Abstract
3-O-(3′,3′-dimethylsuccinyl)betulinic acid (PA-457 or bevirimat) potently inhibits human immunodeficiency virus type 1 (HIV-1) maturation by blocking a late step in the Gag processing pathway, specifically the cleavage of SP1 from the C terminus of capsid (CA). To gain insights into the mechanism(s) by which HIV-1 could evolve resistance to PA-457 and to evaluate the likelihood of such resistance arising in PA-457-treated patients, we sought to identify and characterize a broad spectrum of HIV-1 variants capable of conferring resistance to this compound. Numerous independent rounds of selection repeatedly identified six single-amino-acid substitutions that independently confer PA-457 resistance: three at or near the C terminus of CA (CA-H226Y, -L231F, and -L231M) and three at the first and third residues of SP1 (SP1-A1V, -A3T, and -A3V). We determined that mutations CA-H226Y, CA-L231F, CA-L231M, and SP1-A1V do not impose a significant replication defect on HIV-1 in culture. In contrast, mutations SP1-A3V and -A3T severely impaired virus replication and inhibited virion core condensation. The replication defect imposed by SP1-A3V was reversed by a second-site compensatory mutation in CA (CA-G225S). Intriguingly, high concentrations of PA-457 enhanced the maturation of SP1 residue 3 mutants. The different phenotypes associated with mutations that confer PA-457 resistance suggest the existence of multiple mechanisms by which HIV-1 can evolve resistance to this maturation inhibitor. These findings have implications for the ongoing development of PA-457 to treat HIV-1 infection in vivo.
The emergence of drug-resistant virus isolates is having an increasingly detrimental effect on disease outcome. Therefore, the identification of new drugs targeting novel sites of action remains a high research priority. One such new drug, PA-457 [3-O-(3′,3′-dimethylsuccinyl)betulinic acid], has been described by us (23) and others (17, 48). PA-457 potently inhibits diverse human immunodeficiency virus type 1 (HIV-1) isolates and retains full activity against strains of HIV-1 that are resistant to currently approved reverse transcriptase (RT), protease (PR), and fusion inhibitors (23, 48; unpublished data). PA-457 blocks HIV-1 maturation, and hence disrupts virus infectivity, by inhibiting a specific step in the maturation pathway (23, 48).
The HIV-1 Gag proteins are synthesized as a polyprotein precursor, known as Pr55Gag, that is cleaved by PR during or immediately after virus release from the infected cell. The PR-mediated Pr55Gag processing generates the four major Gag proteins matrix (MA), capsid (CA), nucleocapsid (NC), and p6, as well as spacer peptide 1 (SP1) (formerly known as p2) and spacer peptide 2 (SP2) (formerly referred to as p1) (5, 8, 42), and follows a sequential series of events that is kinetically controlled by the rate of processing at individual cleavage sites (7, 21, 29, 36, 41-43). Completion of the Gag processing cascade is essential for virus maturation and infectivity (3, 18, 20, 23, 36, 42, 43, 48). During maturation, the CA protein reorganizes to form a conical, condensed core in which the viral RNA genome in a complex with NC, RT, and integrase is located (5, 8, 42). PA-457 inhibits HIV-1 infectivity by blocking a late stage in PR-mediated Gag processing, specifically, the release of SP1 from the C terminus of CA (23, 48). Blocking CA-SP1 cleavage, either by site-directed mutagenesis (3, 43) or by PA-457 treatment (23, 48), prevents proper virion maturation. Particles generated in the absence of CA-SP1 processing fail to form conical cores and display an electron-dense layer of Gag adjacent to the viral membrane (3, 23, 43). Although the mechanism by which PA-457 prevents cleavage of CA-SP1 has not been fully defined, recent data suggest that the compound binds directly to the CA-SP1 region of an oligomeric form of Gag within the immature particle (38, 47).
A number of studies have indicated that Gag-Gag interactions during HIV-1 assembly are driven by a region of Gag spanning the C-terminal domain of CA, SP1, and the highly basic N terminus of NC (5, 8). Although structures of the two major domains of CA have been solved by X-ray crystallography (12, 13), the structure of the C-terminal ∼10 residues of CA and SP1 has not been defined. This region of Gag has been observed to be unstructured and highly flexible (13, 32, 45) but may adopt an α-helical conformation (3, 30). Single-amino-acid substitutions within SP1 as well as deletion of this peptide severely disrupt HIV-1 particle assembly and release (3, 20, 24, 43). Mutations near the C terminus of CA also inhibit virus particle production (1, 24, 25, 27). These observations have led to the proposal that the C terminus of CA and SP1 form a continuous assembly domain that promotes Gag multimerization. Some studies have also observed that mutations in this region disrupt the binding of Gag to membrane (14, 15, 25), supporting a link between Gag membrane binding and Gag multimerization mediated by CA and SP1 (16, 25, 34, 40). Despite the numerous studies addressing this issue, the precise role of the C terminus of CA and SP1 in HIV-1 assembly remains to be fully elucidated.
