Abstract
The CTLA4 gene is important for T lymphocyte-mediated immunoregulation and has been associated with several autoimmune diseases, in particular, type 1 diabetes. To model the impact of natural genetic variants of CTLA4, we constructed RNA interference (RNAi) “knockdown” mice through lentiviral transgenesis. Variegation of expression was observed in founders but proved surmountable because it reflected parental imprinting, with derepression by transmission from male lentigenics. Unlike the indiscriminate multiorgan autoimmune phenotype of the corresponding knockout mice, Ctla4 knockdown animals had a disease primarily focused on the pancreas, with rapid progression to diabetes. As with the human disease, the knockdown phenotype was tempered by genetic-modifier loci. RNAi should be more pertinent than gene ablation in modeling disease pathogenesis linked to a gene-dosage variation.
Keywords: autoimmune diabetes, imprinting, lentiviral transgene, RNAi, variegation
Common human diseases are thought to be determined by complex genetic variations. Advances in genetics and genomics, such as whole-genome scans of single-nucleotide polymorphisms (SNP) and genome-wide expression profiling, have led to the identification of many potential disease-susceptibility alleles, but definitively establishing that a candidate gene is truly causal remains a challenge. Often, the genetic linkage of an allele to a disease condition is manifested as a modest change in mRNA and/or protein expression. For instance, many studies have linked the CTLA4 locus to human autoimmune disorders, including type 1 diabetes (1), a disease caused by T lymphocyte-mediated destruction of pancreatic-islet β cells (2). No variation in the regions coding for mature CTLA4 protein has been implicated, but several polymorphisms in the promoter or 3′ untranslated region (UTR) have been shown to affect promoter efficacy (3–6) and/or the ratio of alternatively spliced forms (7), thus reducing the level of functional CTLA4 protein. It is these polymorphisms that most closely associate with autoimmune risk (5, 7).
The Ctla4 gene also has been linked to disease susceptibility in the nonobese diabetic (NOD) mouse model of type 1 diabetes. Some reports suggested a defect in overall expression or in an alternative splice form (liCtla4) unable to bind B7.1/B7.2, the conventional ligands of Ctla4 (7–10). However, for both humans and mice, definitive assignment of susceptibility to an individual SNP (or even to Ctla4 proper, as opposed to combinatorial haplotypes involving Ctla4 and the immediately adjacent genes encoding members of the costimulatory family) is still lacking.
Consistent with its function as an inhibitory receptor (11), recombinational inactivation of Ctla4 results in a devastating lymphoproliferative and autoimmune phenotype, with inflammatory infiltration of multiple organs and death of the homozygous knockout mice within a few weeks (12–14). However, such a complete abrogation of gene function is a gross overrepresentation of most natural variation. In this study, we generated “knockdown” mice with a partial reduction in Ctla4 expression, employing RNAi derived from short hairpin RNAs (shRNA), delivered into the mouse germ line by lentiviral transgenesis (15, 16). RNAi has been harnessed extensively, by using shRNA precursors of interfering RNAs, to manipulate gene expression in vitro (17, 18), and it seemed most appropriate to mimic the variation of expression levels associated with the CTLA4 polymorphisms.
Results
Lentiviral Vector and shRNAs to Target Ctla4.
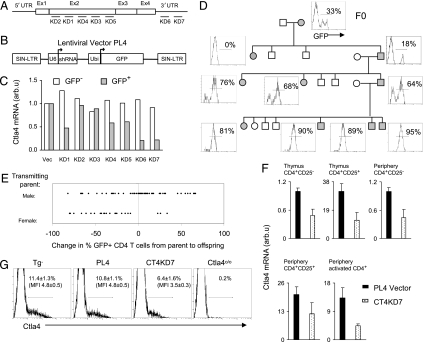
A series of shRNA sequences was designed to target different regions of Ctla4 mRNA (Fig. 1A); each sequence was cloned into PL4, a derivative of the PLL3.7 lentivirus vector (16), which drives shRNA expression under a ubiquitously expressed U6 promoter and carries a green fluorescence protein (GFP) reporter gene (Fig. 1B). The efficacy of the shRNA was tested by transducing T lymphocytes activated in vitro, and the level of Ctla4 transcripts was quantified by PCR. Several shRNAs proved efficacious, the best of which, knockdown (KD)6 and KD7, mapped to the 3′ UTR, reducing the level of Ctla4 mRNA ≈4- to 5-fold (Fig. 1 A and C).
