Abstract
Rhizobial lipo-chitooligosaccharides (LCOs) are signaling molecules involved in host-range recognition for the establishment of the symbiosis with leguminous plants. The major LCO of Rhizobium meliloti, the symbiont of Medicago plants contains four or five N-acetylglucosamines, O-acetylated and N-acylated with a C16:2 fatty acid on the terminal nonreducing sugar and O-sulfated on the reducing sugar. In this paper, the ligand specificity of a high-affinity binding site (Nod factor binding site 2 or NFBS2), enriched in a plasma membrane-enriched fraction of Medicago cell suspension cultures, is reported. By using chemically synthesized LCOs, the role of structural elements, important for symbiotic activities, as recognition motifs for NFBS2 was determined. The results show that the substitutions on the nonreducing sugar of the LCOs (the O-acetate group, the fatty acid, and the hydroxyl group on the C4 of the sugar) are determinants for high-affinity binding to NFBS2. In contrast, the sulfate group, which is necessary for all biological activities on Medicago, is not discriminated by NFBS2. However, the reducing sugar of the LCO seems to interact with NFBS2, because ligand binding is affected by the reduction of the free anomeric carbon and depends on the number of N-acetyl glucosamine residues. These results suggest that the recognition of the LCOs by NFBS2 is mediated by structural elements in both the lipid and oligosaccharidic moities, but not by the sulfate group.
Rhizobia are bacteria that can elicit the formation of nodules on the roots of legumes in which they fix dinitrogen for the benefit of the plant. Nodule formation is very specific, and a given strain of Rhizobium can infect a limited number of species of the legume family. The establishment of the symbiosis depends on a particular class of signaling molecules produced by the prokaryotic partner: the Nod factors. Nod factors are lipo-chitooligosaccharides (LCOs) consisting of a N-acetylglucosamine backbone, N-acylated on the terminal nonreducing sugar. Variations in the number of glucosamine residues (three to five), in the structure of the fatty acid chain, and the presence of different substitutions on the oligosaccharidic backbone are characteristics of each bacterial strain. At subnanomolar concentrations, purified Nod factors provoke the early symbiotic responses in the plant and in certain species they can induce the morphogenesis of bacteria-free nodules (reviewed in refs. 1–4). The fact that most of the biological responses requires a very low concentration of Nod factor with a precise structure strongly suggests that Nod factors may be perceived by specific receptors on the roots of the host plants. Indeed in vivo studies suggest that there may be multiple perception mechanisms with different stringencies for the Nod factor decorations (5, 6).
In vitro, a Nod factor binding site, termed NFBS1, has been identified in a particulate fraction of Medicago truncatula root extracts, by using the major signal [NodRm-IV(Ac,S,C16:2)], labeled with tritium, of its symbiotic bacterium Rhizobium meliloti (7). Several characteristics of this site suggest that it may not play an exclusive role in the symbiosis; it has a relatively low affinity (Kd = 86 nM) and low specificity for LCOs and a similar site exists in a particulate fraction of tomato roots, a nonleguminous plant. To search for other binding sites for Nod factors, a probe with a high specific radioactivity was produced by an enzymatic sulfation with 35S (8). Probing a microsomal fraction prepared from Medicago varia cell suspension cultures with this ligand resulted in the identification of a high-affinity binding site, NFBS2, which was sensitive to proteases (9). In the present paper, we report on the binding requirements of NFBS2; by using differently substituted LCOs, our aim was to evaluate the role of structural elements important for symbiotic responses, in the recognition by NFBS2.
MATERIALS AND METHODS
Plant Material.
M. varia cell suspension cultures were grown as described (9). Cells reaching the stationary growth phase (14 days) were harvested by filtration on a scintered glass filter (size 1), and the cells were washed with 1 vol of 20 mM KCl and stored at −80°C until use.
Microsomal Fraction Preparation and Membranes Isolation.
