Abstract
Membrane fusion of the alphaviruses is mediated by the E1 protein, a class II virus membrane fusion protein. During fusion, E1 dissociates from its heterodimer interaction with the E2 protein and forms a target membrane-inserted E1 homotrimer. The structure of the homotrimer is that of a trimeric hairpin in which E1 domain III and the stem region fold back toward the target membrane-inserted fusion peptide loop. The E1 stem region has a strictly conserved length and several highly conserved residues, suggesting the possibility of specific stem interactions along the trimer core and an important role in driving membrane fusion. Mutagenesis studies of the alphavirus Semliki Forest virus (SFV) here demonstrated that there was a strong requirement for the E1 stem in virus assembly and budding, probably reflecting its importance in lateral interactions of the envelope proteins. Surprisingly, however, neither the conserved length nor any specific residues of the stem were required for membrane fusion. Although the highest fusion activity was observed with wild-type E1, efficient fusion was mediated by stem mutants containing a variety of substitutions or deletions. A minimal stem length was required but could be conferred by a series of alanine residues. The lack of a specific stem sequence requirement during SFV fusion suggests that the interaction of domain III with the trimer core can provide sufficient driving force to mediate membrane merger.
The nucleocapsid of an enveloped virus is encapsulated in a lipid bilayer that is derived from a host cell membrane during virus budding. The viral genome is delivered into the cytoplasm of the target cell via fusion of the virus membrane with the cell membrane, a process driven by the conformational changes of viral membrane fusion proteins. Functional and structural studies have classified many viral proteins as members of the class I and class II fusion proteins (reviewed in references 6, 16, and 20).
The members of class I include the trimeric transmembrane fusion proteins of the orthomyxoviruses, paramyxoviruses, retroviruses, filoviruses, and coronaviruses. Upon triggering of the fusion reaction, the N-terminal parts of the class I fusion proteins form extended trimeric α-helical coiled coils, leading to the insertion of the fusion peptides into the target membrane. This conformation, termed the “prehairpin intermediate,” thus bridges the viral and target membranes. The folding back of the C-terminal part of the fusion protein then induces a membrane merger (8). The postfusion structure is in a conformation termed the “trimer of hairpins,” in which the N-terminal regions of the ectodomain form the inner “trimer core” and the C-terminal regions form the “outer layer.” Synthetic “C-peptides” derived from the outer layer of several class I proteins potently inhibit virus fusion and infection, presumably by binding to the trimer core formed in the prehairpin intermediate (7, 10, 40). T20, a C-peptide from the human immunodeficiency virus type 1 (HIV-1) fusion protein, successfully suppresses the replication of HIV-1 in patients (22) and is in clinical use.
The alphaviruses and flaviviruses are small, enveloped plus-strand RNA viruses whose fusion is mediated by the E1 and E proteins, respectively. These proteins are the inaugural members of the class II viral fusion proteins. The ectodomains of E1 and E are folded into three domains composed predominantly of β-strands and are connected to the transmembrane (TM) domain by a region termed the “stem” (reviewed in references 16, 20, and 31). On the virus surface, these proteins form an icosahedral protein lattice composed of heterodimers (E1-E2 in alphaviruses) or homodimers (E-E in flaviviruses). The internal fusion peptide loop is buried in the dimer contacts, and the fusion protein is oriented tangentially to the virus membrane. During fusion, class II proteins dissociate from the dimer interaction, reorient perpendicularly to the viral surface, insert into target membranes via the fusion loops, and trimerize. Similar to class I proteins, in the postfusion conformation the class II proteins form a “trimer of hairpins,” in which the inner trimer core is composed of domains I and II and the outer layer consists of domain III and the stem region (2, 14, 30). The addition of recombinant domain III proteins during the alphavirus or flavivirus fusion reaction potently inhibits virus fusion and infection (26). The exogenous domain III stably binds to an intermediate trimeric conformation of E1, presumably a class II “prehairpin intermediate” that is formed before the fold-back of domain III and the stem, and thus prevents formation of the final hairpin. Together, these results indicate that the fold-back of domain III observed in the class II homotrimer (HT) structures is required for membrane fusion.
While the fold-back of domain III plays a critical role in forming the “hairpin,” the function of the stem region in fusion is not well understood. In flaviviruses, the stem can facilitate the trimerization of the ectodomain of the E protein (1) and acts to stabilize the E protein HT (37). In alphaviruses, the presence of the stem in the recombinant domain III protein significantly enhances its inhibitory activity and its binding affinity to the trimeric E1 intermediate (26). Our recent studies using stem-directed antibodies reveal a tight correlation between stem packing and the final membrane merger (27). Taken together, these lines of evidence and the similarities of the postfusion conformations of class I and class II proteins suggest that the class II stem region could drive membrane fusion by packing onto the trimer core and that synthetic stem peptides could act as dominant-negative fusion inhibitors (2, 30). However, tests of four alphavirus E1 stem peptides showed no inhibition of virus fusion (27). Although there are many potential reasons for this lack of inhibition, one possibility is that E1 stem packing is not critical for fusion. A direct test of the functional requirements for the specific stem sequence during the virus life cycle is important to understand the class II fusion mechanism and to design better stem-based antiviral reagents.
Here we addressed the functional importance of the alphavirus stem region by mutagenesis of the Semliki Forest virus (SFV) E1 protein. Unexpectedly, neither the conserved length nor any specific residue of the stem was required for the membrane fusion activity of SFV E1, and only a minimum length of the stem appeared to be essential.
(The data in this paper are from a thesis submitted by M. Liao in partial fulfillment of the requirements for a Ph.D. at the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University, Bronx, N.Y., 2006.)