Fig. 1.
Generation of Ctla4 knockdown mice. (A) Diagram of the full-length Ctla4 mRNA and regions targeted by shRNA probes KD1–7. (B) The lentiviral vector used to express shRNA. Ubi, ubiquitin. (C) In vitro testing of the efficacy of the shRNA in silencing Ctla4 expression in activated T lymphocytes. Data are representative of two experiments. (D) Transmission of the Ctla4 KD7 (CT4KD7) lentigene and expression of the GFP reporter through generations. Filled and open symbols indicate, respectively, presence and absence of the lentigene. Histogram shows GFP expression by CD4+ peripheral blood cells. (E) Expression of the lentigene inherited from the male versus female parent. Data are pooled from two PL4 vector and two CT4KD7 lentigenic lines. Each dot represents one animal. (F) Quantitative RT-PCR assessment of Ctla4 mRNA in T cells from the thymus or periphery (spleen and lymph nodes) (mean ± SD). (G) Quantification of Ctla4 protein by intracellular flow cytometric staining of CD4+CD25+ lymph nodes cells ex vivo. Percentage of Ctla4+ population and mean fluorescence intensity (MFI) are indicated (mean ± SD).
Variegation and Imprinting of Lentiviral Transgene (Lentigene) Expression.
The KD7 construct was packaged into high-titer viruses, which were used to infect mouse zygotes by microinjection into the perivitelline space (15). Unlike homologous recombination in ES cells, lentigenesis can be performed on any genetic background, so we chose to microinject NOD zygotes. The microinjections were highly successful for KD7, because six of seven offspring were positive for integration of viral DNA when tested by PCR. These founders were examined for expression of the GFP reporter in blood lymphocytes, revealing a variegated pattern of expression, ranging from 0% to 36% of CD4+ T cells (Fig. 5, which is published as supporting information on the PNAS web site). Two KD7 founders (hereafter referred to as CT4KD7 for cytotoxic T lymphocyte antigen 4 KD7), as well as founders carrying a control lentigene devoid of shRNA (the PL4 vector), were bred to test for transmission of the lentigene. Transmission of the CT4KD7 lentigene from both founders was largely Mendelian, as illustrated for the CT4KD7–26 line (Fig. 1D), indicating that the founders carried only one copy of the lentigene. On the other hand, the founders bearing the PL4 vector exhibited multiple-copy integration (data not shown). Variegation of reporter expression improved significantly through breeding, essentially all cells scoring positive after one to three generations (Fig. 1D; Table 1).
Table 1.
Variegated expression of lentigenes transmitted by females (F) versus males (M)
| Lines | Generation | Parent sex and % GFP+* |
Offspring % GFP+* |
|---|---|---|---|
| KD7–26 | F0 | 33 | |
| F1 | F, 33 | 0, 18; 0 | |
| F2 | M, 18 | 64, 68, 73, 76 | |
| F3 | F, 76 | 10, 1, 36, 33, 15 | |
| M, 64 | 90, 89, 81, 95; 96, 96, 93 | ||
| F4 | F, 81 | 30, 72, 42, 20 | |
| M, 90 | 99, 95 | ||
| KD7–20 | F0 | 31 | |
| F1 | M, 31 | 97, 1, 2, 96, 97, 98; 0, 96 | |
| F2 | M, 98 | 94, 96 | |
| M, 96 | 91, 91, 92; 97, 96 | ||
| F3 | M, ND | 96, 98, 97, 100 |
Litters by the same parent were separated with a semicolon. ND, not determined.
*GFP marker expression was monitored in each generation of lentigenic mice by flow-cytometric analysis of peripheral blood mononuclear cells. The numbers are percentages of GFP+ cells in the CD4+ subset of individual animals. Patterns also reflect CD8+ cells.
