then were used in thioflavin T assays, as described (9). Light-scattering kinetics were fitted to give an apparent half-time (t1/2) and elongation time constant (τ) for fibrillation (13). The lag time (tlag) was calculated from tlag = t1/2 − 2τ.
Electron Microscopy.
Negatively stained samples were made from the stock protein solutions at 0.1-mM concentration. A 4-μl drop was applied to the surface of carbon-coated grids and after 2 min, the grid surface was flushed with successive drops of 2% (wt/vol) aqueous uranyl acetate solution, blotted, and allowed to dry. Images of frozen samples (9) were acquired on Kodak SO163 and Teitz F415 (TVIPS, Gauting, Germany) 4k × 4k CCD cameras on an FEI (Eindhoven, The Netherlands) G2 F20 field emission gun electron microscope at 200 keV. A 626 liquid nitrogen cold stage (Gatan, Pleasanton, CA) held the cryosamples in the microscope at −180°C. All micrographs were recorded under low-dose conditions at a microscope magnification of ×50,000.
Solution NMR.
Backbone relaxation data were acquired for wild-type CA150.WW2 at 285 K in 20 mM potassium phosphate, pH 6.5/30 mM NaCl. The 15N/1H/HSQC spectrum was assigned from published data (Biomagresbank entry 47146) and a 15N/1H 1H TOCSY spectrum. 15N R1, R2, and heteronuclear NOE relaxation data were acquired on a Bruker (Billerica, MA) DRX 500 by using standard pulse sequences (14). Model-free analysis (31) was performed in the program Tensor2 (32). The errors on relaxation rates and I/I0 (heteronuclear NOE) were conservatively estimated to be ≤5% of the average values determined for the structured region of the protein. The isotropic rotational correlation time was estimated as 4.1 ns using R2/R1 values from residues with I/I0 > 0.55 and R2/R1 within 1.5 standard deviations of the mean for the structured part of the molecule. The data were mostly fitted to the simplest models of internal motion [model 1 or 2 (32)]; the S2 values reflect trends clearly visible in the raw R1 and I/I0 data.
Solid-State NMR.
Samples contained ≈15 mg of amyloid fibers formed by the Y19F mutant of CA150.WW2 that reproducibly formed homogeneous amyloid fibers with better spectral dispersion than the wild type. Solid-state MAS triple-resonance NMR spectra were acquired at 284 K with a spinning frequency of 10.5 kHz. A proton radiofrequency field of 80 kHz with the two-pulse phase modulation scheme (33) was used for decoupling in the direct and indirect dimensions. 13C-13C correlations were recorded at several mixing times by using PDSD (34), dipolar-assisted rotational resonance (35), and radiofrequency-driven recoupling (36) for mixing. For 13C-15N spectra, band-selective magnetization transfer from 15N to 13C was performed by using SPECIFIC-CP (37) or adiabatic-passage Hartmann–Hahn cross-polarization (38).
Model Building.
A model of a fibril was built comprising six monomeric units. Hydrogen bonds were introduced between the units to connect the β-strands A2-E12 in a parallel fashion, as well as the strands F19-S28. We used a repeat distance of 4.7 Å between units. The initial model was consistent with results from alanine scanning of fibrillation kinetics and MAS NMR data. Three different distance constraints classes were defined before the calculations (2.5–5.0, 2.5–6.5, and 2.5–7.5 Å), which were assigned to each class depending upon the mixing time at which a cross-peak first appeared. The H-bonds between the β-strands, 27 long-range, and 10 sequential MAS NMR constraints were used in a simulated annealing run (30 cycles of heating for 2,000 fs up to 1,000 K, cooling in 10,000 fs down to 0 K) applying a full force-field AMBER 7.0. The lowest-energy conformation was embedded in water and subjected to a short molecular dynamics simulation for 1 ns to allow relaxation of the structure. The rms deviation of the backbone atoms for the A2-S28 region of the inner four monomers from the five lowest-energy structures was 1.23 Å. No distance constraint was violated.
Supplementary Material
Acknowledgments
We thank Dr. M. D. Allen for assistance in model building. This work was funded by the Medical Research Council (A.R.F.) and by the Deutsche Forschungsgemeinschaft Researcher Group 475 (H.O.) H.T. was supported by a fellowship from Boehringer Ingelheim Fonds.