MATERIALS AND METHODS
Cells.
BHK-21 cells were cultured as previously described (38). CHO cells were grown at 37°C in alpha-minimal essential medium supplemented with 10% fetal calf serum (FCS).
Generation of mutant SFV clones and infectious RNAs.
Mutagenesis of the E1 stem region was performed on the subgenomic DG-1 plasmid as described previously (3). In brief, stem mutations were introduced into DG-1 by circular mutagenesis, and the mutated NsiI/SpeI fragment was then subcloned into the pSP6-SFV4 infectious clone (28). Two independent clones for each mutant were constructed from two different mutagenic PCRs and used to confirm the phenotype. The E1 stem regions from all mutant SFV clones were sequenced (Genewiz, Inc., North Brunswick, NJ) to confirm the presence of the desired mutations and the absence of other mutations. RNAs from the wild type (wt) and the mutant infectious clones were prepared by in vitro transcription and used to infect BHK-21 cells by electroporation (28).
Plaque assay and infectious-center assay.
The production of infectious virus by electroporated cells was quantitated by a plaque assay of the indicator BHK-21 cells contained in the culture medium (21). For some slow-growing mutants, an infectious-center assay was used instead of the plaque assay. BHK-21 cells were incubated with serial dilutions of the culture medium at 37°C for 90 to 120 min. Twenty millimolar NH4Cl was then added to prevent secondary infection, and the cells were incubated at 28°C overnight. The infected cells (termed infectious centers) were then quantitated by immunofluorescence, using Rab, a polyclonal antibody to the SFV envelope proteins.
Virus assembly assay.
Virus assembly was evaluated by pulse-chase assays at 28°C as described previously (3, 5). In brief, BHK-21 cells were electroporated with wt or mutant RNA, plated in 35-mm dishes for 2 h at 37°C, and further incubated at 28°C overnight. The infected cells were then pulse-labeled with [35S]methionine-cysteine at 28°C for 1 h and chased in media without label at 28°C for 7 h. At the end of the incubation, cell lysates and the chase media were harvested and immunoprecipitated as described below.
Immunoprecipitation.
Immunoprecipitation in detergent was performed as previously described (12, 18). In brief, samples were reacted with the indicated antibody, absorbed to zysorbin, washed three times with RIPA buffer (10 mM Tris, pH 8.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 20 μg aprotinin/ml) and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). To recover intact, capsid-containing virus particles from the chase medium for the virus assembly assay, samples of the medium were bound to zysorbin in the absence of detergent, washed three times with detergent-free washing buffer (20 mM Tris [pH 8.5], 150 mM NaCl, 1 mM EDTA, 10 mg bovine serum albumin/ml, 100 μg aprotinin/ml), and then analyzed by SDS-PAGE.
Cell-cell fusion assay.
The fusion activity of viral proteins expressed on the surfaces of infected cells was tested by cell-cell fusion assays as previously described (3, 24). BHK-21 cells infected by RNA electroporation were diluted 1:10 with nonelectroporated cells and incubated at 37°C for 3 h to allow cell attachment to coverslips. The cells were cultured for ∼16 h at 28°C in the presence of 20 mM NH4Cl to prevent secondary infection and then treated with medium at the indicated pH for 1 min at 37°C to trigger cell-cell fusion. After an additional culture at 28°C for 3 h, the cells were fixed in 3% formaldehyde at room temperature for 20 min. The cells were stained using the Rab antibody, and the nuclei were stained with propidium iodide. The numbers of nuclei per envelope protein-positive cell were evaluated by fluorescence microscopy, counting at least 200 nuclei per coverslip. The fusion index was calculated by the equation 1 − (number of cells/number of nuclei).
Lipid-mixing assay using DiI-labeled CHO cells.
BHK-21 cells were electroporated with wt or mutant RNAs and cultured on 22-mm square coverslips in a six-well plate at 28°C for ∼16 h in the presence of 20 mM NH4Cl to prevent secondary infection. To prepare labeled target cells, CHO cells were split onto a 60-mm plate 1 day before the experiment and had just achieved confluence at the time of labeling. The cells were washed twice with 2 ml Dulbecco's modified Eagle's medium (DME) and labeled by incubation with 2 ml DiIC18 staining solution (5 μl Vybrant DiI cell-labeling solution [Molecular Probes/Invitrogen, Carlsbad, CA] in 2 ml DME) at 37°C for 15 min in the dark. The cells were washed three times with 3 ml DME, with a 5-min interval at room temperature between each wash. The cells were then removed from the plate by treatment with 2 ml phosphate-buffered saline (PBS)-EDTA-EGTA (PBS with 0.5 mg EDTA/ml and 0.5 mg EGTA/ml) at room temperature for 5 min. The PBS-EDTA-EGTA was removed, and the CHO cells were resuspended by vigorous pipetting with 3.5 ml DME. Infected BHK-21 cells on the coverslips were washed twice with 2 ml DME. One milliliter per well of the DiI-labeled CHO cells was then added, followed by incubation at 37°C for 45 min to allow CHO cell attachment. Most CHO cells attached to the coverslips and contacted BHK-21 cells. The cocultures were then chilled on ice, washed twice with an ice-cold pH 7 medium to remove unattached cells, and incubated with pH 7 or pH 5 medium at 37°C for 1 min. The coverslips were washed twice with PBS-10 mM glucose, inverted onto microscope slides, and sealed with Cytoseal 60 mounting medium (Richard-Allan Scientific, Kalamazoo, MI). Live cells on coverslips were observed within 40 min of pH treatment, using fluorescence microscopy with a rhodamine filter set. Fluorescence and phase images of selected fields were photographed using an AxioCam (Zeiss, Göttingen, Germany) in the Analytical Imaging Facility, Albert Einstein College of Medicine.
