Abstract
Cereal yellow dwarf virus (CYDV) RNA has a 5′-terminal genome-linked protein (VPg). We have expressed the VPg region of the CYDV genome in bacteria and used the purified protein (bVPg) to raise an antiserum which was able to detect free VPg in extracts of CYDV-infected oat plants. A template-dependent RNA-dependent RNA polymerase (RdRp) has been produced from a CYDV membrane-bound RNA polymerase by treatment with BAL 31 nuclease. The RdRp was template specific, being able to utilize templates from CYDV plus- and minus-strand RNAs but not those of three unrelated viruses, Red clover necrotic mosaic virus, Cucumber mosaic virus, and Tobacco mosaic virus. RNA synthesis catalyzed by the RdRp required a 3′-terminal GU sequence and the presence of bVPg. Additionally, synthesis of minus-strand RNA on a plus-strand RNA template required the presence of a putative stem-loop structure near the 3′ terminus of CYDV RNA. The base-paired stem, a single-nucleotide (A) bulge in the stem, and the sequence of a tetraloop were all required for the template activity. Evidence was produced showing that minus-strand synthesis in vitro was initiated by priming by bVPg at the 3′ end of the template. The data are consistent with a model in which the RdRp binds to the stem-loop structure which positions the active site to recognize the 3′-terminal GU sequence for initiation of RNA synthesis by the addition of an A residue to VPg.
In vitro RNA-dependent RNA polymerase (RdRp) systems have been established for several plus-strand RNA plant viruses and, along with in vivo methods, have been useful in elucidating the mechanisms of the virus RNA replication. Generally the isolated RdRp complexes are comprised of the virus RdRp per se, other virus replication proteins, and host proteins. In many cases, these RdRps are template specific and are able to initiate RNA synthesis de novo at or near the 3′ end of the template (5). Some RdRp complexes or replication-associated proteins have been shown to bind to specific structures in the template that have roles in template selection, assembly of the replication complex, and initiation at the 3′ end of the template, e.g., in Brome mosaic virus (7, 27) and Tobacco mosaic virus (TMV) (20). Most of the studies with in vitro RdRp systems for plant plus-strand RNA viruses have involved viruses with a 7-methylguanosine cap structure at the 5′ end of the genomic RNA. There is no cap structure at the 5′ end of the minus-strand RNA, which is formed as the first stage of the replication process. Furthermore, formation of the cap structure is not required for the initiation of plus-strand RNA synthesis on minus-strand RNA templates in vitro (28), although capping-enzyme active-site mutations suggest roles for capping in RNA replication, in addition to roles in protection from exonuclease degradation and in translation (1).
A number of families of plant viruses, including the Comoviridae, Potyviridae, and some genera of the Luteoviridae, comprise viruses whose plus-strand RNA genomes lack a 7-methylguanosine cap structure but instead have a covalently linked virus-encoded protein (VPg) at the 5′ terminus. Viruses in some families of animal plus-strand RNA viruses, which are evolutionarily related to these plant viruses, also have a 5′-terminal VPg. The best studied of these is poliovirus, a member of the family Picornaviridae (9). During poliovirus replication, VPg acts as the primer for minus-strand RNA synthesis by the virus RdRp, using the plus strand as the template. A uridylylated form of VPg, VPg-UU, acts as the primer for plus-strand RNA synthesis using the minus strand as the template. VPg-UU synthesis is catalyzed by the RdRp, using an internal sequence, cre, in the plus strand as the template in a reaction which also requires a stem-loop structure at the 5′ terminus (15, 17). By analogy with poliovirus and other picornaviruses, it has been assumed that the plant virus VPgs are also needed as primers for RNA synthesis, but no evidence has been produced to support this assumption. It has been shown that the VPg of Potato virus A (family Potyviridae) can be uridylylated in vitro by the virus RdRp (NIb protein) (23), but the reaction did not require a template, and its relevance to virus RNA replication in vivo is not known.
Cereal yellow dwarf virus (CYDV) belongs to the genus Polerovirus in the family Luteoviridae (8). Its 5.7-kb plus-strand genome has a 5′-terminal VPg (16) and encodes six or seven proteins (Fig. 1) (21, 31). Proteins P0 (unknown function), P1 (contains a putative serine protease and the VPg), and P1-P2 (putative fusion frameshift protein with RdRp motifs in P2) are translated from the genomic RNA. The P1 and P1-P2 proteins are probably processed by the P1-encoded protease to produce the VPg, RdRp, and other proteins (12, 14). Proteins P3 (capsid protein), P4 (movement protein), and P3-P5 (putative readthrough aphid transmission protein) are translated from a 2.8-kb subgenomic RNA. P6 is a recently identified putative protein (21). We have recently described the synthesis in vitro of genomic-length double-stranded RNA (dsRNA) and subgenomic dsRNAs of 2.8 kbp and 0.7 kbp by a membrane-bound RNA polymerase isolated from oat plants infected with the New York strain of CYDV serotype RPV (CYDV-RPV-NY) (21). The RNA polymerase completed the synthesis of RNAs already initiated in vivo from endogenous RNA templates. The polymerase did not initiate synthesis de novo from added templates, and attempts to solubilize the enzyme led to loss of activity. Here we describe a CYDV in vitro system which is able to synthesize RNA from added plus-strand or minus-strand CYDV RNA templates and show that this synthesis is template specific and dependent on the addition of an added bacterially expressed VPg protein. Furthermore, synthesis of a minus strand on a CYDV plus-strand RNA template additionally required the presence of a putative stem-loop structure near the 3′ end of the template.
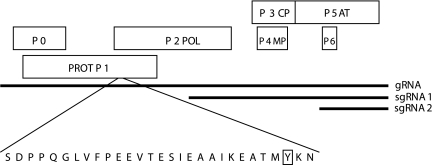
FIG. 1.