Due to the rapid rate of HIV-1 replication and the error-prone nature of HIV-1 RT, resistance to antiretroviral drugs emerges quickly both in culture and in vivo. Although resistance has serious clinical implications for the treatment of patients with antiretroviral drugs, the isolation of drug-resistant viral isolates in culture provides a useful tool for (i) elucidating drug targets and mechanisms of action and (ii) predicting the likelihood and type of resistance that may potentially arise in vivo. Both of these are pertinent to the development of new antiretroviral drugs such as PA-457.
In this study, we sought to identify a broad spectrum of replication-competent HIV-1 variants capable of conferring PA-457 resistance. Six single-amino-acid substitutions, which independently confer PA-457-resistance, were isolated. All the PA-457 resistance-conferring mutations mapped to the CA-SP1 junction of Gag, and no mutations were found in PR. Two of these mutations have been previously identified and were used to help identify the CA-SP1 cleavage site as the target of PA-457 activity (23, 48). The PA-457-resistant isolates were characterized with respect to their effects on virus replication, the kinetics of Gag processing, virus particle assembly and release, and virion morphology. The results demonstrate that acquisition of PA-457-resistance is associated with a variety of viral phenotypes, suggesting multiple possible mechanisms of resistance. This study represents the first comprehensive attempt to isolate and characterize a broad spectrum of mutations conferring resistance to this novel antiretroviral compound.
MATERIALS AND METHODS
PA-457, cell culture, and transfections.
PA-457 was prepared as described previously (11) and used at the concentrations indicated. The Jurkat T-cell line was maintained in RPMI 1640 supplemented with 10% (vol/vol) fetal bovine serum (FBS). HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 5% (vol/vol) FBS. All media were supplemented with l-glutamine (2 mM), penicillin, and streptomycin. Plasmid DNA was purified with the plasmid purification maxiprep kit (QIAGEN) and adjusted to 1 μg/μl. HeLa cells were transfected by calcium phosphate coprecipitation (9) or with ExGen500 (Fermentas), and Jurkat T cells were transfected using DEAE-dextran (19).
Selection for PA-457 resistance in vitro.
PA-457-resistant viral isolates were selected by serial passage of Jurkat T cells transfected with wild-type (WT) HIV-1 pNL4-3 (4) at 50 ng/ml PA-457. Virus replication during the selection process was monitored by RT activity as previously described (9). Cell pellets and virus supernatants were harvested on the days of peak RT activity. Following the first round of selection, virus supernatants were normalized for RT activity and used to infect fresh Jurkat T cells. The infected cells were cultured as described above to confirm acquisition of PA-457 resistance. To identify mutations conferring PA-457 resistance, genomic DNA was extracted from cells on the day of peak RT activity by using a whole-blood DNA purification kit (QIAGEN), and the entire Gag-PR-coding region was amplified by PCR using the forward and reverse primers NL516F (5′-TGC CCG TCT GTT GTG TGA CTC-3′) and NL2897R (5′-AAA ATA TGC ATC GCC CAC AT-3′), respectively. The 2.3-kb PCR product was purified using the QIAquick PCR purification kit (QIAGEN) and sequenced using the primers NL645F (5′-AAC AGG GAC TTG AAA GCG-3′), NL1155F (5′-AGG AAA CAA CAG CCA GGT-3′), NL1410F (5′-GGA AGC TGC AGA ATG GGA TA-3′), NL1754F (5′-TGG TCC AAA ATG CGA ACC-3′), and NL2135F (5′-TTC AGA GCA GAC CAG AGC CAA-3′). Resistance-conferring mutations were introduced into the parental pNL4-3 backbone by subcloning the 500-bp SphI-ApaI gag fragment (nucleotides 1443 to 2006) from the PA-457-resistant virus PCR product into pNL4-3, creating six derivatives of pNL4-3: CA-H226Y, CA-L231M, CA-L231F, SP1-A1V, SP1-A3T, and SP1-A3V. The identities of all plasmids generated were confirmed by sequencing.
Selection of second-site mutations compensating for the SP1-A3V mutation was carried out in Jurkat T cells as described above, except that Jurkat cells were transfected with pNL4-3-SP1-A3V and the cultures were maintained in either 50 ng/ml or 1.0 μg/ml PA-457.
Site-directed mutagenesis.
