Abstract
Studies to localize the herpes simplex virus 1 portal protein encoded by UL6, the putative terminase components encoded by UL15, UL 28, and UL33, the minor capsid proteins encoded by UL17, and the major scaffold protein ICP35 were conducted. ICP35 in B capsids was more resistant to trypsin digestion of intact capsids than pUL6, pUL15, pUL17, pUL28, or pUL33. ICP35 required sectioning of otherwise intact embedded capsids for immunoreactivity, whereas embedding and/or sectioning decreased the immunoreactivities of pUL6, pUL17, pUL28, and pUL33. Epitopes of pUL15 were recognized roughly equally well in both sectioned and unsectioned capsids. These data indicate that pUL6, pUL17, pUL28, pUL33, and at least some portion of pUL15 are located at the external surface of the capsid.
Capsids form in the nuclei of cells infected with all herpesviruses. Herpes simplex virus (HSV) capsid pentons and hexons form spontaneously from five and six molecules of ICP5, respectively; these capsomeres are linked by triplexes consisting of two molecules of VP23 and one molecule of VP19C to form a porous procapsid (23, 36, 43). ICP5 is also associated with ICP35, which forms an internal shell or scaffold within the procapsid. The procapsid is believed to give rise to the three other types of capsids seen in HSV-infected cells, designated types A, B, and C. All of these capsids differ internally but contain identical outer shells, as determined by cryoelectron microscopy (21, 35, 49). Type B capsids retain the scaffold internal to the outer shell, type A capsids contain only the outer shell, and type C capsids lack the internal scaffold but contain viral DNA (14). Type C capsids then bud from the nuclear membrane in a reaction termed primary envelopment (19, 32).
One of the vertices of A, B, and C capsids is biochemically and structurally unique and has been designated the portal vertex. Thus, the UL6-encoded protein (pUL6) forms a dodecameric ring with an internal diameter of at least 65 Å, i.e., sufficiently wide to accommodate DNA as it is packaged into the capsid (44). Critical to the discovery of the portal was the observation that an antibody to the C terminus of pUL6 recognized epitopes on a single vertex of type B capsids, thus showing that at least the C terminus of pUL6 is located at the capsid exterior in a position to access incoming viral DNA (22, 39).
It has also been shown that HSV-1 B capsids contain a number of capsid proteins in addition to triplexes, pUL6, ICP5, and ICP35. These proteins include approximately 1.2 copies of pUL15, 2.4 copies of pUL28, 27 to 42 copies of pUL25, 19.2 copies of pUL17, and an undetermined amount of pUL33 (6, 7, 15, 25, 26, 33, 41, 42, 48). By analogy to extensive studies of bacteriophage capsid assembly, it might be predicted that some of these minor capsid proteins would be involved in processing concatameric DNA and threading the DNA into the portal through the hydrolysis of ATP (9). Such a complex, termed the terminase, remains somewhat enigmatic in HSV, but a variety of indirect evidence suggests that it comprises at least the UL15, UL28, and UL33 proteins. Specifically, (i) all three proteins are among seven proteins required for viral DNA packaging (3, 29, 40); (ii) like other terminases, the UL15 protein contains a conserved P-loop ATPase motif, and mutation of this motif precludes DNA packaging (12, 20, 47); (iii) the UL28 protein can specifically bind DNA sequences known to be required for the correct cleavage of concatameric viral DNA (2); (iv) the UL15 and UL28 proteins interact directly, whereas pUL33 binds pUL28 and enhances the pUL15-pUL28 interaction in coimmunoprecipitation assays (1, 8, 17, 46); and (v) in vitro, both pUL28 and pUL15 can interact with the portal protein encoded by UL6 (45).