Flow cytometry analysis of cell surface E1 expression.
BHK-21 cells were electroporated with wt or mutant RNA, incubated at 37°C for 3 h, and cultured at 28°C for ∼16 h in the presence of 20 mM NH4Cl. After the cells were removed from the plate by treatment with PBS-EDTA-EGTA as described above, they were incubated with 20 μg/ml of the E1-specific monoclonal antibody (MAb) E1-1 in PBS-1% FCS on ice for 30 min. The cells were then washed and incubated with Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (Molecular Probes) in PBS-1% FCS on ice for 30 min, followed by fixation in 1% formaldehyde in PBS at 4°C. Data were acquired for 10,000 events by flow cytometry using a FACScan in the Fluorescence-Activated Cell Sorter Facility, Albert Einstein College of Medicine.
RESULTS
Sequence analysis of the alphavirus E1 stem and construction of stem mutants.
The stem region of SFV E1 starts after domain III at Lys384 and connects the ectodomain to the TM domain (14, 23). The N-terminal residue of the TM domain was predicted to be Ile413 by the TMpred program (17), leaving the positively charged Lys412 as the last residue of the stem. Similarly positioned N termini were also predicted for the TM domains of other alphaviruses. Sequence alignment of E1 proteins from representative alphaviruses showed a totally conserved length for the stem and identified several highly conserved amino acids (Fig. 1A). This conservation pattern is consistent with the requirements for a critical stem length to bridge the distance from domain III to the fused membrane in the postfusion structure and for specific stem residues to mediate important interactions with the trimer core.
FIG. 1.
Sequence alignment and mutants of the alphavirus E1 stem region. (A) Sequence alignment of the E1 stem regions from representative alphaviruses was performed using the program ClustalW. Bars at the top of the panel show the consensus strength for each amino acid based on sequence identity. Four highly conserved amino acids are indicated in red and numbered as in the SFV E1 sequence (His386, His394, Ser403, and Trp409). Virus abbreviations and GenBank accession numbers: SFV, GenBank accession no. CAA01423; ONNV, O'nyong-nyong virus, accession no. P22056; RRV, Ross River virus, accession no. P13890; CHIKV, Chikungunya virus, accession no. AAR84279; SIN, Sindbis virus, accession no. NP_740677; WEEV, Western equine encephalitis virus, accession no. NP_818942; AURAV, Aura virus, accession no. NP_819019; VEEV, Venezuelan equine encephalitis virus, accession no. P09592; EEEV, Eastern equine encephalitis virus, accession no. P27284; BFV, Barmah Forest virus, accession no. NP_819002; IOV, Igbo Ora virus, accession no. NP_741964; SDV, sleeping disease virus, accession no. NP_740661; SPDV, salmon pancreas disease virus, accession no. NP_740643; MIDV, Middelburg virus, accession no. AAO33343; NDUV, Ndumu virus, accession no. AAO33345; TROV, Trocara virus, accession no. AAL55092. (B) Amino acid sequences of the E1 stem mutants constructed in this study. Dots indicate amino acids identical to wt SFV E1, and dashes indicate sequence deletions. Note that in the Δ20 mutant, the 20 residues after Ala391 were deleted, thereby connecting the sequence of E1* to the TM domain via only one lysine residue (Lys412).
To test the function of the specific stem sequence, mutations at four highly conserved residues (Fig. 1A, red letters) were introduced into the SFV infectious clone to generate six virus mutants: the H386A, H386E, S403A, W409A, HSW/3A (an H386A S403A W409A triple mutant), and H394E mutants (Fig. 1B). In addition, the requirements for all residues in the stem and for the conserved length were tested with a series of alanine mutations and/or deletions (DHIV/4A, Δ6, Δ4, Δ8, Δ9, Δ20, and Δ20+11A). Note that the DHIV/4A mutant had substitutions at a cluster of four conserved residues at the N terminus of the stem and that the Δ6 deletion resulted in a shift of all of the more C-terminal residues of the stem toward domain III. The Δ20 mutant contains the sequence of E1 up to the proteolytic cleavage site used to generate the E1* ectodomain (13) and is connected to the TM domain via Lys412. Together, the mutations in these mutants span the entire stem region and allow the evaluation of the roles of specific residues and stem lengths.
Growth properties of the stem mutants.
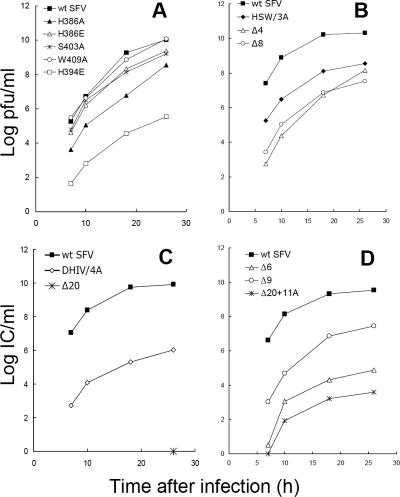
As the first step in the analysis of the stem mutants, wt or mutant SFV RNA was transcribed in vitro and introduced into BHK-21 cells by electroporation. The infectious progeny virus released by the infected cells was measured by plaque assay or infectious-center assay (Fig. 2). The overall growth of the H386E, S403A, and W409A mutants was similar to that of wt SFV, but all other stem mutants showed markedly decreased growth, with virus titers at least 2 logs lower than that of wt SFV. The H394E mutation resulted in a strong inhibition of virus production (a decrease of more than 4 logs) that was more significant than the effects of all other single mutations or of the triple mutation HSW/3A. DHIV/4A and all deletion mutations dramatically inhibited virus growth, producing decreases of 3 to 6 logs. The strongest growth inhibition was observed for the Δ20 mutant, which did not produce any detectable infectious virus even 26 h after RNA electroporation (Fig. 2C). It is important to note that except for the Δ20 mutant, all of the mutants showed growth at early times after electroporation (≤10 h), indicating that the growth properties are intrinsic to the specific mutant and not due to compensatory mutations arising during virus growth (4).