The 500-bp SpeI-ApaI fragment from pNL4-3-CA-G225S/SP1-A3V (nucleotides 1507 to 2006) was subcloned into Bluescript SK(+) (Stratagene) and mutagenized to generate CA-G225S alone or CA-G225S/SP1-A3T with the following mutagenic primers: for CA-G225S, forward 5′-GCA AGA GTT TTG GCT GAA GCA ATG AGC CAA GTA ACA AAT CC-3′ and reverse 5′-GGA TTT GTT ACT TGG CTC ATT GCT TCA GCC AAA ACT CTT GC-3′; for CA-G225S/SP1-A3T, forward 5′-GCA AGA GTT TTG GCT GAA ACA AGT AGC CAA GTA ACA AAT CC-3′ and reverse 5′-GGA TTT GTT ACT TGG CTC ATT GTT TCA GCC AAA ACT CTT GC-3′. All mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Following confirmation of the mutagenesis by sequencing, the SpeI-ApaI fragment was cloned back into WT pNL4-3 to create the molecular clones pNL4-3-CA-G225S and pNL4-3-CA-G225S/SP1-A3T, which were reconfirmed by DNA sequencing.
Radioimmunoprecipitation analysis.
Methods used for metabolic labeling of HeLa cells, preparation of cell and virus lysates, and immunoprecipitation have been previously described in detail (9, 44). Briefly, media and solutions containing PA-457 at the indicated concentrations were prepared immediately before use and vortexed. PA-457 was maintained throughout the transfection and radioimmunoprecipitation procedures. Transfected HeLa cells were starved in Cys/Met-free medium for 30 min and then metabolically radiolabeled for 2 h with [35S]Cys/Met Pro-mix (Amersham). Virions were pelleted by ultracentrifugation. Cell and virus lysates were immunoprecipitated with pooled immunoglobulin from HIV-1-infected patients (HIV-Ig) obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. The radioimmunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and exposed to X-ray film and a phosphorimager plate (Fuji), and the bands were quantified by using Quantity One software (Bio-Rad).
Pulse-chase analysis of CA-SP1 processing.
HeLa cells were transfected with WT or mutant pNL4-3 molecular clones. Twenty-four hours posttransfection, cells were starved for 30 min at 37°C in Cys/Met-free medium and pulse-labeled in the same medium for 15 min at 37°C using 50 μCi of [35S]Cys/Met Pro-mix (Amersham). The cells were then washed, resuspended in Dulbecco modified Eagle medium supplemented with 10% FBS, divided into four equal aliquots, and incubated at 37°C. Cells were harvested at 0-, 30-, 60-, and 90-min chase time points. The cells were lysed, immunoprecipitated, and analyzed as described above.
Rabbit reticulocyte lysate in vitro assembly system.
Plasmid pDABCh3 has been described previously (37). Plasmids pDABCh3.A3V, pDABCh3.H226Y, pDABCh3.L231F, and pDABCh3.L231M were constructed by subcloning the 499-bp SpeI-to-ApaI fragment from pNL4-3-SP1-A3V, -CA-H226Y, -CA-L231F, and -CA-L231M, respectively, into the pDABCh3 background. Production of assembled Gag substrates was performed as previously described (38). Briefly, [35S]Met-labeled Gag was synthesized and allowed to assemble in rabbit reticulocyte lysates (Novagen). Assembled Gag was separated in sucrose gradients, and peak fractions were pooled for use as the processing substrate. Processing reactions were performed for 3 h in the presence or absence of PA-457, at the indicated concentrations, with recombinant HIV-1 PR (Bachem) according to the standardized protocol described previously (38). Radioactivity in the CA and CA-SP1 bands was quantified and adjusted for the number of methionines in each protein. The proportion of remaining CA-SP1 for each reaction was compared to that seen in the control reaction (no PA-457) and then plotted as the percent change in that proportion.
Replication kinetics.
Jurkat T cells were transfected with WT or mutant pNL4-3 molecular clones. PA-457 was added at the time of transfection and was maintained throughout the course of the experiment. The Jurkat cells were split every 2 days, supernatant collected at each time point, and viral replication monitored by RT activity as previously described (9).
Transmission EM.
HeLa cells were transfected with WT or mutant pNL4-3 molecular clones. PA-457 was added at the time of transfection and was maintained until the cells were fixed. Fixation of cells, preparation of samples, and transmission electron microscopy (EM) were performed as described previously (10).
RESULTS
Selection for PA-457-resistant HIV-1 isolates in vitro.
To select for HIV-1 escape from PA-457, we first identified optimal conditions for the emergence of PA-457 resistance in culture. At a PA-457 concentration of 0.1 or 1.0 μg/ml (approximately 0.2 and 2 mM, respectively), virus replication was completely blocked in Jurkat T cells transfected with the full-length molecular clone pNL4-3 for a period of 3 months (data not shown). In contrast, at 50 ng/ml, virus replication was reproducibly observed, with a significant delay to peak RT activity relative to untreated cultures (Fig. 1A). To identify mutations potentially conferring resistance to PA-457, 10 flasks of pNL4-3-transfected Jurkat T cells were cultured in parallel at 50 ng/ml PA-457, genomic DNA was extracted from cells harvested at the peak of RT activity, and the Gag- and PR-coding regions were amplified by PCR and sequenced. Six individual mutations were observed in the vicinity of the CA-SP1 cleavage site: CA-H226Y, CA-L231F, CA-L231M, SP1-A1V, SP1-A3T, and SP1-A3V (Fig. 1B). An additional series of selections in 12 independent flasks at 50 ng/ml yielded no additional mutants (data not shown). No mutations were identified in PR. We and others have previously identified the CA-L231F (48) and SP1-A1V (23) mutations in PA-457 selection experiments.