Approximately 50 g of cells were homogenized at 4°C for 6 × 5 s in a blender (Moulinex type 534, Alençon, France) in the presence of 100 ml of extraction buffer (25 mM Tris⋅HCl buffer, pH 8.5/0.47 M sucrose/5 mM MgCl2/10 mM β-mercaptoethanol/2 μM leupeptin/0.1 mM PMSF) and processed as described (7). The microsomal fraction, sedimenting at 45,000 × g, was resuspended in the binding buffer with 50% glycerol and stored at −80°C. Membrane separation was performed by preparative free flow electrophoresis with a Vap-22 FFE-unit (Weber, Kirchheim, Germany). The electrophoresis chamber medium contained 10 mM Tris-boric acid buffer, pH 8.4, 0.25 M sucrose, 1 mM MgCl2, and 10 mM KCl, and the electrode medium contained 100 mM Tris-boric acid buffer pH 8.4. The sample, suspended in the chamber buffer at 2 mg of protein/ml was injected at 3 ml/hr, under 130 mA constant current (about 950 V). The eluted fractions were monitored at 280 nm, and enriched preparations of plasma membrane (PM), endomembranes, and tonoplast were collected separately. Each pooled fraction was centrifuged for 45 min at 45,000 × g, and the corresponding pellets were resuspended in 20 mM Tris⋅HCl buffer, pH 7, 0.25 M sucrose, 1 mM MgCl2, 1 mM CaCl2, 2 μM leupeptin, and 0.1 mM PMSF. The identification and enrichment of the various fractions was checked by immunodetection of marker proteins: H+-ATPase (98 kDa) for the PM, the endoplasmic reticulum resident chaperone BiP (78 kDa) for the endomembranes, and vacuolar (V)-ATPase (60 kDa) for the tonoplast fractions.
Gel Electrophoresis and Immunodetection of Proteins.
Samples (10 μg protein) were separated on 8% polyacrylamide resolving gels by using the buffer system of Laemmli (10), and the resolved polypeptides were electro-transferred to Hybond-C membranes (Amersham Pharmacia). After transfer, PM H+-ATPase and V-ATPase were detected with the corresponding antibodies by using an ECL technique as described (11). BiP was identified with a 1/15,000e dilution of polyclonal antibody as in ref. 12.
Radiolabeling of NodRm Factors.
Biologically produced nonsulfated factors were purified and labeled with 35S according to ref. 8 with a slight modification in the synthesis of phosphoadenosine phosphosulfate (PAPS). Briefly, PAPS synthesis was performed overnight at 37°C, in 100 mM Tris⋅HCl buffer, pH 8.5 in the presence of 2 mCi of 35S carrier-free sodium sulfate (1,600 Ci/mmol), 28 mM ATP, 8 mM MgCl2, 5 mM GTP, and 30 μl of yeast extract in a total volume of 53 μl. At the end of the synthesis, the labeled ligand, termed the 35S-NodRm factor [NodRm-IV(Ac,35S, C16:2Δ2,9)] had a specific radioactivity of about 800 Ci mmol−1.
Synthesis of LCOs.
The sulfated LCOs were chemically synthesized according to the previously reported sequence (13). Tetra-N,NII,NIII,NIV-acetyl-chitopentaose (CO-V) was prepared by a recombinant Escherichia coli strain harboring the nodBC genes of Azorhizobium caulinodans (14). Tri-N,NII,NIII-acetyl-chitotetraose (CO-IV), di-N,NII-acetyl-chitotriose (CO-III), and N-acetyl-chitobiose (CO-II) were obtained as reported (15). For the acylation procedure, the corresponding CO (10 μmol) and Et3N (50 equiv., 70 μl) in 240 μl of water were mixed at 0°C in a plastic vial with a dioxane solution (480 μl) of anhydride (240 μmol), commercially available or prepared as described (16). The reaction mixture was mixed vigorously from time to time at room temperature for 24 hr. The evolution of the reaction was monitored by TLC using CH3CN/H2O/28% NH3 in H2O (70:30:2, vol/vol) as eluent. HCl (1 M) was added to bring the solution to pH 1, and then the aqueous phase was extracted by vigorous stirring with EtOAc (0.5 ml, 3×). The insoluble pellet in water was sonicated in H2O (1 ml, 3×) and freeze-dried. The absence of residual acid and Et3N⋅HCl was checked by TLC (eluent ButOH/EtOH/H2O, 5:3:2 vol/vol) and 1H-NMR; when traces were still present, an additional extraction was performed with water/dioxane mixture (1:1 vol/vol). The target acylated compounds were obtained in 30–50% yields. The reduced derivative of LCO-IV(S,C16:1) was obtained by treatment with NaBH4 at room temperature in dimethylformamide, followed by precipitation in acetone and HPLC purification. The purity and the structure of each compound was checked by HPLC analysis as in ref. 7, mass spectroscopy, and 1H-NMR. The low-resolution mass measurements were obtained on a Nermag (Houston) R-1010C spectrometer model 200 equipped with a M-Scan Wallys-type gun. The LCOs were analyzed by fast atom bombardment (positive ion mode) on a glycerol matrix. High-resolution mass measurements were recorded at the Service Central d’Analyse, (Centre National de la Recherche Scientifique, Vernaison, France). 1H-NMR spectra were recorded on a Bruker AC300 spectrometer (Wissenburg, France) at 300 MHz.
LCOs Quantification.
LCOs, analyzed by reverse-phase HPLC on a C18 column, were detected by their absorbance at 210 nm. Their quantities were determined by the measurement of the peak area, by reference to known quantities of pure chemically synthesized LCOs used as standards: LCO-IV(S,C16:2Δ2,9) for the LCOs containing a biunsaturated fatty acid or LCO-IV(S,C16:1Δ9) for the monounsaturated compounds.