Recent immunogold analysis of pUL17 and pUL25 supports their location on the external surface of the viral capsid on more than one vertex (24, 41). Although it is also required for DNA packaging, the precise function of UL17 is unknown (34). Analysis of a UL17 deletion mutant revealed an alteration of the normal intranuclear distributions of capsids and a number of viral proteins including pUL6, ICP35, and ICP5 (39). These observations suggest that the UL17 protein is involved in ensuring proper capsid assembly, the reorganization of the infected cell nucleus, or, directly or indirectly, capsid or protein transport within the nucleus. Relevant to this last possibility is the observation that HSV capsids are actively transported in the nucleus and that this transport is both energy and actin dependent (13). UL25 is believed to enhance the stability of capsids and is required for the retention of full-length genomic DNA in the capsid (18, 24, 37, 41).
The hypotheses that pUL6 serves as the portal, the UL15, UL28, and UL33 proteins form the HSV terminase, and pUL17 acts directly or indirectly to mediate the transport of capsids within the nucleoplasm predict that at least portions of these proteins would localize on the external surfaces of capsids. This study was undertaken to test these possibilities.
Production and specificity of a novel chicken antiserum against pUL17.
Because a previously described UL17 antibody produced using a DNA vaccine did not recognize small quantities of UL17 protein (34) (data not shown), UL17 was fused to DNA encoding a His6 tag, and a recombinant baculovirus that expressed the fusion protein was generated. The UL17 fusion protein was purified from lysates of insect cells infected with the recombinant baculovirus by affinity chromatography on Ni2+-containing Sepharose beads as described previously (7). Immunization of chickens with the purified fusion protein was followed by purification of immunoglobulin Y (IgY) from the eggs of immunized hens as described previously (31).
To test the antisera for specificity, Hep-2 cells were mock infected or infected with HSV-1(F) or a UL17 null virus at a multiplicity of infection of 5 PFU per cell. Lysates from approximately 1.3 × 106 cells were prepared by denaturation and boiling in 1% sodium dodecyl sulfate (SDS) and 5 mM β-mercaptoethanol. Denatured proteins were electrophoretically separated on an 8% SDS-polyacrylamide gel and transferred electrically to a nitrocellulose membrane. The nitrocellulose membrane was blocked overnight at 4°C in phosphate-buffered saline (PBS) supplemented with 5.0% milk and 0.2% Tween 20. The membrane was rinsed twice with room-temperature PBS and 0.2% Tween 20 and was further blocked by immersion in a 1:10 dilution of Block Hen (Aves Labs) for 15 min. Polyclonal anti-UL17 IgY was diluted 1:500,000 into PBS with 1% bovine serum albumin and 0.2% Tween 20 and applied overnight. After extensive washes in PBS containing 0.2% Tween 20, the membrane was incubated with horseradish peroxidase-conjugated anti-chicken antibody diluted 1:5,000 in PBS containing 5.0% milk and 0.5% Tween 20. Bound IgY was detected using a 5-min incubation with ECL Plus reagents (Amersham) and flash exposure to Fuji autoradiographic film.
As shown in Fig. 1, the anti-pUL17 antiserum recognized a protein with an apparent Mr of 79,000 that was not present in lysates of mock-infected Hep-2 cells or cells that were infected with the UL17 deletion virus. This size is consistent with a previous study reporting an apparent Mr of 77,000 in virions (34). We did not detect the protein with an apparent Mr of 72,000 that was previously identified in virion lysates by mass spectrometry, suggesting that the smaller protein may be highly enriched in virion preparations (34). The antibody also recognized pUL17 protein expressed by a recombinant baculovirus (data not shown).
FIG. 1.