Genome organization of CYDV showing the location of the N-terminal amino acid sequence of the VPg (29 amino acids) with the most-5′-proximal tyrosine residue of the putative cleaved VPg in a box. The numbered boxes indicate ORFs. PROT, putative protease domain; POL, RNA-dependent RNA polymerase; CP, coat protein; MP, movement protein; AT, aphid transmission factor; gRNA, genomic RNA; sgRNA, subgenomic RNA.
MATERIALS AND METHODS
Virus source.
CYDV-RPV-NY (31) was obtained from S. Gray, USDA, Ithaca, New York. Oat plants (Avena bizantina cv. Coast Black) infected with CYDV-RPV-NY were supplied by J. Morris and I. Barker, Central Science Laboratory, York, United Kingdom.
Expression of the CYDV VPg region in Escherichia coli.
cDNA corresponding to the VPg coding region of open reading frame 1 (ORF1) was synthesized from CYDV RNA using primer VPg1 and Superscript III reverse transcriptase according to the manufacturer's instructions (Invitrogen) and amplified by PCR using Pfu Turbo DNA polymerase and primers VPg1 and CY10 (Table 1) as described by Osman and Buck (20). The PCR product was gel purified, cleaved with NdeI and XhoI, and cloned into the corresponding site of the vector pET28b in E. coli BL21 cells. The protein, containing an N-terminal His tag and designated bVPg, was expressed (29) and purified on a Ni2+-nitriloacetic acid column (32).
TABLE 1.
CYDV oligonucleotides used in this study
| Primer | CYDV nucleotide positionsa | Sequenceb |
|---|---|---|
| CY1 | c5723-5703 | CCCAAAGCTCCCAAGAGATCG |
| CY2 | c5723-5703 | GCCAAAGCTCCCAAGAGATCG |
| CY3 | c5723-5703 | TCCAAAGCTCCCAAGAGATCG |
| CY4 | c5723-5703 | AGCAAAGCTCCCAAGAGATCG |
| CY5 | c5723-5703 | AACAAAGCTCCCAAGAGATCG |
| CY6 | c5723-5703 | ATCAAAGCTCCCAAGAGATCG |
| CY7 | c5723-5703 | GGCAAAGCTCCCAAGAGATCG |
| CY8 | c5723-5703 | TCGAAAGCTCCCAAGAGATCG |
| CY9 | c5723-5703 | --CAAAGCTCCCAAGAGATCG |
| CY10 | c2285-2264 | TCGAGGCTCGAGGTTCAGCGGATGCCCCGACTTG |
| CY27 | 5401-5421 | CGAAATTAATACGACTCACTATAGAAGAGAGCTCAGCGTCCTCC |
| CY28 | c5723-5703 | ACCAAAGCTCCCAAGAGATCG |
| CY50 | c323-301 | CGAAATTAATACGACTCACTATAGTTTGGGTGTATCGGAAATCTGTT |
| CY51 | 1-25 | ACAAAGATTACCGAGGGGTGTCTTC |
| CY100 | c5723-5659 | ACCAAAGCTCCCAAGAGATCGGATAGAGAACCCTTCGAAATTCTTTTAAAGACAAGAACCTAGCC |
| CY101 | c5723-5659 | ACCCTTCGAGGACTCTCTACGGATAGAGAACCCTTCGAAATTCTTTTAAAGACAAGAACCTAGCC |
| CY102 | c5723-5659 | ACCAAAGCTCCCAAGAGATCGGAATCTCTCAAG-AGCTTCTTCTTTTAAAGACAAGAACCTAGCC |
| CY106 | c5723-5659 | ACCAAAGCTCCCAAGAGATTTTTATCTCTCAAGTAGCTTCTTCTTTTAAAGACAAGAACCTAGCC |
| CY108 | 5681-5702 | CGAAATTAATACGACTCACTATAGAAGAAGCTACTTGAGAGATTC |
| VPg1 | 1533-1554 | CAGCAGCTCCATATGTCCGACCCCCCCCAGGGATTG |
The nucleotide positions were taken from the complete CYDV sequence (EMBL/GenBank accession number L25299). Complementary sequences are prefixed by “c.”
Mutations in CYDV sequences are shown in bold. Deletions are indicated by dashes. The T7 promoter sequence is underlined. Non-CYDV sequences are shown in italics.
Template-dependent CYDV RdRp reactions.
A membrane-bound CYDV RNA polymerase was isolated from CYDV-infected oat plants as described previously (21) and treated with BAL 31 exonuclease followed by EGTA as described by Osman et al. (19). RNA polymerase reactions were carried out in the presence of [α-32P]UTP, plus unlabeled ATP, CTP, and GTP, and, when present, bVPg (2 μg), and the reaction products were extracted and analyzed by polyacrylamide gel electrophoresis (PAGE) in gels containing 8 M urea as described by Osman and Buck (18). When required, the RNA polymerase reaction products were treated with S1 nuclease as described by Bates et al. (4). Reactions to analyze the initiation of RNA synthesis by the addition of nucleotides to bVPg were carried out in the presence of [α-32P]ATP and, in some cases, unlabeled CTP and analyzed by sodium dodecyl sulfate (SDS)-PAGE as described previously (22). For use as templates, RNA was isolated from purified virions as described previously, namely, virions of CYDV (21), Cucumber mosaic virus (CMV)-Queensland strain (10), and Red clover necrotic mosaic virus (RCNMV)-Australian strain (4). Other templates were transcribed using T7 RNA polymerase from DNA amplified by reverse transcription-PCR from CYDV RNA using pairs of primers, one of which contained a T7 promoter (Table 1), as described previously (20, 21). The 323 3′-terminal nucleotides of CYDV plus- and minus-strand RNA were amplified using primer pairs CY27/CY28 and CY50/CY51, respectively. Mutants A, B, C, D, E, F, G, H, I, J, K, L, and M were amplified using CY27 as the forward primer and CY1, CY2, CY3, CY4, CY5, CY6, CY7, CY8, CY9, CY100, CY101, CY102, and CY106 as the reverse primers, respectively. For mutant N, the primer pair CY108/CY28 was used. The 278 3′-terminal nucleotides of the plus strand and 271 nucleotides of the minus strand of TMV strain L RNA, the two 3′ mutants of the TMV strain L plus strand (mutants 1 and 2), and the RNA transcript from the LITMUS 28 vector were produced as described previously (20). The template competition experiments were conducted as described previously (18, 20).