Binding Assays.
Microsomal fraction (containing 10–50 μg protein) was incubated in the presence of 0.2 nM of 35S-NodRm factor in 20 mM Na-cacodylate buffer, pH 6.0 containing 0.25 M sucrose, 1 mM MgCl2, 1 mM CaCl2, 0.1 mM PMSF, and 2 μM leupeptin in a total volume of 200 μl. The nonspecific binding component was determined in the presence of 1 μM NodRm-IV(S,C16:2). Incubations were performed 1 hr at 0°C in 96-well microtiter plates (Nunc), and the following steps were as described (7). Binding data were analyzed by radlig software, version 4 (Biosoft, Cambridge, U.K.).
Protein Determination.
Protein was measured by the bicinchoninic acid procedure developed by Pierce with BSA as reference.
RESULTS
NFBS2 Has a pH Optimum of 6.0 and Involves an Essential Lysine Residue for Ligand Binding.
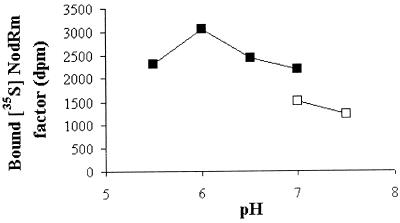
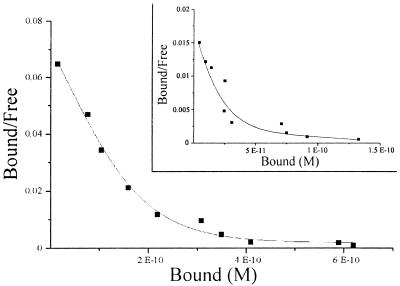
By using the 35S-NodRm factor (NodRm-IV(Ac,35S,C16:2Δ2,9) and the microsomal fraction of M. varia cell cultures, the pH optimum of specific Nod factor binding was determined. The data reported in Fig. 1 show that the binding activity, initially measured at pH 7.0 in Tris buffer (9), was 2-fold higher at pH 6 in Na-cacodylate buffer. Moreover a 6-fold enhancement was obtained if the microsomal fraction was allowed to equilibrate in this buffer, at pH 6, before binding (data not shown). Equilibration at pH 6 in phosphate or Bis-Tris buffer produced only a 2-fold enhancement, suggesting that the effect of the pH 6 Na-cacodylate buffer was not only caused by the pH. This finding prompted us to reassess the thermodynamic constants that previously were determined at neutral pH with Tris buffer. The Scatchard analysis of a saturation experiment with the 35S-NodRm factor at pH 6 revealed a biphasic plot (Fig. 2), as observed before at pH 7 (ref. 9 and Fig. 2, Inset), indicating the presence of two types of binding sites. However, in the new conditions, the relative abundance of both sites was magnified, with the highest stimulation being observed for the high-affinity binding site, NFBS2 (Bmax = 1.5 pmol/mg protein, up from 0.2 pmol/mg protein) whereas the abundance of the low-affinity binding site was increased by about 3-fold (Bmax = 3.5 pmol/mg protein). In comparison to the initially reported value, the affinity of NFBS2 was slightly lower (Kd = 4 nM), which probably reflects a more accurate determination of this parameter, because of the magnification of the specific binding measured in these optimized conditions, whereas the Kd of the low-affinity site remained at about 90 nM. At the concentration of labeled ligand used in all further experiments (0.2 nM) the observed binding was almost exclusively to NFBS2.
Figure 1.
Influence of pH on the specific binding activity. The microsomal fraction of M. varia cell cultures was resuspended in 20 mM Tris⋅HCl buffer, and aliquots corresponding to 30 μg proteins were incubated with 0.2 nM NodRm-IV(Ac,35S,C16:2), either in Tris⋅HCl (□) or in Na-cacodylate (■) binding buffers at different pH. The specific binding to 35S-NodRm factor was measured.
Figure 2.
Scatchard plot of 35S-NodRm factor binding to the microsomal fraction either at pH 6 in Na-Cacodylate buffer or at pH 7 in Tris⋅HCl buffer (Inset).
Amino acid-modifying reagents then were incubated in the presence of the microsomal fraction preparation to determine the residues involved in ligand binding to NFBS2. As demonstrated in Table 1, only compounds reacting with lysine residues affected the binding activity. Analysis of the 35S-NodRm factor by HPLC after incubation either with the microsomal fraction or with the various reagents did not reveal any changes in its elution profile, indicating that the ligand structure had not been modified.
Table 1.