FIG. 2.
Growth kinetics of wt SFV and SFV mutants in BHK-21 cells. BHK-21 cells were electroporated with wt or mutant SFV RNA, diluted 1:1 with nonelectroporated BHK-21 cells, and incubated at 37°C. The cell media were collected at the indicated time points after electroporation, and the progeny virus was measured by plaque assays (A and B) or infectious-center assays (C and D) of BHK-21 cells.
In these growth assays, the level of progeny virus reflected the cumulative effects of the mutations on virus production from electroporated cells and virus entry into nonelectroporated cells. We therefore separately evaluated the exit properties and membrane fusion phenotypes of the stem mutants.
Effect of the stem mutations on virus assembly.
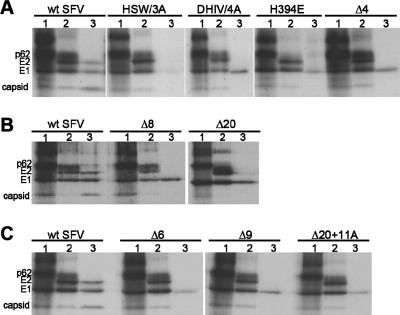
Virus assembly was evaluated by pulse-chase analysis of BHK-21 cells infected by wt SFV or stem mutants (Fig. 3). We tested virus assembly at 28°C, a temperature that is more favorable for the assembly of various envelope protein mutants than 37°C (5). The wt and stem mutants produced comparable levels of structural proteins during the 1-h labeling period (Fig. 3, lanes 1), and p62 was efficiently processed to E2 during a 7-h chase period in all of the samples (Fig. 3, lanes 2). Thus, the overall biosynthesis and transport of the envelope proteins did not appear to be adversely affected by the stem mutations. In fact, the cell surface expression level of viral glycoproteins for all the stem mutants was somewhat higher than that of wt SFV, as determined by immunofluorescence (data not shown) and flow cytometry (see Fig. 5). The analysis of the wt SFV chase medium demonstrated that virus particles budded efficiently from the wt SFV-infected cells, with approximately equal recoveries of E1, E2, and capsid proteins (Fig. 3, lanes 3). In contrast, cells infected with the stem mutants released E1s, a soluble truncated form of E1 (29), but little detectable E2 or capsid protein. Thus, the stem mutants showed significant decreases in virus assembly, in keeping with the observed reduction in virus growth.
FIG. 3.
Effect of the stem mutations on SFV assembly. BHK-21 cells were electroporated with wt or mutant RNA, incubated overnight at 28°C, pulse-labeled with [35S]methionine-cysteine at 28°C for 1 h, and then chased at 28°C for 7 h. (A to C) SFV proteins in the cell lysates before the chase (lanes 1) and in the cell lysates (lanes 2) and media (lanes 3) after the chase were immunoprecipitated with Rab, a polyclonal antibody to the SFV E1 and E2 proteins, and analyzed by SDS-PAGE and fluorography. The medium samples (lanes 3) were immunoprecipitated in the absence of detergent to retrieve intact virus particles containing the viral nucleocapsid. The positions of the viral proteins are indicated. The truncated E1 form is observed in lanes 3 of the mutant samples but shows detectably faster migration than E1 only when the stem is not deleted (e.g., the DHIV sample). Results in each panel are from the same experiment.
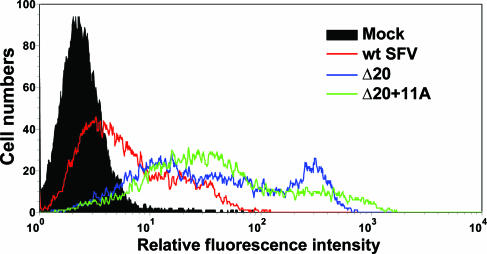
FIG. 5.
Analysis of cell surface expression of wt and mutant E1 proteins. BHK-21 cells were electroporated with wt, Δ20, or Δ20+11A RNA and cultured as described in the legend to Fig. 4A. Cell surface levels of E1 protein were determined by flow cytometry using the E1-specific MAb E1-1 and Alexa Fluor 488-conjugated secondary antibody. The “mock” sample represents wt SFV RNA-electroporated cells for which MAb E1-1 was omitted and is also the position of stained uninfected cells.
Structural studies of alphaviruses have shown that the icosahedral symmetry of the outer glycoprotein shell is maintained by lateral contacts between the glycoproteins and that both the membrane-distal and membrane-proximal ends of E1 interact with E2 (23, 32, 34, 41). It is likely that the effects of the stem mutations on virus budding occur through alterations of the lateral interactions of the glycoproteins such as the E1-E2 heterodimer contacts. Indeed, coimmunoprecipitation analysis of the E1-E2 heterodimer (39) showed that all of the stem mutants tested (the Δ8, Δ9, Δ20, and Δ20+11A mutants) had less-stable heterodimer interactions than those of the wt proteins (data not shown).
Membrane fusion properties of the stem mutants.