Abbreviations
- CA150.WW2
second WW domain of human CA150
- MAS
magic angle spinning
- PDSD
proton-driven spin-diffusion.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jahn TR, Radford SE. FEBS J. 2005;272:5962–5970. doi: 10.1111/j.1742-4658.2005.05021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holbert S, Denghien I, Kiechle T, Rosenblatt A, Wellington C, Hayden MR, Margolis RL, Ross CA, Dausset J, Ferrante RJ, et al. Proc Natl Acad Sci USA. 2001;98:1811–1816. doi: 10.1073/pnas.041566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macias MJ, Gervais V, Civera C, Oschkinat H. Nat Struct Biol. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson N, Johnson CM, Macias M, Oschkinat H, Fersht AR. Proc Natl Acad Sci USA. 2001;98:13002–13007. doi: 10.1073/pnas.221467198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrovich M, J. A.L., Ferguson N, Daggett V, Fersht ARF. J Mol Biol. 2006;360:865–881. doi: 10.1016/j.jmb.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson N, Berriman J, Petrovich M, Sharpe TD, Finch JT, Fersht AR. Proc Natl Acad Sci USA. 2003;100:9814–9819. doi: 10.1073/pnas.1333907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holbert S, Dedeoglu A, Humbert S, Saudlou F, Ferrante RJ, Neri C. Proc Natl Acad Sci USA. 2003;100:2712–2717. doi: 10.1073/pnas.0437967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arango M, Holbert S, Zala D, Brouillet E, Pearson J, Regulier E, Thakur AK, Aebischer P, Wetzel R, Deglon N, Neri C. J Neurosci. 2006;26:4649–4659. doi: 10.1523/JNEUROSCI.5409-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arango M, Holbert S, Zala D, Brouillet E, Regulier E, Thakur AK, Aebischer P, Wetzel R, Deglon N, Neri C. J Neurol Neurosurg Psychiatry. 2005;76(Suppl 4) [Google Scholar]
- 13.Pedersen JS, Christensen G, Otzen DE. J Mol Biol. 2004;341:575–588. doi: 10.1016/j.jmb.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 15.Ventura S, Zurdo J, Narayanan S, Parreno M, Mangues R, Reif B, Chiti F, Giannoni E, Dobson CM, Aviles FX, Serrano L. Proc Natl Acad Sci USA. 2004;101:7258–7263. doi: 10.1073/pnas.0308249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Paz ML, Goldie K, Zurdo J, Lacroix E, Dobson CM, Hoenger A, Serrano L. Proc Natl Acad Sci USA. 2002;99:16052–16057. doi: 10.1073/pnas.252340199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Proc Natl Acad Sci USA. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiti F, Taddei N, Baroni F, Capanni C, Stefani M, Ramponi G, Dobson CM. Nat Struct Biol. 2002;9:137–143. doi: 10.1038/nsb752. [DOI] [PubMed] [Google Scholar]
- 19.Sikorski P, Atkins EDT, Serpell LC. Structure (London) 2003;11:915–926. doi: 10.1016/s0969-2126(03)00149-7. [DOI] [PubMed] [Google Scholar]
- 20.Fandrich M, Dobson CM. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 22.Cornilescu G, Delaglio F, Bax J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 23.Hennetin J, Jullian B, Steven AC, Kajava AV. J Mol Biol. 2006;358:1094–1105. doi: 10.1016/j.jmb.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 24.Uversky VN, Fink AL. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson N, Pires JR, Toepert F, Johnson CM, Pan YP, Daggett V, Oschkinat H, Fersht AR. Proc Natl Acad Sci USA. 2001;98:13008–13013. doi: 10.1073/pnas.221467398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jager M, Nguyen H, Crane JC, Kelly JW, Gruebele M. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 27.Wetzel R. Structure (London) 2002;10:1031–1036. doi: 10.1016/s0969-2126(02)00809-2. [DOI] [PubMed] [Google Scholar]
- 28.Kobe B, Kajava AV. Trends Biochem Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson N, Sharpe TD, Schartau PJ, Sato S, Allen MD, Johnson CM, Rutherford TJ, Fersht AR. J Mol Biol. 2005;353:427–446. doi: 10.1016/j.jmb.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 30.Gill SC, von Hippell PH. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 31.Lipari G, Szabo AJ. J Am Chem Soc. 1982;104:4546–4558. [Google Scholar]
- 32.Dosset P, Hus JC, Blackledge M, Marion D. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 33.Bennett AE, Rienstra CM, Auger M, Lakshami KV, Griffin RG. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 34.Szeverenyi NM, Sullivan MJ, Maciel GE. J Magn Reson. 1982;47:462–475. [Google Scholar]
- 35.Takegoshi K, Nakamura S, Terao T. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 36.Bennett AE, Ok JH, Griffin RG, Vega S. J Chem Phys. 1992;96:8624–8627. [Google Scholar]
- 37.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Mol Phys. 1998;95:1197–1207. [Google Scholar]
- 38.Baldus M, Geurts DG, Hediger S, Meier BH. J Magn Reson. 1996;118:140–144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.