Effect of amino acid modifying reagents on the binding activity
| Reagent and concentration | Target | Inhibition of binding, % | |
|---|---|---|---|
| DIDS | 100 μM | Lys | 37 |
| 1 mM | 80 | ||
| Pyridoxal | 2 mM | Lys | 37 |
| phosphate | 20 mM | 64 | |
| Phenyl glyoxal | 1 mM | Arg | 5 |
| 10 mM | 8 | ||
| N-ethyl maleimide | 2 mM | Cys | 5 |
| 20 mM | 0 | ||
| Phenylmethyl- | 100 μM | Ser | 0 |
| sulphonyl fluoride | 1 mM | 0 | |
| Diethyl | 1 mM | His | 0 |
| pyrocarbonate | 10 mM | 3 |
DIDS, 4-4′-diisothiocyanatostilbenne-2,2′-disulfonic acid. The different reagents were incubated during the binding assay (2 h, 0°C).
The Affinity of LCOs for NFBS2 Greatly Depends on the Structure of the Acyl Moiety.
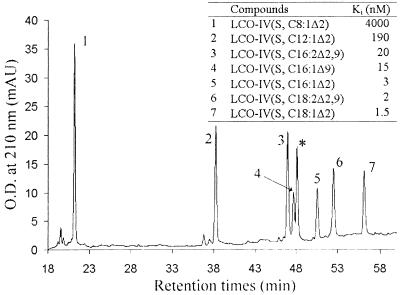
Chemically synthesized tetrameric LCOs, sulfated and non-O-acetylated, differing only in the nature of the acyl moiety (length of the fatty acid chain (C8 to C18), number (one or two), or position (Δ2 or Δ9) of double bonds), were analyzed by reverse-phase chromatography on a C18 column. On this chromatographic support, the retention times of the LCOs are related to the hydrophobicity of the acyl chain, depending mainly on the length of the fatty acid chain and the number of double bonds. These LCOs then were used in competition experiments with radiolabeled NodRm-IV(Ac,S,C16:2Δ2,9) to determine their affinity for NFBS2. The results reported in Fig. 3 show clearly that increasing the length of the fatty acid chain increases dramatically the ability of LCOs to compete with the 35S-NodRm factor for its high-affinity binding site with a plateau for C16 and C18 fatty acids. Thus, LCO-IV(S,C8:1Δ2) with the shortest fatty acid chain was a poor competitor of ligand binding (Ki = 4 μM). In contrast, LCO-IV(S,C18:1Δ2) with the most hydrophobic fatty acid chain was the most potent competitor in preventing 35S-NodRm factor binding (Ki = 1.5 nM). Furthermore, the number or the position of the double bonds, apart from their role in the hydrophobicity of the fatty acid chain, did not seem to exert any particular effect on the affinity for LCOs. It is noteworthy that a mixture of Na-palmitate and tetra-N-acetyl-chitotetraose (molar ratio 1:1) even at high concentrations (1 μM) were unable to inhibit 35S-NodRm factor binding (data not shown).
Figure 3.
Elution of LCOs differing by the structure of the acyl chain from a C18 HPLC column and determination of their affinities (Ki) for NFBS2 by competitive inhibition of 35S-NodRm factor binding. * corresponds to NodRm-IV(Ac,S,C16:2Δ2,9). The RP18 column was eluted at 1 ml/min for 60 min, with a 10–55% acetonitrile linear gradient in 10 mM K2SO4, pH 4.6.
O-Acetylation of the Nonreducing End Sugar, But Not Sulfation of the Reducing Sugar, Influences the Affinity of LCOs for NFBS2.
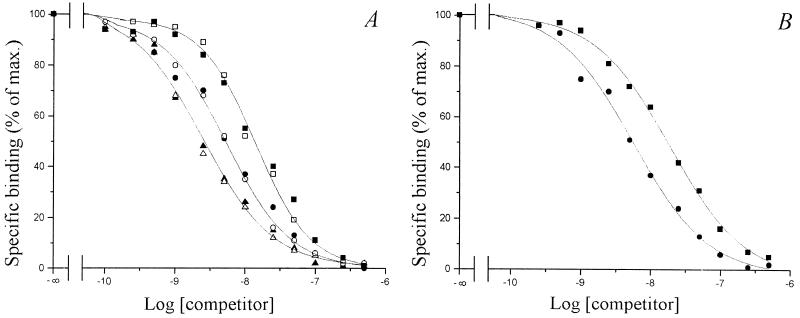
The role of the decorations of the oligosaccharidic backbone, i.e., the sulfate group on the reducing sugar and the O-acetate on the nonreducing end sugar, was examined. Non-O-acetylated LCOs harboring different structures of acyl chain (C16:l or C18:1) and the original O-acetylated Nod factor [NodRm-IV(Ac,C16:2)], either sulfated or nonsulfated, were used in three separate sets of competition experiments with the 35S-NodRm factor. The results reported in Fig. 4A revealed that in each case the sulfated and the corresponding nonsulfated compound have the same ability to inhibit ligand binding, thus NFBS2 does not seem to be selective for the sulfate group. In contrast, the competitive inhibition curves obtained for the O-acetylated NodRm-IV(Ac,S,C16:2) and its non-O-acetylated analog were very different (Fig. 4B); NodRm-IV(S,C16:2) required higher concentrations (5-fold) than NodRm-IV(Ac,S,C16:2) to achieve 50% inhibition of binding of the radiolabeled ligand. A Scatchard analysis of a saturation experiment using the 35S-labeled non-O-acetylated NodRm factor confirmed that its affinity for NFBS2 was lower (20 nM) than that of the O-acetylated compound (4 nM) (data not shown). These results indicate that the decoration carried at the nonreducing end of the molecule, namely the O-acetate group, seems to be an important structural element for the recognition by NFBS2.