Antibodies and immunoblotting.
An antiserum against the CYDV bVPg protein was raised in a rabbit as described by Bates et al. (4). Membrane-bound extracts from CYDV-infected and healthy oat leaves were generated as described above, boiled in loading buffer plus SDS, and microcentrifuged for 10 min. The supernatants were subjected to electrophoresis in a 15% SDS-PAGE system alongside a protein molecular weight marker mixture (Sigma; ColorBurst). The proteins were then transferred to Hybond C Extra (GE Healthcare), the membranes were stained with Ponceau S, and the marker proteins were pencil marked. The membranes were incubated with the CYDV bVPg antiserum (1:5,000), and proteins to which antibodies had bound were detected and sized with a Renaissance chemiluminescence kit (Dupont-NEN).
RESULTS
Detection of free VPg in extracts of CYDV-infected oat plants.
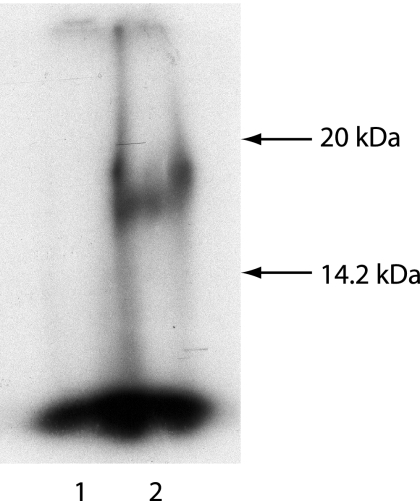
The CYDV VPg has been isolated from CYDV RNA by hydrolysis of the RNA and characterized as a protein with a mass of 17.5 kDa (16). However, there are no reports of the existence of free VPg in CYDV-infected plants, which would be expected if VPg acts as a primer for CYDV RNA synthesis in vivo. In order to detect free VPg, we first raised antibodies to the CYDV VPg. The proteolytic cleavage sites in the precursor P1 protein have not been determined, and therefore the precise N-terminal and C-terminal sequences of the VPg protein are not known. However, the N-terminal sequence of the CYDV VPg has been predicted to start at amino acid 403 of the P1 protein, by sequence alignment of the CYDV P1 protein with those of the VPgs of Potato leafroll virus and other related members of the genus Polerovirus whose N-terminal sequences have been determined (30). Therefore, a region of the P1 ORF starting at a position corresponding to amino acid 403 and terminating at the end of the P1 ORF was cloned into the vector pET28b and expressed in E. coli as a His-tagged protein. The purified bacterially expressed protein, designated bVPg, was used to raise an antiserum in a rabbit. When this antiserum was used in an immunoblot assay to probe a total-protein extract from CYDV-infected oat plants, a protein with a mass of about 17 kDa (Fig. 2, lane 2), which is similar in size to the VPg obtained from CYDV RNA (16), was detected. No partial cleavage products were detected in these experiments. No such protein was detected in protein extracts from healthy oat plants (Fig. 2, lane 1) or in CYDV-infected or healthy oat plants using preimmune serum (not shown).
FIG. 2.
Detection of VPg in protein extracts from CYDV-infected oat plants by immunoblotting. The total protein from healthy (lane 1) or CYDV-infected (lane 2) oat plants was subjected to SDS-PAGE and then blotted onto a membrane and probed with a CYDV bVPg antiserum. The migration positions and sizes of two proteins with known molecular sizes (Sigma; ColorBurst) which were coelectrophoresed in an adjacent lane are indicated by arrows.
Production of a CYDV template-dependent CYDV RdRp: requirement for added bVPg as a primer.
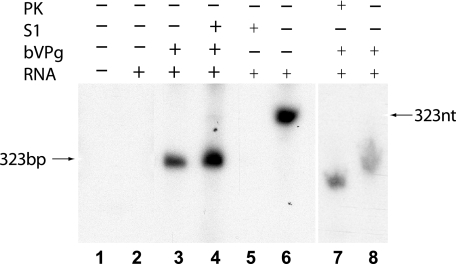
A membrane-bound CYDV RNA polymerase, extracted from CYDV-infected oat plants as described previously (21), was treated with the Ca2+-dependent nuclease BAL 31 to remove the endogenous RNA template. The nuclease was then inactivated by treatment with EGTA, which specifically chelates the Ca2+ ions. When the BAL 31-treated RNA polymerase was incubated with [32P]UTP, unlabeled ATP, CTP, and GTP, and Mg2+ ions and the extracted RNA was analyzed by PAGE, no labeled bands were detected, showing that the endogenous template had been removed (Fig. 3, lane 1). To test if the nuclease-treated RNA polymerase could copy added templates, the enzyme was programmed with an RNA corresponding to the 323 3′-terminal nucleotides of CYDV plus-strand RNA and incubated with [32P]UTP, unlabeled ATP, CTP, and GTP, and Mg2+ ions. Analysis of the extracted RNA by PAGE revealed that no radiolabeled RNA had been synthesized (Fig. 3, lane 2). When the nuclease-treated CYDV RNA polymerase was incubated with the RNA corresponding to the 323 3′-terminal nucleotides of the CYDV plus-strand RNA together with [32P]UTP, unlabeled nucleoside triphosphates, Mg2+, and added bVPg, a clear radiolabeled RNA was produced (Fig. 3, lane 3). The RNA product was resistant to treatment with S1 nuclease (Fig. 3, lane 4). Hence, it appeared that bVPg acted as a primer for minus-strand RNA synthesis on the plus-strand RNA template to form a dsRNA product. To demonstrate the efficacy of the S1 nuclease treatment and the differential mobilities of single-stranded RNA (ssRNA) and dsRNA in these experiments, [32P]UTP-labeled ssRNA transcripts corresponding to the 323 3′-terminal nucleotides of the CYDV minus-strand RNA (21) were coelectrophoresed in the above-described gels before (Fig. 3, lane 6) and after (Fig. 3, lane 5) SI nuclease treatment. A comparison of these lanes with the other lanes shown in Fig. 3 confirms that products of the CYDV RNA polymerase and added bVPg (Fig. 3, lanes 3 and 4) are dsRNA products and that dsRNAs have differential mobilities in the gel system employed compared to ssRNAs. When urea-containing gels are electrophoresed at room temperature, as in our experiments, they are not fully denaturing and dsRNA products migrate faster (Fig. 3, lanes 3 and 4) than the ssRNA equivalent (Fig. 3, lane 6) since they possess twice the charge, even though the molecules are similar in size. The addition of VPg to one RNA strand in the dsRNA product will retard the molecule in a neutral gel containing no SDS but insufficiently for it to have the same mobility as the ssRNA equivalent or to reverse the effect.