FIG. 1.
Selection for PA-457 resistance. (A) Ten flasks of pNL4-3-transfected Jurkat cells were cultured in parallel in the presence of 50 ng/ml PA-457. As controls, two flasks transfected with either pUC19 or pNL4-3 were cultured without PA-457. Genomic DNA was extracted at peak RT activity and analyzed by PCR amplification and DNA sequencing. (B) Mutations identified in the selection experiments shown in panel A. Pr55Gag is represented at the top, with the MA, CA, NC, and p6 domains and the SP1 and SP2 spacer peptides indicated. The alignment shows each of the six potential PA-457 resistance mutations identified.
Biochemical characterization of PA-457-resistant mutants in HIV-1-expressing cells.
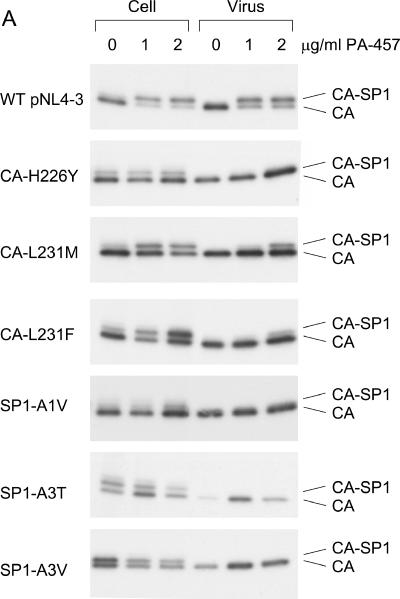
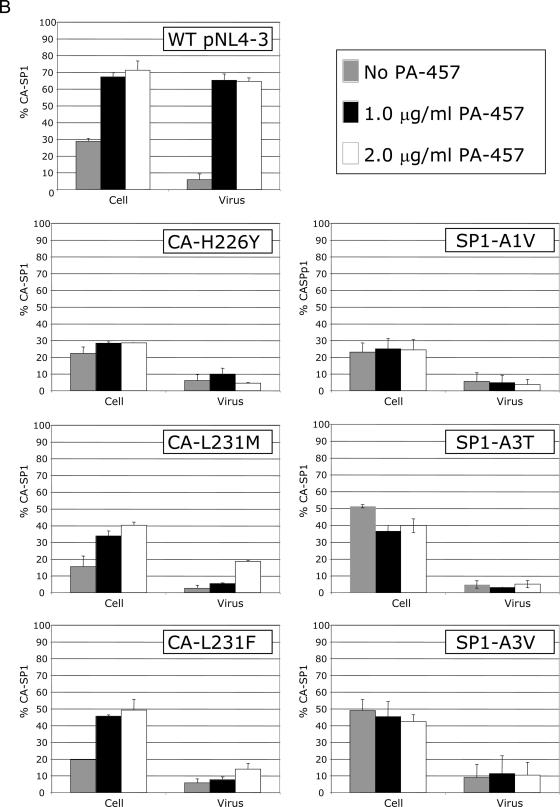
To evaluate the effect of the CA and SP1 substitutions on the sensitivity of HIV-1 to PA-457, we introduced each mutation into the NL4-3 background to generate molecular clones pNL4-3-CA-H226Y, -CA-L231M, -CA-L231F, SP1-A1V, -SP1-A3T, and -SP1-A3V. To determine the effect of these mutations on the ability of PA-457 to disrupt CA-SP1 processing (23, 48), HeLa cells transfected with WT or mutant pNL4-3 molecular clones were cultured either with no drug or with 1.0 or 2.0 μg/ml PA-457. The cells were metabolically labeled with [35S]Met/Cys, and cell- and virion-associated proteins were immunoprecipitated with HIV-Ig. CA-SP1 cleavage was detected (Fig. 2A) and quantified as the percentage of CA-SP1 relative to total CA-SP1 plus CA (Fig. 2B).
FIG.2.
CA-SP1 processing of PA-457-resistant mutants displays insensitivity to PA-457. HeLa cells were transfected with WT pNL4-3 or derivatives containing the indicated mutations and were cultured without PA-457 or in the presence of either 1.0 or 2.0 μg/ml PA-457. Cells were metabolically labeled for 2 h with [35S]Met/Cys, and released virions were pelleted by ultracentrifugation. Cell and virus lysates were immunoprecipitated with HIV-Ig, and processing of CA-SP1 to CA was analyzed by SDS-PAGE and fluorography (A) followed by phosphorimager analysis to quantify the percentage of CA-SP1 relative to total CA-SP1 plus CA (B). Error bars indicate standard deviations (n = 2).