Although virus assembly was significantly inhibited by mutations in the stem, mutants that produced sufficient titers of progeny virus could be tested for their abilities to mediate the low-pH-induced fusion of prebound virus with the plasma membrane of BHK-21 cells (25). As previously observed, wt SFV showed no fusion at pH 6.25, a fusion threshold at ∼pH 6, and maximal fusion at pH 5.75, while by comparison, the H394E, HSW/3A, DHIV/4A, Δ4, and Δ8 stem mutants showed increases in fusion activity at pH 6.25 and pH 6 (data not shown). This increase in fusion at higher pH levels could reflect weaker E1-E2 interactions that dissociate at a higher pH (36).
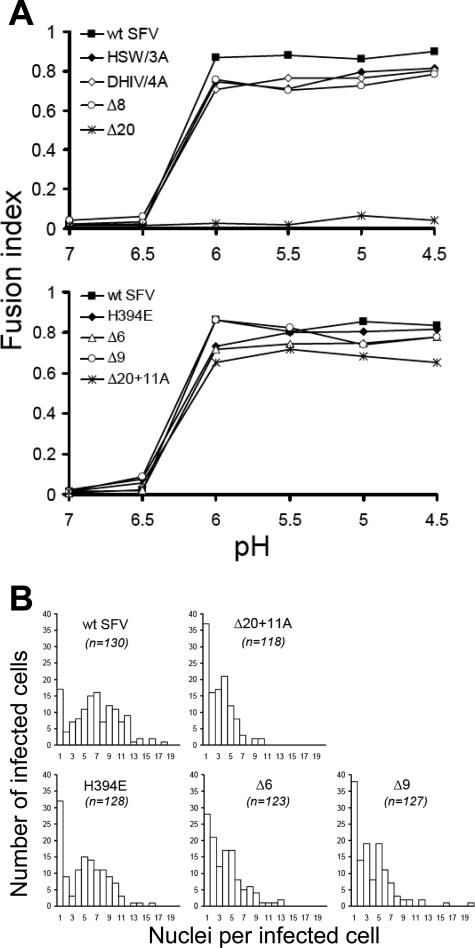
Given the effects of the stem mutations on virus assembly, the mutants' membrane fusion phenotypes were characterized primarily by testing low-pH-induced cell-cell fusion activity, which is independent of virus particle production. In this assay, the viral glycoproteins expressed on the surfaces of infected cells mediate fusion with the neighboring cells when a 1-min low-pH pulse at 37°C is applied. Polykaryon formation can then be measured by determining the number of nuclei per envelope protein-expressing cell and expressed as a fusion index (see Materials and Methods and reference 24). As expected, the wt SFV-infected cells fused efficiently at a threshold of ∼pH 6.0 (Fig. 4A). Cells infected by the DHIV/4A mutant or the various deletion mutants also fused efficiently and with a pH dependence similar to that of wt-infected cells, indicating that neither specific amino acids nor the conserved length of the stem was essential for the fusion capacity of SFV E1. A significant change in the pH threshold was not observed in this assay, perhaps reflecting differences in the organization of E1 proteins at the cell surface compared to that in the virus particle. In contrast to the efficient fusion of all of the other mutants, the deletion of 20 amino acids in the stem completely blocked E1-mediated cell-cell fusion activity, even after treatment with a pH 4.5 buffer (Fig. 4A). Notably, the membrane fusion block of the Δ20 mutant was rescued by adding back an 11-alanine sequence in the Δ20+11A mutant. These fusion phenotypes were not due to alterations in the cell surface density of E1, as flow cytometry demonstrated that cells infected by wt SFV or the Δ20 or Δ20+11A mutant had comparable levels of E1 proteins on the cell surfaces (Fig. 5). In fact, the density of E1 on cells infected by the Δ20 or Δ20+11A mutant was somewhat greater than that on wt SFV-infected cells, presumably reflecting the lack of virus budding. Thus, in contrast to the predication that sequence-specific interactions between the stem and the trimer core drive the SFV fusion reaction, taken together, these data strongly suggest that only a nonspecific stem of minimum length is required for E1-mediated membrane fusion.
FIG. 4.
Cell-cell fusion activity of wt and mutant E1 proteins. (A) pH dependence of cell-cell fusion. BHK cells were electroporated with wt or mutant SFV RNA, diluted 1:10 with nonelectroporated cells, incubated at 37°C for 3 h, and cultured at 28°C for ∼16 h in the presence of 20 mM NH4Cl to prevent secondary infection. The cells were then treated with medium at the indicated pH for 1 min at 37°C to induce fusion and cultured at 28°C for 3 h before fixation. The number of nuclei per viral protein-expressing cell was evaluated, and the fusion index was determined (see Materials and Methods). (B) Size of syncytia produced at pH 5.0. For the pH 5.0 samples shown in the lower graph of panel A, the number of nuclei present in each infected cell from randomly selected microscope fields was determined. The total number of infected cells counted (n) is indicated for each virus. Data are from duplicate coverslips.
The results of the cell-cell fusion assays were expressed as a fusion index (see Materials and Methods), an average value that does not distinguish among individual syncytia with different numbers of nuclei. To compare the fusion activity of wt SFV and those of the stem mutants in more detail, the number of nuclei in each viral protein-expressing cell was individually recorded after cell-cell fusion at pH 5.0 and used to plot the distribution of nuclei per expressing cell (Fig. 4B). wt E1 induced primarily syncytia that contained from 3 to 12 nuclei, while 17 cells (13% of the total) contained only 1 nucleus and were thus unfused. In contrast, the Δ20+11A mutant E1 induced syncytia containing primarily 2 to 6 nuclei, and 37 cells (31% of the total) showed no fusion. The fusion activities of the Δ6 and Δ9 mutants were similar to that of the Δ20+11A mutant and appeared lower than that of the H394E mutant, which formed syncytia containing primarily 2 to 10 nuclei. Thus, the mutant containing the single H394E mutation seemed to affect fusion activity less than the more-drastic deletion mutations. Together, these data suggest that the wt SFV stem sequence does promote the fusion reaction, presumably by providing optimal interactions with the E1 trimer core.