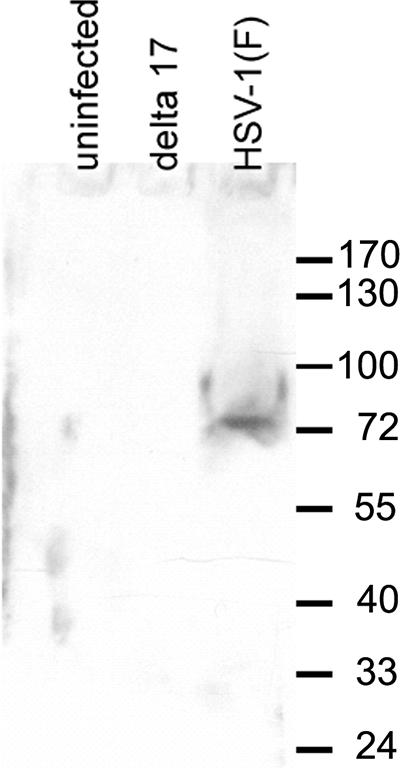
Digitally scanned image of immunoblot of lysates of infected and uninfected cells that reacted with pUL17-specific antiserum. Lysates of cells that were uninfected (left lane) or infected with a UL17 deletion virus (center lane) or wild-type virus HSV-1(F) (right lane) were electrophoretically separated on an SDS-polyacrylamide gel, transferred onto nitrocellulose, and reacted with purified IgY obtained from a chicken immunized with purified pUL17. Bound IgY was revealed as indicated in the text. Sizes of molecular weight markers are indicated to the right in thousands.
Immunogold labeling of wild-type and mutant capsids.
Using standard procedures (28), capsids were purified from nuclear lysates of cells infected with wild-type HSV-1(F) and viruses respectively lacking the UL6, UL15, UL17, UL28, and UL33 genes (5, 11, 27, 34, 40). Briefly, nuclear lysates prepared from 1.5 × 108 Vero cells infected with HSV-1(F) were clarified, and capsids were pelleted through a 35% sucrose cushion in an SW28 rotor. The pellet in resuspended material was subjected to rate-zonal centrifugation through a 20 to 50% (wt/vol) sucrose gradient in a Beckman SW41 rotor at 24,500 rpm for 1 h. A light-refracting band in the middle of the gradient containing B capsids was collected using a Pasteur pipette, and capsids were diluted into a solution containing Tris-HCl (pH 7.8), 150 mM NaCl, and 1 mM EDTA (TNE). Capsids were either attached to Formvar carbon-coated electron microscopic grids or placed into microdialysis tubes (200-μm diameter), subsequently embedded in LRWhite, and sliced with a diamond knife into 60-nm sections that were then placed onto electron microscopy grids.
Previously described rabbit antisera directed against the C terminus of pUL6, the C terminus of pUL15, full-length pUL28, and full-length pUL33 were prepared by adsorption against capsids purified from Vero cells infected with 5.0 PFU/ml of the appropriate viral null mutant (4, 5, 27, 30, 38-40). The adsorbed antisera were then diluted 1:50 in PBS supplemented with 1% Triton-100 and 1% fish gelatin and applied directly to the electron microscopy grids, followed by extensive washing. Experiments performed with the pUL17-specific chicken IgY were similar except that the antibody was not preadsorbed and was diluted 1:5,000 for reactions with capsids. As a control, the capsid samples were also reacted separately with a polyclonal antiserum directed against the internal scaffold protein ICP35 (10) (NC 3-4) (kindly provided by Roselyn Eisenberg and Gary Cohen). Bound immunoglobulins remaining after the washing were recognized by goat anti-rabbit immunoglobulin conjugated to 12-nm gold beads or goat anti-chicken IgY conjugated to 12-nm gold beads. After further washing, the grids were viewed using a Philips 201 electron microscope after counterstaining with 2% aqueous uranyl acetate and 0.5% Reynold's lead citrate. Only capsids that were visually verified as intact B capsids were included in the data. The B capsids were scored as positively immunolabeled only when a gold bead was observed in direct association with the capsid shell or interior.