Consistent with our previous results (23), treatment of WT pNL4-3-transfected cells with PA-457 resulted in a marked accumulation of CA-SP1 in both cell and virion fractions (Fig. 2A and B). In contrast, in the presence of PA-457, no accumulation of CA-SP1 in either the cell or virus fractions was observed for mutant CA-H226Y, SP1-A1V, SP1-A3T, or SP1-A3V (Fig. 2A and B). For CA-H226Y and SP1-A1V, the level of CA-SP1 processing in the absence of PA-457 was similar to that of the WT under these conditions. However, in cells transfected with mutant SP1-A3T or SP1-A3V, the percentage of CA-SP1 was elevated even in the absence of PA-457 (Fig. 2A and B), suggesting that the A3V and A3T mutations themselves decrease the extent of CA-SP1 processing. Expression of CA-L231F and CA-L231M resulted in a low-level accumulation of CA-SP1 in the presence of PA-457 (Fig. 2A and B). Although the accumulated CA-SP1 band for these mutants can be clearly seen in the virus fraction at 2.0 μg/ml PA-457, the percentage of accumulated CA-SP1 was four- to fivefold lower than that observed in PA-457-treated WT virus (Fig. 2A and B). These results indicate that each mutation independently confers resistance to PA-457. Restoration of CA-SP1 processing at 2.0 μg/ml was not complete when the terminal CA residue (Leu) was mutated to Phe (L231F) or Met (L231M), indicating that these mutations confer partial resistance at the higher concentration of PA-457.
Effect of the PA-457 resistance mutations on the kinetics of CA-SP1 processing.
The biochemical data presented above suggest that the extent of cell-associated CA-SP1 cleavage is decreased by the SP1-A3V and SP1-A3T mutations independent of drug. We conducted pulse-chase analysis to investigate more precisely the effect of each mutation on the extent of CA-SP1 processing (Fig. 3). The kinetics of CA-SP1 processing for mutants with resistance-conferring mutations in CA (H226Y, L231M, and L231F) were comparable to those of the WT. However, different degrees of CA-SP1 cleavage were observed for those mutants with substitutions in SP1. In agreement with the biochemical data above, mutations in the third SP1 residue (A3T and A3V) resulted in significantly delayed CA-SP1 cleavage compared to the WT. The delay in CA-SP1 processing was more pronounced for SP1-A3T than for SP1-A3V. The SP1-A1V mutation increased the extent of CA-SP1 processing relative to WT at all time points. To verify consistency between pulse-chase samples in this analysis, we monitored gp160-to-gp120 processing in the same lysates used for the measurement of CA-SP1 and CA levels. As expected, we observed that the ratios of gp160 to gp120 were indistinguishable between samples at each time point (data not shown), thus confirming the consistency of the pulse-chase analysis. Importantly, in these assays no correlation between rate of CA-SP1 processing and PA-457 resistance was observed, as various degrees of cleavage were observed within the set of six resistance-conferring mutations.
FIG. 3.
The extent of CA-SP1 processing does not correlate with PA-457 resistance. HeLa cells were transfected with WT pNL4-3 or derivatives containing the indicated mutations and pulse-labeled for 15 min with [35S]Met/Cys. Cells were chased for the indicated times in unlabeled medium, and cell lysates were immunoprecipitated with HIV-Ig. The extent of processing of CA-SP1 to CA was analyzed by SDS-PAGE and fluorography (A and B) followed by phosphorimager analysis to quantify the percentage of CA-SP1 relative to total CA-SP1 plus CA (C and D). Error bars indicate standard errors of the means (n = 4 [C] or 3 [D]).
Biochemical characterization of PA-457-resistant mutants in an in vitro assembly system.
To confirm the biochemical data presented above, we tested the effects of several of the PA-457 resistance mutations in an in vitro assembly system (37). This system uses an HIV-1 Gag derivative known as chimera 3 (Ch3), which contains the M-PMV p12 region fused to the C terminus of HIV-1 Gag in the place of p6. Expression of Ch3 in rabbit reticulocyte lysates leads to the assembly of structures that resemble immature HIV-1 capsids (37). Addition of recombinant PR to these gradient-purified, assembled structures results in Gag processing, with both CA-SP1 and mature CA readily detectable (37) (Fig. 4A). In the presence of ∼600 ng/ml (1 μM) or ∼6 μg/ml (10 μM) PA-457, the conversion of CA-SP1 to CA was markedly inhibited in a dose-dependent fashion (Fig. 4A and B). Introduction of mutation CA-H226Y, CA-L231F, CA-L231M, SP1-A1V, or SP1-A3V into Ch3 resulted in a nearly complete loss of the ability of PA-457 to block cleavage of CA-SP1 to CA (Fig. 4A and B). Consistent with the partial resistance observed for CA-L231F in our biochemical assays (Fig. 2A and B), a small reduction in processing of CA-SP1 upon addition of 6 μg/ml PA-457 was observed for this mutant in the reticulocyte lysate system (Fig. 4A and B). We also tested the effect of these mutations on the processing of Gag in the absence of PA-457 (Fig. 4C). Several of the mutations increased the extent of CA-SP1 processing, with SP1-A1V having the most significant effect. In contrast, mutation SP1-A3V inhibited CA-SP1 processing. The increased degree of CA-SP1 processing for SP1-A1V and the decreased degree for SP1-A3V are consistent with the results obtained in the cell-based pulse-chase analysis described above.