Interestingly, the improvement in variegation varied with the gender of the transmitting parent. Offspring of variegated males showed higher frequencies, and those from nonvariegated males remained uniformly GFP-positive. On the other hand, offspring of female lentigenics exhibited reduced expression in virtually all cases. This control of variegation through parent-specific imprinting was observed in several independent lines, whether encoding shRNA or not (Fig. 1E; Table 1) and suggests that germ-line methylation and demethylation is responsible from the stochastic silencing of the lentigenes.
Ctla4 Knockdown in the Lentigenic Mice.
Two lines of CT4KD7 lentigenics (CT4KD7–20 and CT4KD7–26) and two lines of PL4 controls were established. None of them showed any gross anatomical or developmental abnormalities. A previous study reported that expression of some species of shRNAs by lentiviral infection or plasmid transfection into cultured cells could trigger a potent type 1 IFN response, as evidenced by massive induction of 2′5′-oligoadenylate synthetase (2′5′-OAS) (19). We did not detect any induction of 2′5′-OAS in the CT4KD7 lentigenic mice (Fig. 6, which is published as supporting information on the PNAS web site). We then analyzed Ctla4 gene expression in T cells sorted from the thymus and peripheral lymphoid organs of CT4KD7 knockdown and PL4 control lentigenic mice, ex vivo or after 72 h of in vitro anti-CD3/28 stimulation to induce Ctla4 transcription. The CT4KD7 lentigene reduced Ctla4 mRNA levels between 2- and 4-fold (Fig. 1F). The relative reduction observed in conventional CD4+CD25− cells was the same as in CD4+CD25+“regulatory” T (Treg) cells, which express 20-fold more Ctla4 mRNA. Incidentally, we did not detect any reduction in the frequency of this Treg cell subset in CT4KD7 mice (Fig. 7, which is published as supporting information on the PNAS web site). A similar reduction in Ctla4 transcripts was detected in CD3/28-stimulated CD4+ T cells. The KD7 shRNA targets the 3′ UTR region of Ctla4 mRNA (Fig. 1A), a segment shared by both the full-length and short, liCtla4, splice variants; we observed a similar level of knockdown for these two isoforms (data not shown). Consistent with the reduced mRNA, Ctla4 protein also was diminished 2- to 4-fold in lentigenic Treg cells analyzed ex vivo (Fig. 1G).
Autoimmunity.
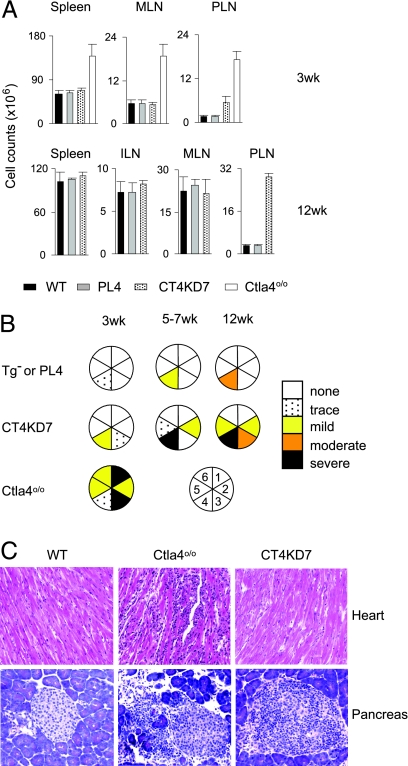
Fully deficient Ctla4o/o mice suffer from systemic lymphoproliferation, with dramatically increased cell counts in the spleen and all lymph nodes of young animals (12–14). This drastic phenotype also was characteristic of the NOD-backcrossed Ctla4 knockout animals (NOD.Ctla4o/o) used for reference (20), with a generalized lymphoproliferation already present at 3 wk (Fig. 2A) and an activated phenotype in most CD4+ T cells (Fig. 8, which is published as supporting information on the PNAS web site). No such lymphoproliferation was observed in CT4KD7 mice, where lymphocytes remained at normal numbers, although with a subtle increase in the frequency of activation/memory cells (Figs. 2A and 8). The only exception was the pancreatic lymph nodes (PLN), where a 3-fold increase in cellularity relative to control littermates was detected at 3 wk, 5-fold by 7 wk, and 9-fold by 12 wk (Fig. 2A and data not shown).