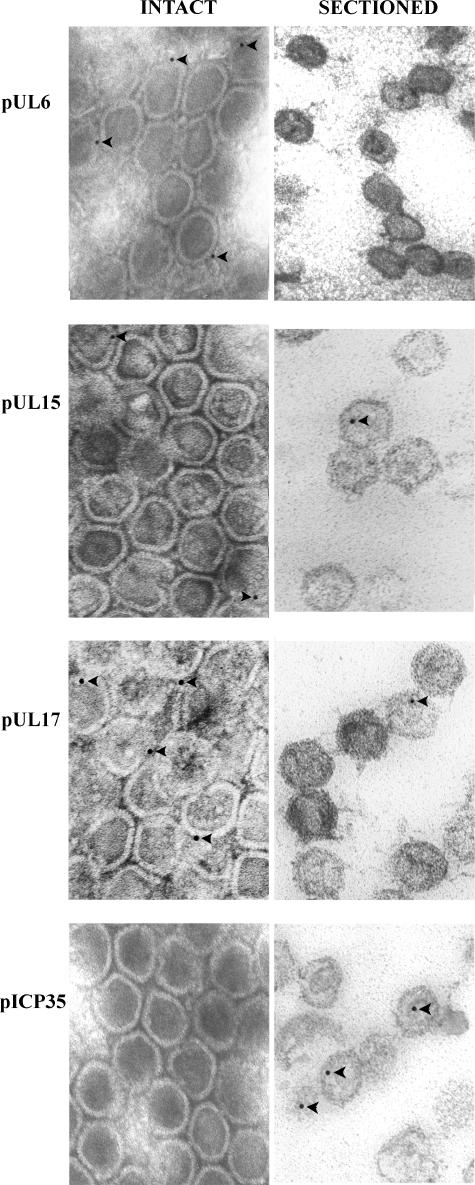
The results are summarized in Table 1, and representative examples of immunostained capsids are shown in Fig. 2.
TABLE 1.
Percentages of capsids immunolabeled with monospecific antiseraa
| Antibody | Virus | % of capsids immunolabeled with antiserum (no. of immunolabeled capsids/no. of capsids examined)
|
|
|---|---|---|---|
| Intact | Sectioned | ||
| ICP35 | HSV-1(F) | 1.8 (18/1007)c | 27.8 (115/413)c |
| pUL6b | HSV-1(F) | 16.3 (53/325)d | 4.0 (48/1181)d |
| pUL6 | UL6− | 0.4 (4/945) | 0.3 (3/929) |
| pUL17b | HSV-1(F) | 13.5 (97/714)d | 5.4 (134/2,447)d |
| pUL17 | UL17− | 0.0 (0/1,000) | 0.2 (2/969) |
| pUL33b | HSV-1(F) | 8.8 (25/281)d | 2.1 (35/1,644)d |
| pUL33 | UL33− | 0.8 (9/1,155) | 0.3 (4/1,042) |
| pUL28b | HSV-1(F) | 8.7 (33/378)e | 4.4 (43/960)e |
| pUL28 | UL28− | 0.7 (8/1,074) | 0.3 (3/973) |
| pUL15Cb | HSV-1(F) | 4.7 (40/847)f | 5.8 (35/595)f |
| pUL15C | UL15− | 1.0 (6/571) | 0.7 (2/1,000) |
Capsids were purified from cells infected with wild-type viruses or viruses lacking the indicated open reading frames (−). In some experiments (intact), capsids were attached to grids and reacted with the indicated antibodies. In other experiments, capsids were embedded and sectioned, followed by reaction of the thin sections with the indicated antibodies. The number of immunolabeled capsids versus the number of capsids examined is indicated in parentheses, and the resulting percentage of labeled capsids is shown. All P values were obtained with Fisher's exact t test.
The amount of immunoreactivity with a given antibody was greater (P < 0.001) in wild-type capsids than in the corresponding deletion mutant capsids.
The immunoreactivity of sectioned capsids was greater than that of intact capsids (P < 0.001).
The immunoreactivity of intact capsids was greater than that of sectioned capsids (P < 0.001).
The immunoreactivty of intact capsids was greater than that of sectioned capsids (P = 0.01).