Figure 4.
Competitive inhibition of 35S-NodRm factor binding to NFBS2 by unlabeled LCOs differing by the presence of the sulfate group (A) or the O-acetate group (B). The tested compounds were (A) NodRm-IV(Ac,S,C16:2) (●) NodRm-IV(Ac,C16:2) (○), LCO-IV(S,C18:1Δ2) (▴), LCO-IV(C18:1Δ9) (▵), LCO-IV(S,C16:1Δ9) (■), and LCO-IV(C16:1Δ9) (□), and (B) NodRm-IV(Ac,S,C16:2) (●) and NodRm-IV(S,C16:2) (■); 100% corresponds to a specific binding activity of approximately 6,000 dpm/assay.
The Length of the Chitin Oligomer Backbone Is Important for the Binding of LCOs to NFBS2 and Modifications of the Sugar at Both Ends of the Molecule Decrease the Affinity for NFBS2.
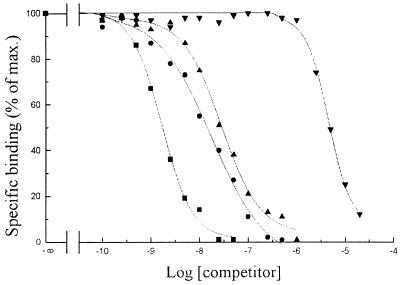
The Nod factors produced by R. meliloti vary in the length of their carbohydrate backbone, from three to five residues of N-acetylglucosamine (17). To evaluate the importance of the oligosaccharidic moiety in the binding to NFBS2, the affinity of LCOs with a number of sugar residues ranging from two to five (termed dimeric-pentameric factors) were examined: all were non-O-acetylated and nonsulfated compounds with the same acyl chain, C16:1Δ9. The results reported in Fig. 5 show that the affinity for NFBS2 was the highest for a LCO-V (Ki = 0.7 nM) and decreased as the number of N-acetyl-glucosamine decreased: indeed a LCO-II was a poor competitor of ligand binding (Ki = 5 μM). The highest affinity, for LCO-V, was confirmed by a saturation experiment with a 35S-LCO-V; this experiment also showed that NFBS2 does not recognize the sulfate group on pentameric factors. Recent data have shown that the nodulation of vetch by its symbiotic partner Rhizobium leguminosarum bv. viciae, could be partially inhibited (50%), by an exogenous addition of a LCO for which the nonreducing glucosamine residue was substituted by a galactosamine residue (18). The nodule formation in the presence of the unsubstituted LCO [LCO-IV(C18:1Δ9)] was identical to that elicited by the bacteria alone. Röhrig et al. (18) suggest that the GalN-containing LCO could compete with the LCO produced by the rhizobia for perception by a putative receptor. Thus the ability of such a modified LCO to inhibit 35S-NodRm factor binding to NFBS2 was determined. The results reported in Table 2 showed that in comparison to LCO-IV(C18:1Δ9) the GalN-containing analog had a significantly lower affinity (Ki = 11 nM against 2 nM).
Figure 5.
Competitive inhibition of 35S-NodRm factor binding to NFBS2 by unlabeled LCOs differing by the number of N-acetyl glucosamine residues. The tested compounds were LCO-V(C16:1Δ9) (■) (Ki = 0.7 nM), LCO-IV(C16:1Δ9) (●) (Ki = 15 nM), LCO-III(C16:1Δ9) (▴) (Ki = 30 nM), and LCO-II(C16:1Δ9) (▾) (Ki = 5 μM).
Table 2.