FIG. 3.
Requirement of added bVPg for template-dependent CYDV RdRp and demonstration of addition of bVPg to an S1-resistant dsRNA product. All RdRp reactions in lanes 1 to 4, 7, and 8 contained BAL 31-treated CYDV RdRp, [32P]UTP, unlabeled ATP, CTP, and GTP, and no further additions (lane 1), a 3′-terminal 323-nucleotide CYDV plus-strand RNA template (RNA) (lane 2), RNA plus bVPg (bVPg) with no treatment (lane 3) and with treatment with S1 nuclease (S1) (lane 4), and RNA plus bVPg and the product with no treatment (lane 8) and with treatment with proteinase K (PK) (lane 7). To confirm the efficacy of the SI nuclease treatment, [32P]UTP-labeled 323-nucleotide (323nt) 3′-terminal CYDV ssRNA transcripts generated as described in the text were left untreated (lane 6) or were treated (lane 5) with S1 nuclease. The reaction products were subjected to electrophoresis in 8% polyacrylamide gels containing 8 M urea and detected by autoradiography. The products of these reactions, which have differential mobilities as described in the text, are indicated by arrows.
To show that the bVPg primer is covalently bound to the RNA, the RNA polymerase reaction product was treated with proteinase K prior to PAGE. As expected, this led to an increase in the mobility of the product RNA (Fig. 3, compare lanes 7 and 8). To show that the priming was specific to bVPg, RNA polymerase reactions were set up with the 323-nucleotide CYDV plus-strand template using (i) bovine serum albumin and (ii) the bacterially expressed alpha subunit of chrysanthemum plastid-encoded RNA polymerase (kindly donated by I. C. Livieratos). No product could be detected in either case (not shown).
Template specificity of the CYDV RdRp.
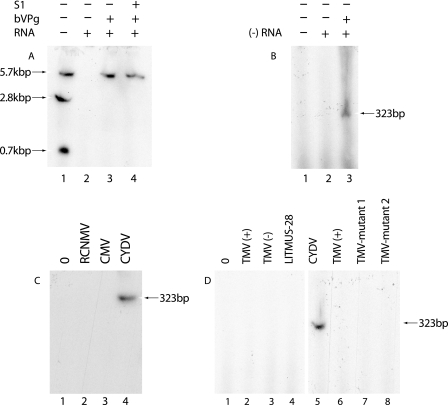
To determine if the CYDV RdRp could copy a full-length template, RNA polymerase reactions were set up in the presence of bVPg and CYDV virion RNA. A single S1 nuclease-resistant band was formed which had the same mobility as the 5.7-kbp genomic-length dsRNA formed by the non-BAL 31-treated CYDV RNA polymerase which utilizes the endogenous template (Fig. 4A, lanes 1 to 4). The 2.8- and 0.7-kbp subgenomic dsRNAs which were formed by the non-BAL 31-treated CYDV RNA polymerase (Fig. 4A, lane 1) (21) were not detected (Fig. 4A, lane 3). As in the previous experiments, [32P]UTP-labeled ssRNA transcripts corresponding to the 323 3′-terminal nucleotides of the CYDV minus-strand RNA (21) were simultaneously treated and analyzed in a separate gel before and after SI nuclease treatment, to confirm the efficacy of the S1 nuclease treatment (not shown). The CYDV dsRNA products in the gels were sized as in our previous investigation (21) with dsRNAs of Aspergillus foetidus and Penicillium stoloniferum viruses as the markers (4). A 323-nucleotide RNA corresponding to the 3′-terminal sequence of the CYDV minus strand was also an efficient template for the template-dependent CYDV RNA polymerase, and the reaction also required added bVPg (Fig. 4B, lanes 1 to 3).
FIG. 4.
(A) Template specificity of CYDV RdRp. RdRp reactions containing [32P]UTP were carried out either with a membrane-bound CYDV RNA polymerase containing endogenous CYDV RNA template (lane 1) or with BAL 31 nuclease-treated CYDV RNA polymerase with various added templates in the absence or presence of bVPg (lanes 2 to 4). The reactions were carried out as described in the text and included no added CYDV virion RNA (RNA) (lane 1), RNA only (lane 2), RNA plus bVPg (lane 3), or RNA plus bVPg and treatment with S1 nuclease (S1) prior to PAGE (lane 4). (B) Lane 1, no additions; lane 2, 323 3′-terminal nucleotides of CYDV minus-strand template RNA [(−) RNA]; lane 3, (−) RNA plus bVPg. Lane 1 in both panels C and D contained no RNA. Lanes 2 to 4 and 2 to 8, respectively, in panels C and D all contained bVPg and various virion RNAs—plus-stranded (+) or minus-stranded (−) virion RNA or DNA fragment templates as described in the text and shown in their abbreviated forms. All RNA products were analyzed by electrophoresis in 4% (A, C) or 8% (B, D) polyacrylamide gels containing 8 M urea. The sizes and mobilities of the products are described in the text and are indicated by arrows.