FIG. 4.
Biochemical characterization of PA-457-resistant mutants in an in vitro assembly system. [35S]Met-labeled assembled Gag was used as substrate for proteolytic processing by 3 h of incubation with purified PR in the presence or absence of PA-457 at the indicated concentrations. (A) Portions of gels from representative experiments showing CA-SP1 and CA. The concentration of PA-457 is given at tops of gel lanes (0.6 μg/ml ∼ 1 μM; 6 μg/ml ∼ 10 μM). (B) Quantitative representation of the gel data from panel A normalized to the maximum amount of processing as seen in the no-drug control. (C) Extent of cleavage achieved under standard reaction conditions in the absence of drug, presented as percentage of CA-SP1 relative to total CA-SP1 plus CA. For panels B and C, error bars indicate the standard deviations for six replicate experiments. Note that the data for SP1-A1V were reported in a previous study (38) and are recapitulated here for comparison.
Replication kinetics of PA-457-resistant mutants.
To confirm that the CA and SP1 mutations confer PA-457 resistance and to investigate any effects of these mutations on virus replication capacity, we evaluated virus replication kinetics in the Jurkat T-cell line. Each WT or mutant molecular clone was transfected into Jurkat cells and passaged either without drug or in the presence of the suboptimal (50 ng/ml) or fully inhibitory (1.0 μg/ml) concentration of PA-457 (Fig. 5). Similar results were obtained from several independently repeated experiments. In the absence of PA-457, WT NL4-3 replication peaked at approximately 1 week posttransfection. At 50 ng/ml, virus replication emerged between 2 to 3 weeks posttransfection, presumably upon acquisition of a resistance-conferring mutation(s). At 1.0 μg/ml PA-457, no virus replication was detected. The CA mutants replicated with essentially WT kinetics in the absence of PA-457, and their replication was not significantly affected at 50 ng/ml PA-457. A several-day delay was repeatedly observed for these mutants at 1.0 μg/ml PA-457, indicating a residual level of drug sensitivity. The SP1 mutants exhibited three distinct phenotypes (Fig. 5). The SP1-A1V mutant consistently replicated with WT kinetics in the absence of PA-457, and its replication was not affected by even high drug concentrations. In contrast, the A3T and A3V mutations severely impaired virus replication. SP1-A3T failed to display detectable replication for 1 month in culture under any conditions. SP1-A3V exhibited a less severely impaired replication phenotype than SP1-A3T, with RT levels peaking at 2 to 3 weeks posttransfection both without PA-457 and at a 50-ng/ml concentration of PA-457. Interestingly, the peak of virus replication for SP1-A3V occurred earlier at the higher (1.0-μg/ml) drug concentration than in the absence of the compound (Fig. 5). This enhanced replication of SP1-A3V at high PA-457 concentrations was observed in numerous independent replication assays (data not shown; see below).
FIG. 5.
Replication kinetics of PA-457-resistant viruses. Jurkat T cells were transfected with WT pNL4-3 or derivatives containing the indicated mutations. Cultures were maintained either without PA-457 or with 50 ng/ml or 1.0 μg/ml PA-457. Cells were split every 2 days, and supernatants were reserved at each time point for RT analysis.
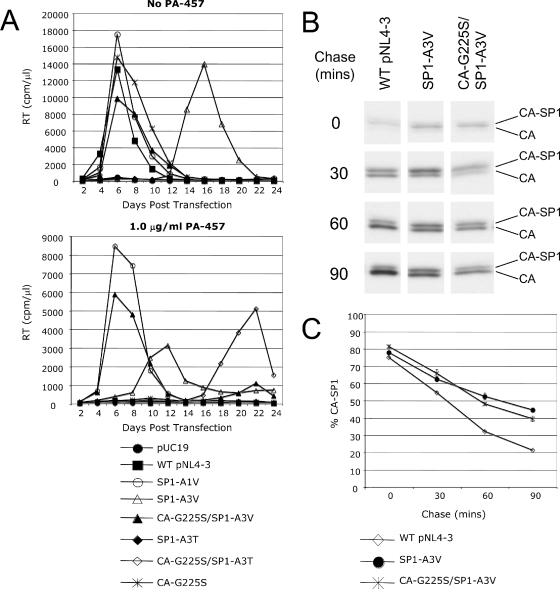
SP1-A3V compensatory mutations and PA-457 dependence.