Influence of modifications of the reducing or the nonreducing sugar of the LCO on the affinity for NFBS2, as measured by competitive inhibition of 35S-NodRm factor binding
| Compound | Ki, nM |
|---|---|
| LCO-IV(C18:1Δ9E) | 2 |
| LCO-GaIN (GlcNAc)3(C18:1Δ9E) | 11 |
| LCO-IV(S, C16:1Δ9) | 15 |
| LCO-IV(S, C16:1Δ9) reduced | 140 |
A more dramatic decrease in affinity occurred (Ki = 140 nM) for a LCO resulting from the reduction of the free anomeric carbon of the reducing sugar, indicating that the reducing end of the molecule constituted a recognition motif for the binding protein.
NFBS2 Is Enriched in Plasmalemma-Enriched Fractions.
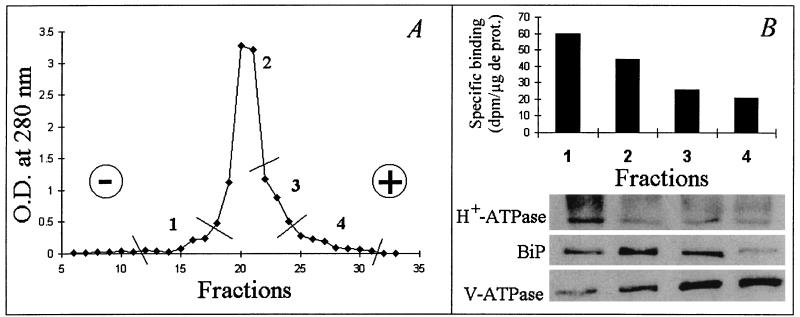
The microsomal fraction was analyzed by free-flow electrophoresis, a technique widely used for plant membrane purification (11). The distribution pattern was divided into four pooled fractions as reported in Fig. 6A, and for each pooled fraction, the immunodetection of protein markers was carried out (Fig. 6B, Lower). The endoplasmic reticulum marker BiP was mainly located in fraction II, which represents 85% of total protein recovered after electrophoresis. The PM H+-ATPase was most concentrated in fraction I (4% of total protein), and the tonoplastic V-ATPase was enriched in fractions III and IV (9% and 2% of total protein respectively). The results indicate that the minor fractions numbered I and IV corresponded to PM- and tonoplast-enriched fractions, respectively, whereas the bulk of membranes migrating in fractions II and III was enriched in endomembranes. The specific binding activity for the 35S-NodRm ligand (Fig. 6B, Upper) was the highest in fraction I and decreased in each of the pooled fractions, indicating that the PM-enriched fraction also was enriched in binding sites. The binding activity associated to fraction II could reflect either a contamination of this fraction by the PM as suggested by the presence of H+-ATPase (Fig. 6B, lane 2) or a dual localization of the binding sites. It is noteworthy that the binding sites detected in fractions I and II exhibited the same affinity for 35S-NodRm and did not discriminate the sulfate group (data not shown).
Figure 6.
Separation of M. varia microsomal fraction membranes, monitored by absorbance at 280 nm, by free-flow electrophoresis (A) and distribution of NFBS2 into the different pooled membrane fractions (B, Upper) whose composition was checked by immunodetection of protein markers (B, Lower). For the determination in B, the same amount of protein was used for each fraction. H+-ATPase, BiP, and V-ATPase are markers for PM, endomembrane, and tonoplast, respectively.
DISCUSSION
Nod factors are the bacterial determinants of nodule organogenesis and host-range in the symbiosis with leguminous plants. However despite their importance, the mechanisms by which these bioactive LCOs exert their effects on their legume hosts are totally unknown. Nod factors provoke many responses in the roots of their host plant similar to rhizobia-induced responses, including root hair deformation, cortical cell division, and expression of specific genes. Genetic studies on both rhizobia and legumes and studies using purified LCOs show that some responses have different structural requirements than others in a given plant (4–6). From these results, it has been proposed that Nod factors may be perceived by more than one receptor with different stringencies for the Nod factor structure. In addition to receptors, Nod factors may encounter other binding proteins, and indeed there is direct evidence for the production of Nod factor-degrading enzymes by legume roots (19) and indirect evidence for transporters (20, 21). Only by the complete characterization of all such binding proteins will the mode of action of Nod factors be fully understood.
In our search for Nod factor binding proteins, an examination of Medicago roots previously had revealed a relatively low-affinity binding site (NFBS1) with a poor selectivity toward specific LCOs (7). A higher affinity site also may be present in these symbiotic tissues but its low abundance and partial masking by NFBS1 prohibited further characterization (F.G. and J.-J.B., unpublished work). As an alternative, we have used cell cultures to characterize NFBSs, as they represent an abundant source of plant material and have been used from a number of species for the biochemical characterization of many putative receptors (22, 23). By using M. varia cell cultures (9), a high-specific activity ligand (8) and a range of pure, modified, and differentially substituted LCOs (13, 14, 18, 24), we have characterized a high-affinity, protease-sensitive binding site for Nod factors (NFBS2) that exhibits a Kd of 4 nM for the major Nod factor of R. meliloti, the symbiont of Medicago.