Lipid-mixing assays of the Δ20 mutant E1 protein.
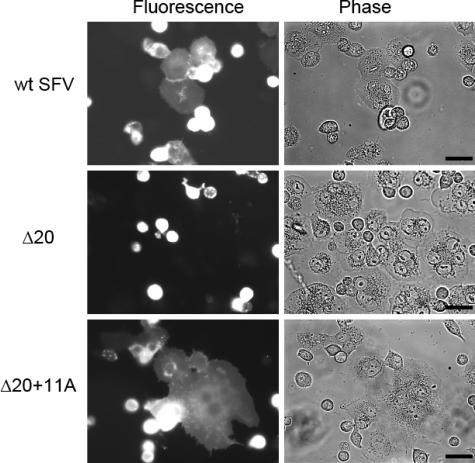
The Δ20 mutant produced no infectious virus and was completely inactive in cell-cell fusion, a content-mixing assay that detects complete fusion. It was possible that the Δ20 mutant could still mediate the initial mixing of the outer leaflets of the virus and target membranes, a step termed “hemifusion.” To test this, BHK-21 cells were infected as described above for the cell-cell fusion assay and mixed with noninfected CHO cells prelabeled with the lipophilic tracer DiI, and the cocultures were treated at pH 5.0 to trigger fusion (Fig. 6). Using this assay, fusion was readily observed for BHK-21 cells infected with either wt SFV or the Δ20+11A mutant, as shown by the spread of DiI from the rounded CHO cells to the larger and flatter BHK-21 cells. In contrast, no fusion was observed for the Δ20 mutant after pH 5.0 treatment, indicating that the initial lipid-mixing step was also blocked due to the shortened stem. Parallel experiments using treatment at pH 7.0 showed no fusion for wt SFV or the Δ20 or Δ20+11A mutant (data not shown), confirming the absence of nonspecific dye transfer in this assay.
FIG. 6.
Effects of the Δ20 and Δ20+11A mutations on the lipid-mixing step of membrane fusion. BHK-21 cells were electroporated with wt SFV RNA or Δ20 or Δ20+11A mutant SFV RNA and cultured on coverslips as described in the legend to Fig. 4A. CHO cells stained with DiIC18 were then added to the BHK-21 cell cultures and incubated at 37°C for 45 min to allow CHO cell attachment. The cocultures were treated at 37°C for 1 min at pH 7 (data not shown) or pH 5.0 and visualized by fluorescence microscopy within 40 min of pH treatment. Representative microscope fields were selected, and phase and fluorescence images of the same field are shown. Bar = 20 μm.
Low-pH-induced conformational changes of the mutant E1 proteins.
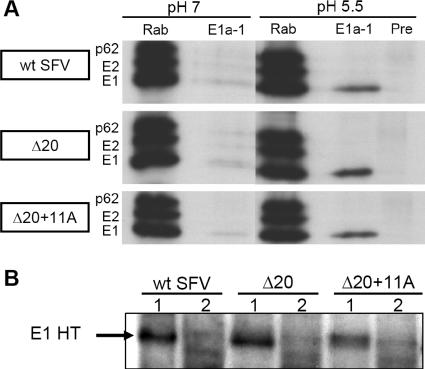
Previous studies have shown that the formation of the E1 HT is a critical step in the SFV fusion reaction (19, 26). While the truncated E1 ectodomain forms an HT that is biochemically comparable to the full-length E1 HT (13), the ability of the protein to trimerize when it is tethered to the membrane without the full stem was unknown. We therefore analyzed the low-pH-dependent conformational changes of E1 proteins expressed on wt- or mutant-infected cells. BHK-21 cells were infected with wt SFV or the Δ20 or Δ20+11A mutant, pulse-labeled, and chased at 37°C for 30 min to permit delivery of labeled E1 to the plasma membrane (18). The E1 proteins on the cell surface were then treated at pH 7 or 5.5, and the cells were lysed and immunoprecipitated with the acid conformation-specific MAb E1a-1 (Fig. 7A). Similar to results for the wt and the Δ20+11A mutant E1 proteins, the Δ20 mutant E1 efficiently changed conformation at a low pH to a form that reacted with MAb E1a-1. The wt E1 HT has been well characterized for its resistance to trypsin digestion and to dissociation by SDS sample buffer at 30°C (11, 13). As shown in Fig. 7B, low-pH-treated wt E1, Δ20 E1, and Δ20+11A E1 were resistant to trypsin digestion and showed the typical electrophoretic mobility of the E1 homotrimer in SDS-PAGE when solubilized at room temperature. Thus, the general conformation and stability of the E1 HT were not altered by stem deletion.
FIG. 7.