To determine if bVPg-primed RNA synthesis occurred on templates of unrelated viruses, CYDV RdRp reactions were set up using different virus RNAs. No bands were formed when the genomic RNA of two unrelated viruses, RCNMV and CMV, was added to the RNA polymerase reactions in the presence of bVPg, indicating that, as anticipated, the RNAs of these two viruses do not act as templates for the bVPg-primed CYDV RNA polymerase reactions (Fig. 4C, lanes 1 to 4). Similarly, TMV RNAs corresponding to the 278 3′-terminal nucleotides of the plus-strand RNA or the 271 3′-terminal nucleotides of the minus-strand RNA, which have been shown to act as efficient templates for a template-dependent TMV RNA polymerase (18), did not act as a template for the bVPg-primed CYDV RNA polymerase (Fig. 4D, lanes 1 to 3, 5, and 6). Further evidence to support the template specificity of the CYDV RNA polymerase was provided by the fact that a 273-nucleotide nonviral RNA corresponding to nucleotides 2466 to 2738 of the cloning vector LITMUS 28 (New England Biolabs) did not act as a template for the VPg-primed CYDV RdRp reaction (Fig. 4D, lane 4).
The 3′-terminal GU sequence is necessary but not sufficient for bVPg-primed template-dependent RNA polymerase reactions.
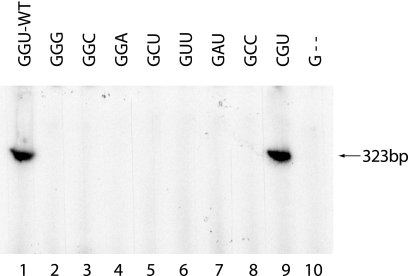
The 3′-terminal sequences of the CYDV plus and minus strands are GGU and UGU, respectively. In contrast, the 3′-terminal sequences of CMV, RCNMV, and TMV are CCA, CCC, and CCA, respectively. Since we have shown that the bVPg-primed CYDV template-dependent RNA polymerase reaction is template specific, we considered that the 3′-terminal sequence GU might be a structural requirement in the template. In order to test this possibility, the 3′-terminal GGU sequence of the RNA corresponding to the 323 3′-terminal nucleotides of the CYDV plus strand was mutated to GGG (mutant A), GGC (mutant B), GGA (mutant C), GCU (mutant D), GUU (mutant E), GAU (mutant F), GCC (mutant G), CGU (mutant H), and G (3′ GU deleted) (mutant I). These nine mutant RNAs were tested for their ability to act as templates in bVPg-primed CYDV RdRp reactions. The results are shown in Fig. 5. The mutation of GGU to CGU had no effect on the ability of the RNA (mutant H) to act as a template for the bVPg-primed RdRp reaction (Fig. 5, compare lanes 1 and 9). All the other mutations completely abolished the ability of the RNA to act as a template. Hence it appears that the 3′-terminal GU sequence is important for the CYDV plus strand to act as a template for the RdRp reaction.
FIG. 5.
Effects of 3′-terminal mutations on the template activity of a 323-nucleotide CYDV 3′-terminal plus-strand RNA. RdRp reactions were carried out in the presence of [32P]UTP and bVPg and various CYDV RNA templates. Lanes 1 to 10 contain wild-type (WT) or mutant CYDV 323-nucleotide RNA templates A to I (lanes 2 to 10, respectively) as shown above the lanes and as described in the text. The labeled RNA products were analyzed by PAGE in 8% gels and detected by autoradiography. The products and their size are indicated by the arrow.
In order to determine if the 3′-terminal GU sequence is the only structural requirement for an RNA to act as a template in the bVPg-primed RdRp reaction, two experiments were carried out. First, the 3′-terminal CCA sequence of the RNA corresponding to the 278 3′-terminal nucleotides of TMV RNA were mutated to CGU (mutant 1)and GGU (mutant 2). Neither of these mutants was able to act as a template for the CYDV bVPg-primed RdRp reaction (Fig. 4D, lanes 7 and 8). Second, poly(GU) was tested as a possible template. Reactions were carried out with the BAL 31 nuclease-treated CYDV RdRp in the presence of bVPg, [32P]ATP, and unlabeled CTP. The formation of the labeled product was monitored by PAGE and autoradiography and by trichloroacetic acid precipitation and scintillation counting. In none of the assays could any product be detected (not shown). Hence, a 3′-terminal GU sequence is not sufficient on its own for an RNA to act as a template for the CYDV bVPg-primed RdRp reaction.
A putative stem-loop structure at the 3′ end of CYDV RNA is required for the bVPg-primed RdRp reaction.
Folding analysis of the 323 3′-terminal nucleotides of CYDV plus-strand RNA with the mfold program, version 3.2 (11, 34), predicted a stable stem-loop structure (Fig. 6A). The structure has a UCCG tetraloop, three GU base pairs, and a bulge at an A residue. Although we have not verified the structure experimentally, the prediction appears to be robust, as the structure is predicted to occur in several alternative forms with similar free energies and in CYDV 3′-terminal sequences of lengths ranging from 40 to 500 nucleotides. The calculated ΔG value of the 43-nucleotide structure shown in Fig. 6A is −18.9 kcal/mol. To determine if this predicted stem-loop structure is necessary for the VPg-primed CYDV RdRp structure, several mutations to disrupt various features of the structure within the RNA corresponding to the 323 3′-terminal nucleotides of CYDV plus-strand RNA were made and their effect on the ability of the RNA to act as a template in RdRp reactions was tested. The mfold program did not predict any significant secondary structures for the 3′ terminus of the CYDV minus-strand RNA.
FIG. 6.
Predicted structures of the 43 3′-terminal nucleotides of the CYDV plus-strand wild-type RNA (A) and the compensated CYDV RNA mutant K (B). Both structures were produced using the mfold program, version 3.2 (11, 34).