Fig. 2.
Contrasting phenotypes between Ctla4 knockdown and knockout mice. (A) Cellularity in lymphoid organs of NOD. Ctla4o/o, CT4KD7, and PL4 mice at 3 or 12 wk of age (mean ± SD). MLN, mesenteric; ILN, inguinal. (B) A summary of histology survey. 1, heart; 2, lung; 3, exocrine pancreas; 4, islet; 5, liver; 6, skeletal muscle. (C) Representative hematoxylin and eosin sections of pancreas and heart from 3- to 5-wk-old mice. (Original magnification: ×100.)
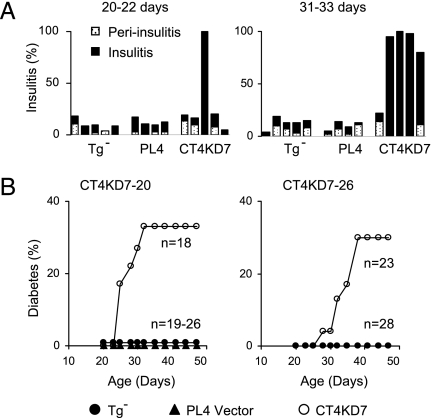
Another prominent phenotype of Ctla4-deficient mice is multiorgan infiltration and damage, most severe in the heart, liver, lung, and exocrine pancreas, and lethal by 3–4 wk of age (12–14) (Fig. 2 B and C). No such dramatic phenotype was apparent in CT4KD7 animals (Fig. 2 B and C; Table 2, which is published as supporting information on the PNAS web site). Of 24 mice examined between 3 and 12 wk of age, only 1 showed any heart involvement. Infiltration of other organs was detected sporadically, mainly in aged animals. The one exception, however, was the pancreas: by 3 wk of age, one of the five animals had severe insulitis, and by 5 wk, most animals in the knockdown group were so affected (Fig. 3A). In contrast to the severe inflammatory destruction in the exocrine pancreatic tissue but minimal damage to the islets in NOD.Ctla4o/o mice, the infiltration in the CT4KD7 pancreas largely was limited to the endocrine tissue (Figs. 2 B and C and 3A; Table 2). As a consequence, ≈30% of the animals in both CT4KD7 lines developed diabetes between 4 and 6 wk of age (Fig. 3B), strikingly faster than in standard NOD mice, which become diabetic mainly between 15 and 25 wk of age. On the other hand, CT4KD7 animals that resist early diabetes remain free of disease until the end point of monitoring at 14 wk of age. Thus, CT4KD7 mice are spared from the multiorgan damage of the corresponding knockout mice, with a focusing and enhancement of anti-islet autoimmunity, resulting in diabetes.
Fig. 3.
Impact of Ctla4 down-modulation on insulitis and diabetes development. (A) Percentage of pancreatic islets affected by inflammatory infiltration in mice at 20–22 days or 31–33 days of age. Each bar represents one animal. (B) Accumulative incidence of diabetes in the two independent lines of CT4KD7 knockdown mice, compared with lentigene-negative littermates and PL4 vector controls.
MHC-Dependent Penetrance.