Characterization of low-pH-induced conformational changes of wt and mutant E1 proteins. (A) BHK-21 cells were electroporated with wt SFV RNA or Δ20 or Δ20+11A mutant SFV RNA and incubated at 37°C for 6 h. The infected cells were pulse-labeled with [35S]methionine-cysteine at 37°C for 15 min, chased at 37°C for 30 min, and then treated with pH 7 or pH 5.5 medium for 1 min at 37°C. The viral envelope proteins in the cell lysates were immunoprecipitated by Rab, MAb E1a-1 (specific for the low-pH-induced conformation of E1), or rabbit preimmune serum (Pre) and analyzed by SDS-PAGE and fluorography. (B) BHK-21 cells were infected, radiolabeled, and treated with medium as described for panel A. The cells were then lysed in PBS-1% Triton X-100 and digested with 50 μg/ml trypsin at 37°C for 15 min. The samples were solubilized in SDS sample buffer for 3 min at room temperature (lane 1) or at 95°C (lane 2) and analyzed by SDS-PAGE and fluorography.
DISCUSSION
The stem region is important for the assembly of SFV.
Previous structural studies of E1 and the alphavirus particle indicate that at a neutral pH, E2 interacts with both the fusion loop-containing region of E1 and the membrane-proximal region of E1, including the domain III AB loop and the stem (32, 34, 41). It was suggested that the interaction with E2 changes the direction of the C-terminal residues of the E1 ectodomain, thereby positioning the stem to point downward to the viral membrane (34). The cryoelectron microscopic density of the E1 stem is not separated from that of E2, and the TM helices of E1 and E2 cross the lipid bilayer in close proximity (41). Thus, the structural studies suggest a close E1-E2 interaction from the E1 sequence at the end of domain III through the TM domain. In fact, this heterodimer interaction appears to prevent the binding of an antibody specific to the N-terminal stem sequence, even when SFV is solubilized in detergent (26).
Our present work provided a direct test for the functions of this stem-mediated E1-E2 interaction. The envelope proteins from all of the stem mutants were efficiently transported to the cell surface and processed to the mature E2 form at either 28°C (Fig. 3) or 37°C (Fig. 7). However, the budding of the SFV envelope proteins from the cell surface was very sensitive to the alteration of either the length or the specific residues of the E1 stem, and the stem deletions and many of the point mutations significantly decreased virus growth. The inhibition of budding is most likely due to the effects of the stem mutations on the E1-E2 interactions within each trimeric spike, in keeping with the observed decreases in mutant heterodimer stability. The heterodimer interactions are known to be directly important in virus assembly (9) and may cause additional effects on the E1-mediated icosahedral lattice (34). Since all of the stem mutants except the Δ20 mutant are fusion active, the relative effects of the mutations on budding can be roughly evaluated from the overall growth kinetics of the mutants (Fig. 2). These kinetics thus suggest the possible effects of specific stem sequences on critical lateral interactions. Based on this, the N-terminal E1 stem sequence appears to be more important for E2 interaction than the C-terminal E1 stem region: the S403A and W409A mutations produced very little growth retardation, and the DHIV/4A, H394E, and Δ6 mutations inhibited virus growth more significantly than the Δ8 and Δ9 mutations. These data therefore imply that the E1 stem interacts strongly with E2 to form lateral interactions that are important during budding. These interactions are suggested to occur predominantly via the highly conserved N-terminal stem residues (Fig. 1A), while the interactions of the C-terminal stem appear less significant.
The specific stem sequence is not essential for SFV membrane fusion.
A very plausible model for alphavirus fusion would be that domain III initially folds back and then the stem region drives membrane fusion via sequence-specific interactions with the grooves of the trimer core. Such a model was supported by the observed conservation of the length and specific residues of the stem (Fig. 1A) and by the increased binding and inhibition observed with domain III proteins containing the stem (26). Surprisingly, however, we found that neither the conserved length nor any specific residue of the stem was necessary for E1-mediated membrane fusion and virus infection. Only a minimum length of the stem appeared to be required to span the distance between the C terminus of the folded-back domain III and the fusing membranes. These unexpected results strongly suggest that SFV membrane fusion can proceed without specific stem packing and that the fold-back of domain III per se provides enough energy for the membrane merger. However, if the stem associates with the groove via significant main chain interactions, stem packing might be functionally preserved, even in the face of drastic stem mutations. Our data do suggest that, at the least, the packing of the SFV E1 stem is significantly different from the packing of the outer layer of the class I proteins during fusion. For example, membrane fusion by influenza virus HA2 is completely blocked by the mutation of specific residues in the “stem-like” C-terminal ectodomain region just prior to the TM domain, presumably reflecting essential sequence-specific interactions between the outer layer and the trimer core (33).
Although the wt SFV E1 stem was not required for fusion, it does appear to be optimized for the highest fusion activity, since several stem mutants induced smaller syncytia (Fig. 4B). This decrease in fusion activity might be caused not only by the suboptimal packing of the stem onto the trimer core during hairpin formation but also by the suboptimal regulation of fusion by the E2 protein. A key regulatory step in alphavirus fusion is the low-pH-induced dissociation of the E1-E2 dimer, which releases E1 for its subsequent interaction with the target membrane and trimerization (15, 35). Previously described SFV E1 TM domain mutants showed less-stable heterodimers, E1 trimerization at a higher pH threshold, and decreased fusion activity (36). Similarly, tests of several of our stem mutants showed less-stable heterodimers, virus-cell fusion at an elevated pH threshold, and a higher sensitivity to acid inactivation (data not shown). These properties are in keeping with a role for the E1 stem-E2 interaction in the proper regulation of membrane fusion.