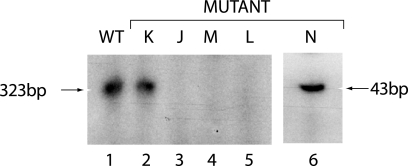
First, all the bases on the left side of the stem (nucleotides 5705 to 5720) were mutated to UUUCGAAGGGUUCUCUA, which the mfold program predicted would completely disrupt the stem structure (mutant J). This mutation completely abolished the template activity of this RNA (Fig. 7, lane 3). Compensatory mutations were then introduced into mutant J to reform the stem structure, but with bases on each side of the stem which are complementary to those present in the wild-type sequence, to produce mutant K. The ΔG value of the 43-nucleotide structure shown in Fig. 6B, calculated with the mfold program, is −18.3 kcal/mol, similar to that of the wild-type structure. In the bVPg-primed CYDV RdRp reaction, the amount of product formed with mutant K as the template was similar to that formed by the wild-type RNA (Fig. 7, lane 2). A further mutation to delete the A bulge to produce a completely base-paired stem (mutant L) completely abolished the template activity of the RNA (Fig. 7, lane 5). Similarly, mutation of the UCCG tetraloop from UCCG to AAAA (mutant M) also abolished the template activity of the RNA (Fig. 7, lane 4). Finally, nucleotides 5401 to 5680 were deleted to generate a 43-nucleotide RNA which retains the stem-loop structure (mutant N) (Fig. 6A). This RNA acted as an efficient template for the bVPg-primed CYDV RdRp (Fig. 7, lane 6). When the product of this reaction was coelectrophoresed with a labeled ssRNA transcript corresponding to the 3′-terminal 43 nucleotides of the CYDV minus-strand RNA (21) in a 12% polyacrylamide gel, the ssRNA migrated more slowly than its dsRNA equivalent (not shown). Products of this size have to be electrophoresed in higher-strength acrylamide gels so that they are not electrophoresed off the gels, but a comparison of the migrational characteristics of the ssRNA transcript and the dsRNA product shows that those of the ssRNA transcript are similar to those shown and discussed for the larger products shown in Fig. 3. It was concluded that the predicted 3′-terminal stem-loop structure, along with the 3′-terminal GU sequence, is the required structural element for an RNA to act as a template for the bVPg-primed CYDV RdRp reaction in vitro.
FIG. 7.
Effects of mutations in the predicted 3′-terminal stem-loop structure on the template activities of CYDV plus-strand RNAs. RdRp reactions were carried out and the products analyzed as described in the legend to Fig. 5. Lane 1 contains wild-type (WT) RNA, and lanes 2 to 6 contain mutant RNAs K, J, M, L, and N, respectively, as described in the text. An 8% gel was used for lanes 1 to 5 and a 12% gel for lane 6. The products and their sizes are indicated by arrows.
To determine whether the 3′-terminal GU sequence or the upstream structure is required for the binding of the CYDV RdRp to its template, competition experiments (6, 19, 25, 26) were carried out. Here bVPg-primed RdRp reactions were carried out using the RNA corresponding to the 323 3′-terminal nucleotides of CYDV plus-strand RNA as a template in a constant concentration (12.5 nM) and increasing concentrations of competitor RNA (mutant A). The amount of product formed was assayed by PAGE and quantified by phosphorimaging. The molar concentration of the competitor necessary to reduce minus-strand RNA synthesis from the wild-type template by 50% (IC50) for mutant A was calculated to be 12.0 nM. This is similar to the IC50 values of efficient competitors of TMV minus-strand RNA synthesis (19). The results show that mutant A, which has a GGG 3′-terminal sequence and is unable to act as a template for the bVPg-primed CYDV RdRp, is nevertheless able to bind to the RdRp complex. Taken together with the results of the mutational analysis of the stem-loop structure, it may be concluded that the stem-loop structure, not the 3′-terminal GU sequence, is the main structural element responsible for the binding of the RdRp complex to the RNA template.
Evidence that bVPg primes CYDV minus-strand RNA synthesis at the 3′ terminus of the template RNA.
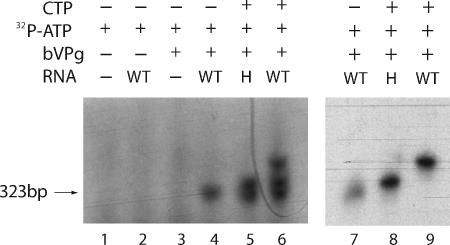
Free poliovirus VPg is believed to act as the primer for minus-strand RNA at the poly(A) tail at the 3′ end of poliovirus plus-strand RNA (15). However, priming of poliovirus plus-strand RNA synthesis requires the formation of VPg-UU, the synthesis of which is carried out using an internal cre sequence as the template. The VPg-UU sequence is then transported to the 3′-terminal sequence (AA) of the negative-strand RNA template to initiate plus-strand synthesis (15, 17). To obtain evidence as to whether bVPg initiates CYDV minus-strand RNA synthesis directly or forms a VPg-A or VPg-AC primer using an internal sequence as the template, bVPg-primed CYDV RdRp reactions were carried out with the RNA template corresponding to the 323 3′-terminal nucleotides of the CYDV plus-strand RNA and [α-32P]ATP as the only nucleoside triphosphate. The reaction product was analyzed by SDS-PAGE. A single radiolabeled band was formed (Fig. 8, lane 4); it migrated slightly more slowly than the free bVPg which was coelectrophoresed in an adjacent lane, stained with Coomassie blue, and spiked with [α-32P]ATP for later comparison with the VPg product (not shown). This band was not formed in the absence of the template or bVPg (Fig. 8, lane 1), the presence of the template and the absence of bVPg (Fig. 8, lane 2), or the absence of the template and the presence of bVPg (Fig. 8, lane 3). This indicates that the CYDV RdRp complex is able to add one or more A residues to bVPg in a template-dependent reaction. To obtain more information, a bVPg-primed RdRp reaction using the same template was carried out with [32P]ATP and unlabeled CTP (10 μM). Analysis of the reaction products by SDS-PAGE showed that three bands had been formed, one in the position of the band synthesized using [32P]ATP alone and two slower bands (Fig. 8, lane 6). The 3′-terminal sequence of CYDV RNA is CUUUGGU. The results are consistent with the formation of VPg-A in reactions containing [32P]ATP only and the formation of VPg-A, VPg-AC (and/or VPg-ACC), and VPg-ACCAAA in reactions containing [32P]ATP and unlabeled CTP with the reactions starting at the 3′ terminus of the template RNA. However, since the template also contained internal sequences containing U followed by G and then further U residues, the results did not prove unequivocally that the reaction did not use an internal sequence as a template.