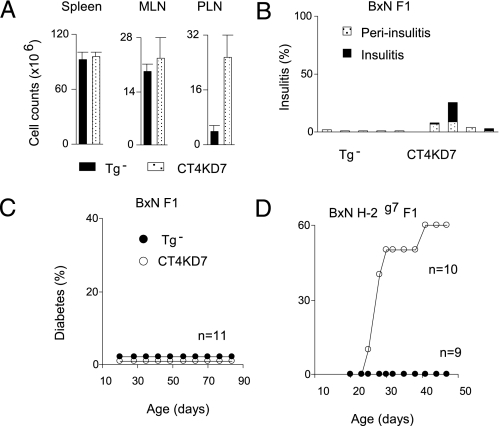
Next, we crossed the CT4KD7 lines with C57BL/6 (B6) mice to address two related questions: first, whether the disease would be influenced by modifier loci (the massive lymphoproliferation in Ctla4 knockouts being essentially background-independent), and second, whether the islet-focused autoimmunity in CT4KD7 mice reflected the reduced expression of Ctla4 itself or resulted from a particular predisposition of the NOD background on which the lentigenes had been integrated. (B6×NOD) F1 animals are normally strongly resistant to insulitis and diabetes. Similar to the animals on the NOD genetic background, 12-wk-old CT4KD7/B×N F1 animals showed an ≈8-fold increase in cellularity in the PLN but not in other lymphoid organs (Fig. 4A). They also had insulitis, albeit much milder than that found on the pure NOD background (Fig. 4B). No early diabetes was observed in these knockdown animals (Fig. 4C), although one of seven CT4KD7/B×N F1 males did become diabetic at 6 months of age. Thus, the protective alleles of the B6 background were able to dampen, but not to eradicate, the pancreas-specific autoimmunity imparted by diminished Ctla4 expression, resulting in a degree of pancreas pathology never observed in (B6×NOD) F1 mice. The B6 background genes did not elicit a retargeting of the autoimmunity: histological examination showed no other pathology besides a persistence of the salivary gland and stomach infiltration (Table 2). In a preliminary exploration into the nature of the modifier loci, we crossed CT4KD7/NOD and B6.H2g7 mice, the latter a congenic line that carries the major histocompatibility complex (MHC) from the NOD genome on the B6 background. The MHC appeared to be a major determinant of the protection offered by the B6 genome, as 6 of 10 CT4KD7/B6×NOD.H2g7 animals became diabetic ≈4–6 wk of age (Fig. 4D).
Fig. 4.
Influence of genetic modifiers. (A) Lymphoid organ cell counts in mice on (B6×NOD) F1 background at 12 wk of age. (B) Insulitis development in this set of animals. Each bar represents one animal. (C and D) Accumulative diabetes incidence in mice on (B6×NOD) F1 (C) or (B6×NOD) H-2g7 F1 (D) background.
Discussion
The tempered reduction of Ctla4 expression by RNAi transgenesis led to autoimmune disease preferentially focused on pancreatic islets, unmasking a specificity that is hidden in the indiscriminate lymphoproliferative and multiorgan autoimmunity of the Ctla4-deficient mice, which actually tends to spare islets. Thus, shRNA transgenesis, in contrast to traditional gene ablation by targeted recombination, better “captured” the human disease phenotypes associated with CTLA4 expression variation.
Harnessing RNAi to study chronic, complex diseases in animal models requires stable and inheritable siRNA expression. Variable success in transgenic RNAi has been documented with traditional approaches through pronuclear injection or engineering of embryonic stem cells (17), but lentiviral transgenesis has been favored because of the high efficiency of transgene integration and, hence, the ability to introduce lentigenes into essentially any genetic background (15, 16, 21); this potential was realized here, with direct delivery into the NOD genome, which has proven largely refractory to ES-based approaches. In addition, it had been suggested that transgenes introduced by lentiviral vectors might not be subject to transcription repression in the germ line, as observed with retroviral vector delivery (22, 23). However, we, as well as others (24), encountered substantial variegation of lentigene expression. Kissler et al. (24) suggested that interference between multiple copies of lentiviral integrants and/or unidentified repressive elements in the original PLL3.7 construct might account for the partial silencing. Our data do not suggest that copy interference is involved (variegation was observed in situations of Mendelian segregation of a single insert); rather, the contrasting expression pattern of paternally vs. maternally inherited lentigenes strongly suggests epigenetic silencing particular to the female germ line. There has been precedent for this phenomenon with a retroviral transgene (25). Because the variegated expression patterns are observed already in the founders, whose lentigenes did not go through gametogenesis, one might hypothesize that early embryonic development coincides with partial silencing but that the paternally inherited copy is preferentially protected, perhaps from the active demethylation that occurs immediately after fertilization (26). A puzzling aspect of this transmission is that the effects appear probabilistic (e.g., a female showing expression in 76% of lymphocytes gives rise to offspring whose lentigene is active in 10% to 36% of cells, full extinction being relatively rare); how is such a probability transmitted? In practice, however, differentially transmitted variegation implies that full expression of lentigene can be stably achieved after one or two generations of crossing from the founder, provided male breeders are used.