The data presented above demonstrate that mutations in the third residue of SP1 confer PA-457 resistance in biochemical assays but markedly decrease viral fitness when cells are cultured without PA-457 or at a low concentration of the compound. The A3V mutation exhibits a less severely impaired replication phenotype than does A3T. To determine whether the virus replicating in the pNL4-3-SP1-A3V-transfected cultures gained a compensatory mutation that reversed the replication defect imposed by A3V, we determined the genotype of the virus from these cultures at the peak of RT activity. Interestingly a Gly-to-Ser change at CA residue 225 (CA-G225S) was identified. To determine whether this mutation compensates for the A3V-imposed replication defect, the CA-G225S/SP1-A3V double mutant was generated. This double mutant displayed WT fitness and a high level of PA-457 resistance in both replication and biochemical assays (Fig. 6A and data not shown). The CA-G225S single mutant did not replicate in the presence of PA-457 (Fig. 6A), demonstrating that this mutation acts as a compensatory mutation to SP1-A3V and does not independently confer PA-457 resistance. This observation was confirmed by biochemical analysis: CA-G225S/SP1-A3V restored CA-SP1 processing in the presence of PA-457, whereas mutant CA-G225S accumulated CA-SP1 to the same levels as PA-457-treated WT virus (data not shown). It is noteworthy that, as seen for SP1-AV3 and -A3T (Fig. 2A and B), the percentage of CA-SP1 in cells expressing CA-G225S/SP1-A3V were elevated relative to WT (data not shown) and that the extent of CA-SP1 processing was reduced to the same extent as for SP1-A3V (Fig. 6B and C). These results indicate that while the CA-G225S mutation reverses the replication defect imposed by SP1-A3V, it does not correct the A3V defect in CA-SP1 processing.
FIG. 6.
CA-G225S compensates for the replication defect imposed by SP1-A3V. (A) Jurkat T cells were transfected with the indicated WT or mutant molecular clones. Cultures were maintained either without or with 1.0 μg/ml PA-457. Cells were split every 2 days, and supernatants were reserved at each time point for RT analysis. (B and C) Effect of the CA-G225S mutation on the extent of cleavage of CA-SP1 to CA. HeLa cells were transfected with WT pNL4-3 or derivatives containing the indicated mutations and pulse-labeled for 15 min with [35S]Met/Cys. Cells were chased for the indicated times, cell lysates were immunoprecipitated with HIV-Ig, and the extent of processing of CA-SP1 to CA was analyzed by SDS-PAGE and fluorography (B) followed by phosphorimager analysis to quantify the percentage of CA-SP1 relative to total CA-SP1 plus CA (C). Error bars indicate standard errors of the means (n = 3).
To investigate whether the CA-G225S mutation also rescues the SP1-A3T replication defect, a second double mutant, CA-G225S/SP1-A3T, was constructed. In this case, the CA-G225S mutation did not rescue virus replication when cells were cultured in the absence of PA-457. Interestingly, however, delayed replication was observed for CA-G225S/SP1-A3T at 1.0 μg/ml PA-457 (Fig. 6A). Replication of CA-G225S/SP1-A3T at a high concentration of PA-457 but not in its absence suggests that, as observed for SP1-A3V, the replicative capacity of CA-G225S/SP1-A3T is enhanced by PA-457.