FIG. 8.
Initiation of CYDV minus-strand RNA synthesis by addition of nucleotides to bVPg. The reactions were carried out using BAL 31 nuclease-treated CYDV RdRp, and the reaction products were analyzed by SDS-PAGE (10% gels) and autoradiography. The template, when present, was an RNA corresponding to the 323 3′-terminal nucleotides of wild-type CYDV RNA plus strand (WT) or mutant H CYDV RNA (H). Lanes 1 to 6 contained 10 μM [α-32P]ATP, and lanes 7 to 9 contained 250 μM [α-32P]ATP. Lanes 5 and 6 contained 10 μM CTP, and lanes 8 and 9 contained 250 μM CTP. The fastest-migrating products are indicated by an arrow.
Further evidence was obtained using mutant H, which has a 3′-terminal CGU sequence and has been shown to act as an efficient template in bVPg-primed CYDV RdRp reactions. In bVPg-primed reactions with mutant H as the template and [32P]ATP and unlabeled CTP (10 μM), only two bands were formed, with similar mobilities to the slower of the two bands formed using the wild-type template (Fig. 8, lane 5). It is likely that these two bands represent bVPg-A and bVPg-AC and resulted from template reactions from the 3′-terminal GU sequence. Since the only difference between the wild-type sequence and mutant H lies in the 3′-terminal sequence, the formation of three bands from the wild-type sequence and only two bands from mutant H is consistent with initiation at the 3′ terminus of the template and cannot easily be explained by initiation at an internal sequence.
The above-described reactions were carried out in low concentrations of ATP and CTP (10 μM) so that reaction intermediates, as well as the longest possible products, could be detected. In order to drive the reactions to completion, the concentrations of [32P]ATP and unlabeled CTP were increased to 0.25 mM. In reactions with [32P]ATP only and the wild-type template, the single faster-moving band was formed, as before (Fig. 8, lane 7). However in reactions with [32P]ATP and unlabeled CTP, only the slowest-moving band was formed with the wild-type template (Fig. 8, lane 9) and only the intermediate band was formed with the mutant H template (Fig. 8, lane 8). The results are consistent with the identities of the three bands as bVPg-A (Fig. 8, lane 7), bVPg-AC (Fig. 8, lane 8), and bVPg-ACCAAA (Fig. 8, lane 9) and provide additional evidence that the bVPg-primed reactions initiate at the 3′ terminus, not on an internal sequence, of the template RNA.
DISCUSSION
The results show that synthesis of CYDV minus-strand RNA by the template-dependent RdRp in vitro requires an added VPg primer, a 3′-terminal GU sequence, and a stem-loop structure near the 3′ terminus of the plus-strand template. The base-paired stem (but not its sequence), a single-nucleotide (A) bulge in the stem, and the sequence of the tetraloop were all essential features of the structure of the stem-loop. The evidence indicates that the initiation of minus-strand RNA synthesis by the addition of nucleotides to VPg requires a template and starts at the non-base-paired 3′ terminus of the plus-strand template by the formation of VPg-A. The binding of the RdRp to the plus-strand template did not require the 3′-terminal GU sequence. The data are consistent with a model in which the CYDV RdRp binds to the stem-loop structure at the 3′ end of the CYDV plus-strand RNA, which positions the catalytically active site of the enzyme close to the essential 3′-terminal GU sequence for the initiation of minus-strand synthesis by the formation of VPg-A and VPg-AC, followed by elongation and copying of the entire template strand.
We have utilized a bacterially expressed VPg (bVPg) for these studies. VPg is likely generated in vivo by cleavage of the P1 polyprotein by a P1-encoded serine protease, but the precise cleavage sites have not been determined. The N-terminal sequence could be predicted with a high degree of confidence based on sequence comparisons with the experimentally determined N-terminal sequences of the VPgs of related poleroviruses, an alignment of which has been shown previously by van der Wilk et al. (30). However, the likely C terminus could only be predicted based on the approximate mass (17.5 kDa) of the VPg isolated from CYDV RNA. To make sure that all of the VPg sequence was present, we extended the sequence in the bVPg to the end of the P1 ORF. We therefore cannot be certain that the priming activity of bVPg lies in viral sequences beyond the end of the VPg formed in vivo. However, this seems unlikely. First, although the VPg could be linked to serine, threonine, or tyrosine residues, the VPgs of picornaviruses and potyviruses are known to be linked to tyrosine residues (2, 3, 24). If the CYDV VPg is also linked by tyrosine, there is only one tyrosine residue between the start of the predicted VPg sequence and the end of the P1 ORF. This is located at amino acid 27 in the predicted VPg sequence (shown in Fig. 1). It is possible that that adenylation of the VPg may occur at this tyrosine residue, but this clearly requires experimental verification. Second, two unrelated proteins which contain several serine, threonine, and tyrosine residues did not act as primers for the CYDV RdRp.