Experiments involving RNAi can be confounded by so-called “off-target” effects, the dampening of transcripts unrelated to the target gene (27). Although we cannot formally rule out such effects in the CT4KD7 lines, the fit between the manifestations elicited by the RNAi lentigene and the suspected role of Ctla4 is a strong argument that the phenotype is indeed provoked by Ctla4 down-modulation. In addition, it would be very difficult to explain how off-target effects would be affected by haplotype differences at the MHC in the (B6×NOD) and (B6.H2g7×NOD) intercrosses. Similarly, the phenotype is unlikely to be attributable to insertional effects of the lentigene, because the same phenotypes were observed with two independent lines.
That a partial reduction in Ctla4 expression is sufficient to elicit islet-specific pathology raises the question of why no such disease was reported for Ctla4 knockout heterozygotes (20), which might be expected to have 50% of the normal level of expression. It is possible that gene-copy number is not rate-limiting in determining Ctla4 transcript levels, and we did find that Ctla4o/+ mice expressed ≈70% of normal Ctla4 message levels (data not shown). There may be finely defined threshold effects, the phenotype requiring >50% reduction, or there may be epigenetic control such that one active genomic copy would be different from two partially active copies. It is interesting to note, however, that no patients have so far been described with loss-of-function mutations for CTLA4, especially given that patients with FOXP3 deficiencies are not uncommon. One might speculate that heterozygosity for a CTLA4 deficiency in humans has a dominant deleterious effect, akin to the mouse knockdown, and thereby is strongly counterselected in human populations.
The preferential focus of autoimmunity on islets typical of the NOD mouse was observed even in the genetic context of the (B6×NOD) F1 mouse, normally refractory to islet autoimmunity. Although a contribution of other NOD alleles in the heterozygous state cannot be ruled out, this result suggests a peculiar connection between Ctla4 and the pancreas target. Interestingly, the increase in PLN cellularity was detected uniformly in CT4KD7 mice, even at a time when their islet infiltration was minimal or absent, perhaps suggesting that the PLN is the source of this particular focus. We have reported recently that there are unusual anatomical connections to the PLN, which drains an endocrine organ but is also a preferential drainage of the peritoneum and intestinal tract and, further, that perturbations in the gut affect the presentation of islet autoantigens (28). Might the islet-specificity of the disease in Ctla4 hypomorphs somehow be a consequence of this particular intersection of self and non-self?
This study serves as a proof-of-principle that RNAi-mediated-knockdown can be a more pertinent genetic tool for defining the impact of variation in expression associated with disease pathogenesis. It also may be worth keeping in mind that gene expression differences highlighted through microarray analysis often fall in the range of a few-fold. So testing their significance is likely to be another important application of RNAi-mediated-knockdown technology. Lastly, the results presented herein also argue that expression variation in the Ctla4/CTLA4 gene itself underlies the genetic susceptibility to type 1 diabetes that maps to the T cell costimulatory region in mice and humans.
Materials and Methods
Lentiviral Vector and Ctla4 shRNA Probes.