The immunoreactivity of sectioned capsids compared to that of intact capsids was not statistically different (P = 0.06).
FIG. 2.
Digital image of representative electron micrographs of capsids labeled with various antibodies. Capsids were purified from cells infected with HSV-1(F). These were attached to copper mesh grids (left column) or were embedded in Lowicryl and sectioned (right column). Each row shows intact capsids and thin sections that reacted with antisera directed against the indicated proteins. Bound immunoglobulin was identified by reactions with appropriate antisera conjugated to 12-nm gold beads. Arrowheads indicate gold beads associated with capsids. Electron micrographs of immunogold analyses performed with pUL33- and pUL28-specific antibodies were similar to those of pUL17 (data not shown). A comprehensive analysis of the data is presented in Table 1.
Examination of at least 400 capsids in each treatment group and data from multiple experiments revealed the following information.
(i) Background levels of immunostaining with the pUL15-, pUL17-, pUL28-, and pUL33-specific antisera, as revealed by the number of appropriate mutant capsids bearing gold beads, were significantly below similarly stained wild-type HSV-1(F) capsids. (All P values were <0.001 as assessed by Fisher's exact t test.)
(ii) As shown previously (22), pUL6-specific epitopes were recognized on the surface of the capsid inasmuch as significantly more (P < 0.001) gold beads were present in intact wild-type capsids reacted with the pUL6-specific antiserum than in capsids lacking pUL6. These epitopes were detected more frequently in intact capsids than in sectioned capsids (P < 0.001), suggesting that the bulk of the epitopes were available primarily for reaction at the capsid surface rather than internal to the capsid shell. Approximately 16.3% (53 of 325) of capsids were labeled, suggesting either that the immunogold staining was insensitive and did not detect portal protein in many capsids or that many capsids lacked portals. Biochemical studies showing that populations of B capsids average 14.8 ± 2.6 copies of pUL6 per capsid (22), coupled with the high likelihood that the portal ring contains 12 copies of pUL6 (44), argue against the latter possibility.
(iii) As expected, the ICP35-specific antiserum did not recognize the external surface of capsids to an appreciable extent inasmuch as only 18 capsids of 1,007 capsids examined (0.018%) were labeled with the NC 3-4 antibody. Upon sectioning of the capsids, however, ICP35-specific epitopes were rendered significantly more immunoreactive with the antiserum (P < 0.001), as revealed by increased labeling of sectioned capsids (115 [28%] of 413 sectioned capsids). These observations indicated that, as expected, ICP35 was present in the capsid interior rather than the capsid surface and verified that the inner surfaces of unsectioned capsids were sequestered from the applied antibodies under the experimental conditions used.
(iv) Epitopes from pUL17, pUL28, and pUL33 localized at the surface of the capsid, as revealed by immunoreactivity of intact capsids, which was significantly above background levels obtained upon reaction with the corresponding deletion virus capsids. (All P values were less than 0.001 by Fisher's exact t test.) In all three cases, although immunoreactivity was present in sectioned HSV-1(F) capsids, the level of immunoreactivity was significantly lower than that obtained using intact capsids, presumably because a given thin section contains only a limited portion of the capsid surface. Another possibility is that embedding capsids could reduce the immunoreactivity of pUL17, pUL28, and pUL33. More capsids (13%) were labeled with the pUL17-specific antibody than with either the pUL28- or pUL33-specific antibody (8.7% and 8.9%, respectively). This could be a consequence of increased affinity of the pUL17-specific antibody relative to the other antibodies or increased amounts of pUL17 in association with capsids. Given the observation that only around two copies of pUL28 are present per B capsid, and the observation that pUL17 can localize to multiple vertices, it seems likely that more pUL17 is associated with capsids than pUL28 (7, 41). In any case, these data are consistent with other studies of pUL17 in HSV capsids showing that the protein is on the external capsid surface but are in contrast with the localization of the pUL17 homolog of pseudorabies virus that has been reported to associate with packaged DNA (16, 41).