It is noteworthy that when presented with a genomic-length CYDV plus-strand template, the RdRp produced only a genomic-length minus strand in the form of a genomic-length dsRNA. Whether genomic-length dsRNA or ssRNA is the true intermediate of replication, and whether dsRNA is an artifact of the in vitro system, has not yet been resolved unequivocally for any plant or animal plus-strand RNA virus (5). However, the absence of any subgenomic minus-strand RNAs as products of the CYDV RdRp reaction, either as dsRNA, ssRNA, or a product that is partially dsRNA and partially ssRNA, is worthy of comment. Several mechanisms for the formation of subgenomic RNAs of plus-strand RNA viruses have been described, including the synthesis of subgenomic plus-strand RNAs from subgenomic promoters in genomic-length minus-strand RNA and the premature termination of minus-strand RNA synthesis to create separate subgenomic minus-strand RNA templates for subgenomic plus-strand RNA synthesis (13, 33). There was no evidence for premature termination of CYDV minus-strand RNA synthesis in the in vitro RdRp system studied here.
This is the first report of VPg-primed in vitro synthesis by a positive-stranded RNA plant virus. It will open the way for further in vitro and in vivo studies of the replication of CYDV and other VPg-containing plant viruses. In poliovirus, it is likely that plus- and minus-strand RNA syntheses take place by different mechanisms (cre dependent and independent, respectively) (15, 17), and in CYDV it would appear that the RNA structural requirements for the initiation of plus- and minus-strand syntheses also differ. Questions that remain to be answered include what structural features in negative-strand RNA templates are required for VPg-primed genomic plus-strand RNA synthesis, what comprises the regulation and mechanism of genomic and subgenomic plus-strand RNA synthesis, and what is the nature of the interactions of the VPg and RdRp with each other and with their RNA templates.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Ahola, T., J. A. den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. [PubMed] [Google Scholar]
- 3.Anindya, R., S. Chittori, and H. S. Savithri. 2005. Tyrosine 66 of Pepper vein banding virus genome-linked protein is uridylylated by RNA-dependent RNA polymerase. Virology 336:154-162. [DOI] [PubMed] [Google Scholar]
- 4.Bates, H. J., M. Farjah, T. A. M. Osman, and K. W. Buck. 1995. Isolation and characterization of an RNA-dependent RNA polymerase from Nicotiana clevelandii plants infected with red clover necrotic mosaic dianthovirus. J. Gen. Virol. 76:1483-1491. [DOI] [PubMed] [Google Scholar]
- 5.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, M. R., and C. C. Kao. 1999. A minimal promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., A. Noueiry, and P. Ahlquist. 2003. An alternate pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J. Virol. 77:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Arcy, C. J., and L. Domier. 2005. Luteoviridae, p. 889-898. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy, VIIIth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 9.Flanegan, J. B., and D. Baltimore. 1977. Poliovirus-specific primer-dependent RNA polymerase is able to copy poly(A). Proc. Natl. Acad. Sci. USA 74:3677-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363-368. [DOI] [PubMed] [Google Scholar]
- 11.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 12.Miller, W. A., S. P. Dinesh-Kumar, and C. P. Paul. 1995. Luteovirus gene expression. Crit. Rev. Plant Sci. 14:179-211. [Google Scholar]
- 13.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 14.Miller, W. A., and L. Rasochova. 1997. Barley yellow dwarf viruses. Annu. Rev. Phytopathol. 35:167-190. [DOI] [PubMed] [Google Scholar]
- 15.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, J. F., C. J. D'Arcy, and J. M. Clark. 1989. Barley yellow dwarf virus has a 5′-terminal genome-linked protein. J. Gen. Virol. 70:2253-2256. [Google Scholar]
- 17.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman, T. A. M., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osman, T. A. M., C. L. Hemenway, and K. W. Buck. 2000. Role of the 3′ tRNA-like structure in tobacco mosaic virus minus-strand RNA synthesis by the viral RNA-dependent RNA polymerase in vitro. J. Virol. 74:11671-11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osman, T. A. M., and K. W. Buck. 2003. Identification of a region of the tobacco mosaic virus 126- and 183-kilodalton replication proteins which binds specifically to the viral 3′-terminal tRNA-like structure. J. Virol. 77:8669-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osman, T. A., J. Morris., R. H. Coutts, and K. W. Buck. 8 June 2006. Synthesis of genomic and subgenomic RNAs by a membrane-bound RNA-dependent RNA polymerase isolated from oat plants infected with Cereal yellow dwarf virus. Arch. Virol. [Epub ahead of print.] [DOI] [PubMed]
- 22.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puustinen, P., and K. Mäkinen. 2004. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 279:38103-38110. [DOI] [PubMed] [Google Scholar]
- 24.Rothberg, P. G., T. J. R. Harris, A. Nomoto, and E. Wimmer. 1978. O4-(5′-uridylyl) tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc. Natl. Acad. Sci. USA 75:4868-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel, R. W., S. Atkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel, R. W., L. Bellon, L. Beigelman, and C. C. Kao. 1998. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 95:11613-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivakumaran, K., S.-K. Choi, M. Hema, and C. C. Kao. 2004. Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J. Virol. 78:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivakumaran, K., C. H. Kim, R. Tayon Jr., and C. C. Kao. 1999. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier, F. W., A. H. Rosenburg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 30.van der Wilk, F., M. Verbeek, A. M. Dullemans, and J. F. J. M. van den Heuvel. 1997. The genome-linked protein of potato leafroll virus is located downstream of the putative protease domain of the ORF1 product. Virology 234:300-303. [DOI] [PubMed] [Google Scholar]
- 31.Vincent, J. R., R. M. Lister, and B. A. Larkins. 1991. Nucleotide sequence analysis and genome organization of the NY-RPV isolate of barley yellow dwarf virus. J. Gen. Virol. 72:2347-2355. [DOI] [PubMed] [Google Scholar]
- 32.Walz, A.-C., B. Demel, B. de Kruijff, and R. Matzel. 2002. Aerobic sn-glycerol-3-phosphate dehydrogenase from Escherichia coli binds to the cytoplasmic membrane through an amphipathic helix. Biochem. J. 365:471-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304:147-154. [DOI] [PubMed] [Google Scholar]
- 34.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]