A modified version of PLL3.7 lentiviral vector (16) was used to express shRNA. To drive a better GFP marker expression in activated T lymphocytes, the CMV promoter of PLL3.7 was replaced by a ubiquitin promoter (yielding the PL4 vector). The Ctla4 shRNA probes were designed based on compatibility with the U6 promoter, ≤84% (16/19) homology to any unintended mouse mRNA targets in the mouse EST database and minimal homology to the human CTLA4 sequence. The following sequences were used for the Ctla4 shRNA constructs: KD1, 5′-GCGGCAGACAAATGACCAA-3′; KD2, 5′-GGTGACCCAACCTTCAGTG-3′; KD3, 5′-GCAGAGTGAACCTCACCAT-3′; KD4, 5′-GGTCTGTGCCACGACATTC-3′; KD5, 5′-GGACTGAGAGCTGTTGACA-3′; KD6, 5′-GGAGCATGAACAGAGAGCT-3′; and KD7, 5′-GCAGCATAAGGATATAGCA-3′. To test the efficacy of these probes, splenic T lymphocytes from a BDC2.5 T cell antigen receptor transgenic mouse were activated with antigenic mimotope peptide for 24 h, and the cells were infected with lentiviral preparations. The cells were harvested 48 h after infection, and RNA was extracted (TRIzol) from cells sorted between GFP− control and GFP+ transduced populations.
Mice.
Ctla4o/o mice, originally generated on the 129 genetic background (29), were backcrossed onto the NOD/Lt genetic background for more than 23 generations. Lentiviral transgenic (lentigenic) mice were generated by injecting a small amount of high-titer lentiviral particles into the perivitelline space of fertilized eggs of the NOD/Lt inbred strain (15) (detailed in Supporting Text, which is published as supporting information on the PNAS web site). The lentigene was detected in DNA from tail biopsies by PCR for the EGFP insert (primers are listed in Supporting Text). Experiments were carried out according to procedures approved by the Joslin Diabetes Center's Institutional Animal Care and Use Committee.
Quantitative RT-PCR.
Ctla4 mRNA levels were measured by quantitative PCR with primers and probes specific for wild-type Ctla4 (7, 9). Quantitative RT-PCR for 2′5′-OAS was performed as described previously (19) with RNA isolated from lymph nodes of 3-wk-old animals. The mRNA levels for Ctla4 and 2′5′-OAS were normalized to hypoxanthine phosphoribosyltransferase (HPRT) mRNA. Sequences of primers are provided in the Supporting Text.
Diabetes Monitoring.
Urine glucose levels were tested every 2 to 3 days with Uristix (Bayer Diagnostics, Tarrytown, NY), and positive reads were confirmed by blood glucose measurement on 2 consecutive days (>300 mg/dl).
Flow Cytometry.
Single-cell suspensions from blood or lymphoid organs were blocked with anti-CD16/32 (2.4G2) and normal mouse sera and stained with antibody conjugates to surface molecules. For intracellular staining of Ctla4, after staining of cell-surface markers, samples were fixed with Cytofix/Cytoperm buffer for 20 min on ice, washed, and then stained on ice for 30 min with phycoerythrin (PE)-conjugated anti-Ctla4 antibody (clone UC10–4F10–11) in Per/Wash buffer (BD PharMingen, San Diego, CA). Intracellular expression of Foxp3 was examined with PE-conjugated monoclonal antibodies (eBioscience, San Diego, CA) according to the manufacturer's protocol. Staining with 7-aminoactinomycin D was used to exclude dead cells.
Supplementary Material
Acknowledgments
We thank C. Dillon for advice on lentiviral shRNA, and E. Hyatt, K. Hattori, G. Buruzula, J. Lavecchia, A. Pinkhasov, and C. Laplace for assistance. This work was supported by Juvenile Diabetes Research Foundation Grant 4-2004-368, National Institutes of Health Grant P01 AI56299-04, grants from the William T. Young Chair (to D.M. and C.B.), and the core services of Joslin's National Institute of Diabetes and Digestive and Kidney Diseases-funded Diabetes Endocrinology Research Center. Z.C. has been supported by postdoctoral fellowships from the Juvenile Diabetes Research Foundation and the Program in AIDS Research (Dana–Farber Cancer Institute/National Institutes of Health).
Abbreviations
- 2′5′-OAS
2′5′-oligoadenylate synthetase
- B6
C57BL/6
- KD
knockdown
- CT4KD
cytotoxic T lymphocyte antigen 4 KD
- NOD
nonobese diabetic
- PLN
pancreatic lymph nodes
- lentigene
lentiviral transgene
Footnotes
The authors declare no conflict of interest.