(v) Antisera directed against C-terminal epitopes of pUL15 were recognized on the external surface of capsids, as revealed by the increased immunoreactivity of intact capsids compared to that of pUL15-negative capsids. Unlike the case with pUL6-, pUL33-, pUL28-, and pUL17-specific antibodies, immunoreactivity of the pUL15-specific antibody remained high in sectioned capsids. A slight increase in immunoreactivity in sectioned capsids compared to that obtained with unsectioned capsids was not statistically significant (P = 0.06). On the other hand, because less external surface area of the capsid is represented in a 40- to 60-nm thin section, the preservation of immunoreactivity in sectioned capsids suggests that more pUL15 C-terminal epitopes were present within the capsid interior than were epitopes of pUL28, pUL17, or pUL33. It is unclear whether these observations represent the possibilities that (a) pUL15 extends from the external surface to the internal surface of the capsid, (b) the pUL15 epitopes are masked less efficiently upon embedding than pUL28, pUL17, or pUL33 epitopes, or (c) multiple copies of pUL15 are present at different locations within the capsid. Assuming that all B capsids are biochemically identical (an assumption that has not been tested), the observation that each B capsid contains only 1.2 copies of pUL15 (7) argues against the latter possibility.
(vi) Very few capsids that reacted with any of the antibodies contained more than one gold bead. This is in contrast to results reported previously by others (41) and may reflect the respective affinities of the different antibodies in the two studies.
Comparative resistance of capsid-associated proteins to protease digestion.
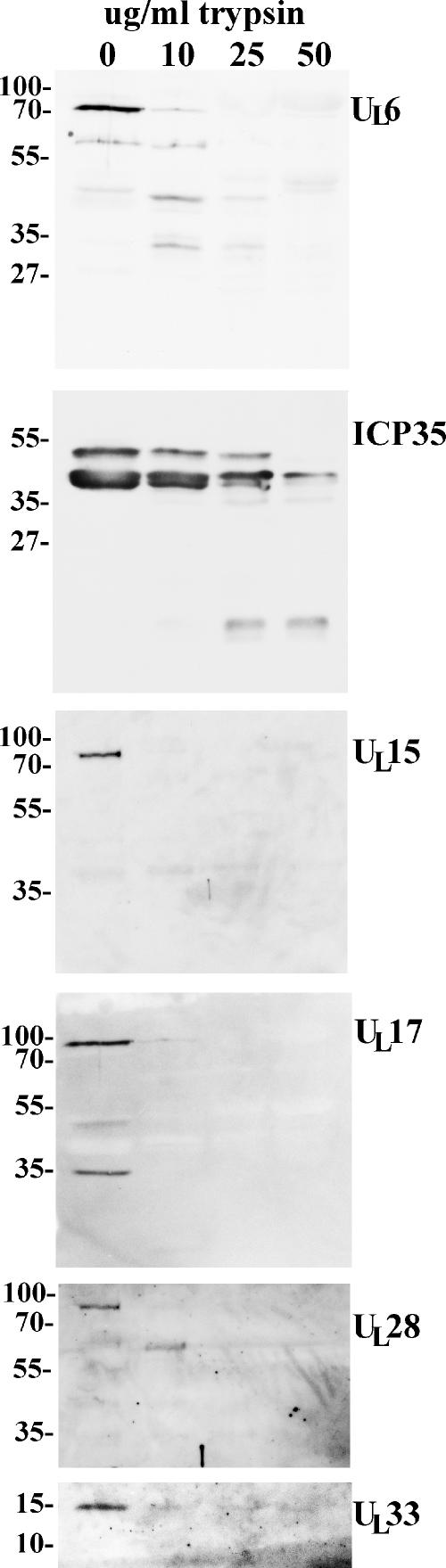
To confirm the results obtained by immunogold labeling, B capsids were purified on continuous sucrose gradients as described above and divided into four equal pools of 250 μl. The aliquots were incubated in the absence of trypsin or in the presence of 10, 25, or 50 μg/ml trypsin (MP Biomedicals) for 45 min at 37°C. The digested capsids were then diluted into 14 ml of ice-cold TNE containing protease inhibitors (1 tablet Complete protease inhibitors [Roche] per 50 ml TNE), and the diluted capsids were pelleted in an SW41 rotor at 35,000 rpm for 2 h. Pelleted capsids were solubilized in approximately 50 μl denaturing buffer containing SDS, mercaptoethanol, and bromophenol blue, and 25 μl of each sample was electrophoretically separated on a single lane of an SDS-polyacrylamide gel, followed by immunoblotting with the indicated antibodies as described above, except that the primary antibodies were diluted as follows: rabbit anti-pUL6, 1/1,000; rabbit anti-ICP35, 1/1,000; rabbit anti-pUL15, 1/1,000; chicken anti-pUL17, 1/50,000; rabbit anti-pUL28, 1/500; and rabbit anti-pUL33, 1/500. Anti-rabbit and anti-chicken secondary antibodies conjugated to horseradish peroxidase were diluted 1/5,000, and bound immunoglobulin was revealed by ECL (Amersham). The results are shown in Fig. 3.
FIG. 3.