High-affinity binding to NFBS2 requires the covalent linkage of both the lipid and the chitooligosaccharide moieties. Thus this site is clearly different to the chitin-fragment binding sites described in rice and tomato, which show high affinities for chitooligosaccharides and (at least in tomato) a lower affinity for a Nod factor (25, 26). To date all oligosaccharide binding sites described in plants appear to be located in the PM. However recently, clones that could correspond to the β-glucan elicitor binding sites have been obtained, which encode a protein located in the PM (as determined by immunocytochemistry), but without clear membrane-spanning domains (27). In this paper we show that NFBS2 is enriched in the PM-enriched fraction, suggesting at least a partial PM localization although the possibility of additional subcellular locations cannot be ruled out.
We have established further that NFBS2 is able to discriminate certain substitutions on the basic LCO structure. On the nonreducing end the structure of the acyl chain, the O-acetylation of C6 and a hydroxyl group in the equatorial rather than axial position on C4 appear to be important for high-affinity binding. The length of the oligosaccharide chain also has a major effect with at least three sugar residues being required for high-affinity binding and the affinity increasing with four and five. At the reducing end of the molecule, the anomeric carbon is important, but surprisingly the presence of O-sulfate on C6 of the reducing sugar of tetrameric or pentameric factors neither hinders nor enhances the binding. This finding is unexpected in view of the contribution that these sugars make to the binding, the bulkiness and charge of the sulfate group, and the predicted inverse orientation (in relation to the acyl chain) of the sulfate on tetrameric versus pentameric factors (because of the β1–4-linkages). Our results are suggestive of a LCO-binding protein, with a hydrophobic pocket, in which a C16/C18 acyl chain and an O-acetate on the nonreducing glucosamine are the most important elements of recognition, the reducing end of the Nod factor molecule being recognized in a more flexible manner.
The question now arises as to the biochemical and physiological roles of NFBS2. Several features of the site would be consistent with a role as a receptor, rather than as a binding site involved in transport, sequestration, or degradation; binding to the membrane vesicles is saturable, reversible, does not modify the ligand, and is of high affinity. However, the specificity of this site does not correspond fully to what would be expected for a putative Nod factor receptor with respect to structure-activity studies in the Medicago symbiosis. In particular, NFBS2 does not discriminate the presence of the sulfate group, which is essential in all Medicago symbiotic bioassays [nodulation, root hair deformation (Had), expression of ENOD12]. However, this substitution is recognized by Medicago plants in Had assays even when the sulfate is presented in a quite different molecular context—linked to a fucose group attached to the reducing sugar of Rhizobium sp NGR234 Nod factors (28)—thus suggesting that the perception of the sulfate group may be more complex than expected.
Thus although NFBS2 cannot at this point be positioned in a symbiotic context, it represents to date the only high-affinity, LCO binding site described in plants, and its PM localization, specificity characteristics, and presence in a legume suggest that it may be related to symbiotic Nod factor receptors. The site is also relatively abundant and has been solubilized with an approximate 40% recovery of activity (F.G. and J.-J.B., unpublished data). Our aim now is to purify the binding protein and then to use antibody or DNA probes to characterize the structure and expression of this or related proteins in the symbiotic tissues. Together with genetic and physiological approaches, the biochemical characterization of NFBSs should lead to a better understanding of Nod factor perception in legumes and perhaps in other organisms.
Acknowledgments
We are grateful to J.-M. Beau for supplying the differently acylated sulfated LCOs and H. Röhrig, J. Schmidt, M. John, and J. Schell for the GalN-containing LCO and its corresponding analog. We thank M. Boutry, R. Ratajczak, and A. Vitale for providing the antisera. This work was supported by grants from the Région Midi-Pyrénées, the Centre National de la Recherche Scientifique Physique et Chimie du Vivant program, and the European Union Training and Mobility of Researchers Program.