References
- 1.Kristiansen OP, Larsen ZM, Pociot F. Genes Immun. 2000;1:170–184. doi: 10.1038/sj.gene.6363655. [DOI] [PubMed] [Google Scholar]
- 2.Tisch R, McDevitt H. Cell. 1996;85:291–297. doi: 10.1016/s0092-8674(00)81106-x. [DOI] [PubMed] [Google Scholar]
- 3.Ligers A, Teleshova N, Masterman T, Huang WX, Hillert J. Genes Immun. 2001;2:145–152. doi: 10.1038/sj.gene.6363752. [DOI] [PubMed] [Google Scholar]
- 4.Wang XB, Zhao X, Giscombe R, Lefvert AK. Genes Immun. 2002;3:233–234. doi: 10.1038/sj.gene.6363869. [DOI] [PubMed] [Google Scholar]
- 5.Anjos SM, Tessier MC, Polychronakos C. J Clin Endocrinol Metab. 2004;89:6257–6265. doi: 10.1210/jc.2004-0881. [DOI] [PubMed] [Google Scholar]
- 6.Baniasadi V, Narain N, Goswami R, Das SN. Tissue Antigens. 2006;67:383–389. doi: 10.1111/j.1399-0039.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- 7.Ueda H, Howson JMM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KMD, Smith AN, Di Genova G, et al. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 8.Lamhamedi-Cherradi SE, Boulard O, Gonzalez C, Kassis N, Damotte D, Eloy L, Fluteau G, Levi-Strauss M, Garchon HJ. Diabetes. 2001;50:2874–2878. doi: 10.2337/diabetes.50.12.2874. [DOI] [PubMed] [Google Scholar]
- 9.Vijayakrishnan L, Slavik JM, Illes Z, Greenwald RJ, Rainbow D, Greve B, Peterson LB, Hafler DA, Freeman GJ, Sharpe AH, et al. Immunity. 2004;20:563–575. doi: 10.1016/s1074-7613(04)00110-4. [DOI] [PubMed] [Google Scholar]
- 10.Lundholm M, Motta V, Lofgren-Burstrom A, Duarte N, Bergman ML, Mayans S, Holmberg D. Diabetes. 2006;55:538–544. doi: 10.2337/diabetes.55.02.06.db05-1240. [DOI] [PubMed] [Google Scholar]
- 11.Thompson CB, Allison JP. Immunity. 1997;7:445–450. doi: 10.1016/s1074-7613(00)80366-0. [DOI] [PubMed] [Google Scholar]
- 12.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, Thompson CB, Griesser H, Mak TW. Science. 1995;270:985–989. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 13.Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 14.Chambers CA, Sullivan TJ, Allison JP. Immunity. 1997;7:885–895. doi: 10.1016/s1074-7613(00)80406-9. [DOI] [PubMed] [Google Scholar]
- 15.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 16.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Zhang M, McManus MT, Gertler FB, Scott ML, et al. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 17.Dillon CP, Sandy P, Nencioni A, Kissler S, Rubinson DA, Van Parijs L. Annu Rev Physiol. 2005;67:147–173. doi: 10.1146/annurev.physiol.67.040403.130716. [DOI] [PubMed] [Google Scholar]
- 18.Hannon GJ, Rossi JJ. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 19.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 20.Luhder F, Chambers C, Allison JP, Benoist C, Mathis D. Proc Natl Acad Sci USA. 2000;97:12204–12209. doi: 10.1073/pnas.200348397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiscornia G, Singer O, Ikawa M, Verma IM. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeifer A, Ikawa M, Dayn Y, Verma IM. Proc Natl Acad Sci USA. 2002;99:2140–2145. doi: 10.1073/pnas.251682798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis J. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 24.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS. Nat Genet. 2006;38:479–483. doi: 10.1038/ng1766. [DOI] [PubMed] [Google Scholar]
- 25.Swain JL, Stewart TA, Leder P. Cell. 1987;50:719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- 26.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 27.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 28.Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Proc Natl Acad Sci USA. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers CA, Cado D, Truong T, Allison JP. Proc Natl Acad Sci USA. 1997;94:9296–9301. doi: 10.1073/pnas.94.17.9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.