Digital images of immunoblots of capsids incubated in the presence and absence of trypsin. B capsids were purified and incubated with the indicated concentrations of trypsin for 45 min at 37°C. The reaction was stopped by immersion in an excess volume containing protease inhibitors, and the capsids were pelleted in an ultracentrifuge, denatured in SDS, and electrophoretically separated, followed by immunoblotting with antisera directed against the products of the genes indicated to the right of the figure. Positions of size standards and their Mrs (in thousands) are indicated.
Unlike all the other proteins examined, ICP35 was not significantly affected by incubation with 10 μg/ml trypsin. Upon digestion with 25 μg/ml trypsin, however, the amounts of full-length ICP35 species were decreased, and a band that ran faster than the 27,000-Mr marker became apparent. In contrast, upon incubation with 10 μg/ml trypsin, pUL6 was partially cleaved to proteins with approximate Mrs of 40,000 and 30,000, and these bands remained detectable even upon digestion with up to 50 μg/ml trypsin.
In contrast to the results obtained with ICP35 and pUL6, immunoreactivity of pUL15 was completely eliminated upon digestion with 10 μg/ml trypsin, whereas digestion at this concentration significantly reduced but did not eliminate reactivity with pUL17-specific and pUL33-specific antibodies. Incubation with concentrations higher than 10 μg/ml of trypsin completely eliminated pUL17 and pUL33 immunoreactivity. Digestion of capsids with 10 μg/ml trypsin cleaved pUL28 into a prominent band containing a protein with an apparent Mr of 60,000, whereas concentrations of trypsin higher than 10 μg/ml precluded the detection of any pUL28-specific immunoreactivity. These data indicate that ICP35, a protein located within the capsid interior, is more resistant to tryptic digestion than pUL6, pUL15, pUL17, pUL28, or pUL33.
Taken together, the data presented herein indicate that at least some epitopes of pUL6, pUL15, pUL17, pUL28, and pUL33 are located at the external surface of the viral capsid. These observations are consistent with the hypotheses that pUL15, pUL28, and pUL33 represent the viral terminase inasmuch as their external location would facilitate an interaction with DNA as it is being packaged. One model that is also supported by the presence of some pUL15 epitopes in sectioned capsids (Table 1) is that pUL15 is more intimately associated with the capsid, whereas pUL28 is located more peripherally. This is consistent with the observation that empty capsids that are believed to have engaged but not retained DNA (A capsids) contain approximately 12 copies of pUL15 but less than 1 copy of pUL28 on average (7). Thus, consistent with its DNA binding activity (2), pUL28 may associate with DNA as it is expelled and may thereby become lost from the A capsid. In contrast, pUL17 may be located externally to either stabilize the capsid or capsomeres or engage molecular motors for capsid transport in the nucleus or cytoplasm. As shown previously by others, the presence of pUL17 at multiple vertices is consistent with these possibilities (41).