The Δ20 mutant was inactive in both lipid-mixing and content-mixing fusion assays. However, the Δ20 mutant E1 protein still responded to a low pH by converting to a conformation that reacted with the acid-specific E1a-1 MAb and that was both trypsin and SDS resistant (Fig. 7). The exposure of the E1a-1 MAb epitope and the formation of the SDS-resistant HT are correlated with the fold-back of domain III (26). Thus, the remaining ∼9-residue-long stem of the Δ20 mutant is apparently sufficient to allow the folding back of domain III but too short to allow the completion of the fusion reaction. Such an uncoupling of the fold-back of domain III from stem packing is observed when E1 HT formation is induced by low-pH treatment in the presence of fusion-inactive cholesterol-deficient target membranes (27). It is also supported by our previous finding that the prebinding of an antibody to the N-terminal stem region does not inhibit the insertion of the E1 ectodomain into the target membrane or the formation of an SDS-resistant HT (27). An alternative explanation for the Δ20 phenotype is that the severely shortened stem prevents even the initial fold-back of domain III and that the observed fold-back actually occurred upon membrane solubilization. The E1 HTs formed by wt SFV and the Δ20 and Δ20+11A mutants show very similar biochemical properties, and thus, we favor the idea that the Δ20 mutant E1 forms a hairpin at low pH but that stem truncation prevents the hairpin from carrying out the fusion reaction.
In summary, while the wt SFV stem sequence may have specific interactions with the trimer core that generate an additional driving force for the alphavirus fusion reaction, these interactions are not absolutely required. The fold-back of domain III appears to be the dominant step that locks the E1 protein into the highly stable hairpin conformation and provides energy for membrane merger.
Role of the stem of class II proteins.
For alphaviruses, our data indicate that membrane fusion and infection can take place in spite of major changes in the stem sequence. The structure and fusion mechanism of the flavivirus fusion protein E are very similar to those of the alphavirus E1 (16, 20). However, the E proteins have a longer stem region (∼50 residues versus ∼30 residues for the alphavirus E1 stem), and the E stem is predicted to be more structured, with two α-helical regions and a central conserved sequence. Compared to alphavirus E1 (14), during hairpin formation the flavivirus domain III moves a shorter distance toward the fusion loop, leaving a longer distance to be spanned by the stem (2, 30). This may mean that flavivirus membrane fusion involves more-critical stem-groove interactions. Further studies will be required to define potential functional differences of the stem regions of these class II proteins.
Acknowledgments
We thank Alice Guo for excellent technical assistance. We thank all of the members of our laboratory for helpful discussions and comments on the manuscript.
This work was supported by a grant to M.K. from the Public Health Service (R01 GM052929) and by Cancer Center Core Support grant NIH/NCI P30-CA13330.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chanel-Vos, C., and M. Kielian. 2004. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J. Virol. 78:13543-13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanel-Vos, C., and M. Kielian. 2006. Second-site revertants of a Semliki Forest virus fusion-block mutation reveal the dynamics of a class II membrane fusion protein. J. Virol. 80:6115-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffus, W. A., P. Levy-Mintz, M. R. Klimjack, and M. Kielian. 1995. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 69:2471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 9.Ekstrom, M., P. Liljeström, and H. Garoff. 1994. Membrane protein lateral interactions control Semliki Forest virus budding. EMBO J. 13:1058-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons, D. L., A. Ahn, P. K. Chatterjee, and M. Kielian. 2000. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 74:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons, D. L., A. Ahn, M. Liao, L. Hammar, R. Holland Cheng, and M. Kielian. 2004. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J. Virol. 78:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, D. L., and M. Kielian. 2002. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 76:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbons, D. L., M.-C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 15.Glomb-Reinmund, S., and M. Kielian. 1998. fus-1, a pH-shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit. J. Virol. 72:4281-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, S. C. 2005. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 64:231-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 18.Kielian, M., S. Jungerwirth, K. Ullrich Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian, M., and F. A. Rey. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kielian, M. C., S. Keränen, L. Kääriäinen, and A. Helenius. 1984. Membrane fusion mutants of Semliki Forest virus. J. Cell Biol. 98:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 23.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 24.Levy-Mintz, P., and M. Kielian. 1991. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J. Virol. 65:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao, M., and M. Kielian. 2005. The conserved glycine residues in the transmembrane domain of the Semliki Forest virus fusion protein are not required for assembly and fusion. Virology 332:430-437. [DOI] [PubMed] [Google Scholar]
- 26.Liao, M., and M. Kielian. 2005. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus-membrane fusion. J. Cell Biol. 171:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao, M., and M. Kielian. 2006. Site-directed antibodies against the stem region reveal low pH-induced conformational changes of the Semliki Forest virus fusion protein. J. Virol. 80:9599-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, Y. E., C. H. Eng, S. G. Shome, and M. Kielian. 2001. In vivo generation and characterization of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 75:8329-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13-22. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay, S., W. Zhang, S. Gabler, P. R. Chipman, E. G. Strauss, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, H. E., J. A. Gruenke, and J. M. White. 2003. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 10:1048-1053. [DOI] [PubMed] [Google Scholar]
- 34.Roussel, A., J. Lescar, M.-C. Vaney, G. Wengler, G. Wengler, and F. A. Rey. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14:75-86. [DOI] [PubMed] [Google Scholar]
- 35.Salminen, A., J. M. Wahlberg, M. Lobigs, P. Liljeström, and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus II: cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sjöberg, M., and H. Garoff. 2003. Interactions between the transmembrane segments of the alphavirus E1 and E2 proteins play a role in virus budding and fusion. J. Virol. 77:3441-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stiasny, K., C. Kössl, and F. X. Heinz. 2005. Differences in the postfusion conformations of full-length and truncated class II fusion protein E of tick-borne encephalitis virus. J. Virol. 79:6511-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlberg, J. M., W. A. M. Boere, and H. Garoff. 1989. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 63:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retrovir. 9:1051-1053. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]