ABBREVIATIONS
- NFBS
Nod factor binding site
- LCO
lipo-chitooligosaccharide
- CO-V
tetra-N,NII,NIII,NIV-acetyl-chitopentaose
- CO-IV
tri-N,NII,NIII-acetyl-chitotetraose
- PM
plasma membrane
References
- 1.Long S R. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dénarié J, Debellé F, Promé J-C. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 3.Heidstra R, Bisseling T. New Phytol. 1996;133:25–43. [Google Scholar]
- 4.Cohn J, Day R B, Stacey G. Trends Plant Sci. 1998;3:105–110. [Google Scholar]
- 5.Ardourel M, Demont N, Debellé F, Maillet F, De Billy F, Promé J-C, Dénarié J, Truchet G. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geurts R, Heidstra R, Hadri A-E, Downie A J, Franssen H, van Kammen A, Bisseling T. Plant Physiol. 1997;115:351–359. doi: 10.1104/pp.115.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bono J-J, Riond J, Nicolaou K C, Bockovich N J, Estevez V A, Cullimore J V, Ranjeva R. Plant J. 1995;7:253–260. doi: 10.1046/j.1365-313x.1995.7020253.x. [DOI] [PubMed] [Google Scholar]
- 8.Bourdineaud J-P, Bono J-J, Ranjeva R, Cullimore J V. Biochem J. 1995;306:259–264. doi: 10.1042/bj3060259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niebel A, Bono J-J, Ranjeva R, Cullimore J V. Mol Plant Microbe Inter. 1997;10:132–134. [Google Scholar]
- 10.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Bardy N, Carrasco A, Galaud J-P, Pont-Lezica R, Canut H. Electrophoresis. 1998;19:1145–1153. doi: 10.1002/elps.1150190715. [DOI] [PubMed] [Google Scholar]
- 12.Pedrazzini E, Giovinazzo G, Bielli A, de Virgilio M, Frigerio L, Pesca M, Faoro F, Bollini R, Ceriotti A, Vitale A. Plant Cell. 1997;9:1869–1880. doi: 10.1105/tpc.9.10.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demont-Caulet, N., Maillet, F., Tailler, D., Jacquinet, J.-C., Promé, J.-C., Nicolaou, K. C., Truchet, G., Beau, J.-M. & Dénarié, J. (1999) Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- 14.Samain E, Drouillard S, Heyraud A, Driguez H, Geremia R A. Carbohydr Res. 1997;302:35–42. doi: 10.1016/s0008-6215(97)00107-9. [DOI] [PubMed] [Google Scholar]
- 15.Drouillard S, Armand S, Davies G J, Vorgias E, Henrissat B. Biochem J. 1997;328:945–949. doi: 10.1042/bj3280945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roulleau F, Plusquellec D, Brown E. Tetrahedron Lett. 1983;24:4195–4196. [Google Scholar]
- 17.Schultze M, Quicklet-Sire B, Kondorosi E, Virelizier H, Glushka J N, Endre G, Gero S D, Kondorosi A. Proc Natl Acad Sci USA. 1992;89:192–196. doi: 10.1073/pnas.89.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Röhrig H, Schmidt J, Wieneke U, Walden R, Schell J, John M. In: Biological Fixation of Nitrogen for Ecology and Sustainable Agriculture. Legocki A, Bothe H, Puhler A, editors. Berlin: Springer; 1997. pp. 59–62. [Google Scholar]
- 19.Staehelin C, Schultze M, Kondorosi E, Mellor R B, Boller T, Kondorosi A. Plant J. 1994;5:319–330. [Google Scholar]
- 20.Philip-Hollingsworth S, Dazzo F B, Hollingsworth R. J Lipid Res. 1997;38:1229–1241. [PubMed] [Google Scholar]
- 21.Timmers A C J, Auriac M C, de Billy F, Truchet G. Development (Cambridge, UK) 1998;125:339–349. doi: 10.1242/dev.125.3.339. [DOI] [PubMed] [Google Scholar]
- 22.Basse C W, Fath A, Boller T. J Biol Chem. 1993;268:14724–14731. [PubMed] [Google Scholar]
- 23.Nümberger T, Nennstiel D, Jabs D, Sacks W R, Hahlbrock K, Scheel D. Cell. 1994;78:449–460. doi: 10.1016/0092-8674(94)90423-5. [DOI] [PubMed] [Google Scholar]
- 24.Tailler, D., Jacquinet, J.-C. & Beau, J.-M. (1994) J. Chem. Soc. Chem. Commun. 1827–1828.
- 25.Baureithel K, Felix G, Boller T. J Biol Chem. 1994;269:17931–17938. [PubMed] [Google Scholar]
- 26.Ito Y, Kaku H, Shibuya N. Plant J. 1997;12:347–356. doi: 10.1046/j.1365-313x.1997.12020347.x. [DOI] [PubMed] [Google Scholar]
- 27.Umemoto N, Kakitani M, Iwamatsu A, Yoshikawa M, Yamaoka N, Ishida I. Proc Natl Acad Sci USA. 1997;94:1029–1034. doi: 10.1073/pnas.94.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price N P J, Relic B, Talmont F, Lewin A, Promé D, Pueppke S G, Maillet F, Dénarié J, Promé J-C, Broughton W J. Mol Microbiol. 1992;6:3575–3584. doi: 10.1111/j.1365-2958.1992.tb01793.x. [DOI] [PubMed] [Google Scholar]