Acknowledgments
These studies were supported by Public Health Service grant R01 GM 50740 from the National Institutes of Health.
We are grateful to Fred Homa, Arvind Patel, and Andrew Davison for recombinant viruses, Gary Cohen and Roselyn Eisenberg for antibody to ICP35, and the Cornell Integrated Microscopy Center.
Footnotes
Published ahead of print on 18 August 2006.
REFERENCES
- 1.Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 4.Baines, J. D., C. Cunningham, D. Nalwanga, and A. J. Davison. 1997. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., A. P. W. Poon, J. Rovnak, and B. Roizman. 1994. The UL15 gene of herpes simplex virus encodes two proteins and is required for cleavage of viral DNA. J. Virol. 68:8118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beard, P. M., and J. D. Baines. 2004. The DNA cleavage and packaging protein encoded by the UL33 gene of herpes simplex virus 1 associates with capsids. Virology 324:475-482. [DOI] [PubMed] [Google Scholar]
- 7.Beard, P. M., C. Duffy, and J. D. Baines. 2004. Quantification of the DNA cleavage and packaging proteins UL15 and UL28 in A and B capsids of herpes simplex virus type 1. J. Virol. 78:1367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. The DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catalano, C. E., D. Cue, and M. Feiss. 1995. Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol. 16:1075-1086. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham, C., and A. J. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 12.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 13.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7:429-431. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goshima, F., D. Watanabe, H. Takakuwa, K. Wada, T. Daikoku, H. Yamada, and Y. Nishiyama. 2000. Herpes simplex virus UL17 protein is associated with B capsids and colocalizes with ICP35 and VP5 in infected cells. Arch. Virol. 145:417-426. [DOI] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, A. Karger, and T. C. Mettenleiter. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J. Virol. 79:13442-13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koslowski, K. M., P. R. Shaver, J. T. Casey II, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenny, and N. E. Pedersen. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell, M. S., S. Matsuzaki, S. Imai, and V. B. Rao. 2002. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 30:4009-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and S. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 24.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel, A. H., and J. B. Maclean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-478. [DOI] [PubMed] [Google Scholar]
- 27.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 28.Perdue, M. L., J. C. Cohen, M. C. Kemp, C. C. Randall, and D. J. O'Callaghan. 1975. Characterization of three species of nucleocapsids of equine herpesvirus type-1 (EHV-1). Virology 64:187-204. [DOI] [PubMed] [Google Scholar]
- 29.Poon, A. P. W., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds, A. E., Y. Fan, and J. D. Baines. 2000. Characterization of the UL33 gene product of herpes simplex virus 1. Virology 266:310-318. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-403. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology. Plenum Press, New York, N.Y.
- 33.Salmon, B., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J. Virol. 72:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrag, J. D., B. V. Prasad, F. J. Rixon, and W. Chiu. 1989. Three-dimensional structure of the HSV1 nucleocapsid. Cell 56:651-660. [DOI] [PubMed] [Google Scholar]
- 36.Spencer, J. V., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 1998. Assembly of the herpes simplex virus capsids: preformed triplexes bind to the nascent capsid. J. Virol. 72:3944-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taus, N. S., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: the UL28 gene product is a minor component of B capsids. Virology 252:443-449. [DOI] [PubMed] [Google Scholar]
- 39.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115-125. [DOI] [PubMed] [Google Scholar]
- 40.Tengelsen, L. A., N. E. Pedersen, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and capsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19C and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 44.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, K., and J. D. Baines. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 80:5733-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32-44. [DOI] [PubMed] [Google Scholar]
- 48.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, Z. H., B. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]