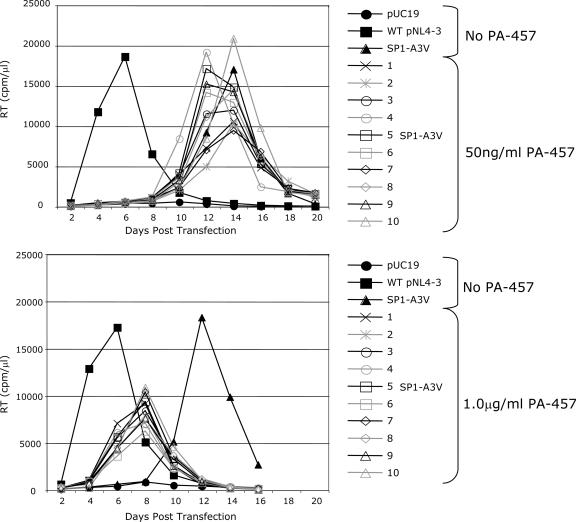
The results presented above indicate that the replication capacity of SP1 residue 3 mutants can be increased by second-site compensatory changes (e.g., CA-G225S) and that the replication of mutants bearing substitutions in the third residue of SP1 can be stimulated by high concentrations of PA-457. To investigate further the observed enhancement of SP1-A3V replication capacity in the presence of PA-457, we transfected Jurkat cells with NL4-3-SP1-A3V and cultured 2 flasks without PA-457, 10 flasks at 50 ng/ml PA-457, and 10 flasks at 1.0 μg/ml PA-457 (Fig. 7). Virus replication peaked 12 to 14 days posttransfection in the flasks cultured without or with 50 ng/ml PA-457. In contrast, at 1.0 μg/ml PA-457, virus replication peaked uniformly at 8 days posttransfection. Upon repassage of these virus populations at the drug concentration in which they were initially cultured, virus replication at 50 ng/ml PA-457 now peaked at the same time or earlier than replication of viruses cultured in 1.0 μg/ml PA-457 (data not shown). The CA-SP1 region of Gag was sequenced to identify potential compensatory mutations. Sequence data were obtained for eight flasks selected at 50 ng/ml PA-457, and mutations were identified in six (75%) of those flasks. The mutations identified were the previously characterized CA-G225S and three new mutations: SP1-V7I, SP1-T8I, and SP1-T8R (data not shown). Sequencing results were obtained for one flask selected without drug, and the SP1-T8I mutation was observed. Sequence information was obtained for seven flasks cultured in 1.0 μg/ml PA-457, and a change (CA-G225S) was observed in only one flask (14%); no mutations were observed in the remaining six flasks. This analysis of the CA-SP1 region indicates that NL4-3-SP1-A3V typically acquires compensatory changes when propagated in the absence of PA-457 or at low concentrations of the compound yet can replicate in the absence of compensatory changes at high concentrations of PA-457. These results support the hypothesis that the replication capacity of SP1 residue 3 mutants is enhanced at high PA-457 concentrations. These data also suggest the possibility that the SP1-V7I, SP1-T8I, and SP1-T8R mutations could, like CA-G225S, act as second-site compensatory changes for SP1-A3V.
FIG. 7.
Replication of SP1-A3V at various concentrations of PA-457. Ten flasks of Jurkat T cells transfected with pNL4-3-SP1-A3V were cultured in parallel in the presence of 50 ng/ml or 1.0 μg/ml PA-457. Cells transfected with pUC19, WT pNL4-3, or pNL4-3-SP1-A3V, cultured without PA-457, served as controls. Cells were split every 2 days, and supernatants were reserved at each time point for RT analysis.
Virus release efficiency of the PA-457-resistant SP1 mutants.
To investigate further the phenotypes associated with SP1-A3T and SP1-A3V, we examined the virus release efficiency of these mutants (Fig. 8A and B). The WT, along with SP1-A1V and CA-G225S/SP1-A3V, both of which are highly fit and drug resistant, were used as controls.
FIG. 8.
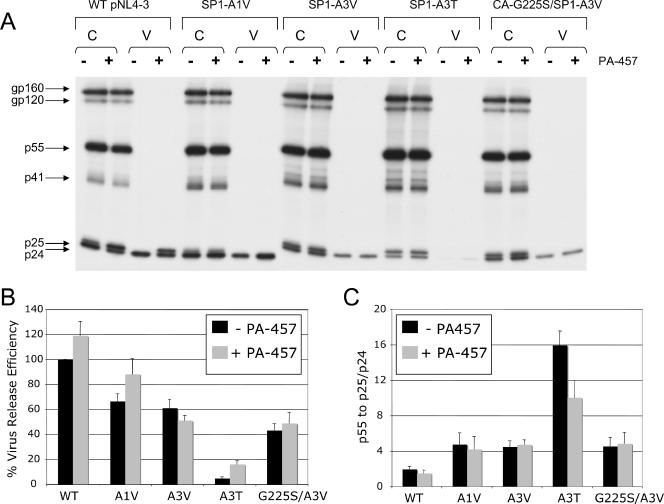
SP1-A3T exhibits inefficient virus release and Gag processing. HeLa cells were transfected with WT pNL4-3 or derivatives containing the indicated mutations and were cultured without PA-457 or in the presence of 1.0 μg/ml PA-457. Cells were metabolically labeled with [35S]Met/Cys, and released virions were pelleted by ultracentrifugation. Cell and virus lysates were immunoprecipitated with HIV-Ig and analyzed by SDS-PAGE and fluorography (A) followed by phosphorimager analysis to quantify virus release efficiency, calculated as the amount of particle-associated Gag as a fraction of total (cell plus virion) Gag (B) and the cellular ratio of Pr55Gag to total CA-SP1 plus CA (C). Error bars indicate standard errors of the means (n = 6 [B] or 5 [C]).
The SP1-A3T mutation imposed a significant defect in virus release, with particle production reduced 20-fold in the absence of PA-457 (Fig. 8A and B). This virus release defect was less severe in the presence of PA-457, as virus production was increased approximately threefold. We also noted that the ratio of Pr55Gag to CA-SP1 plus CA in cell lysates was significantly elevated (Fig. 8C), and again, this defect was partially corrected by PA-457. For SP1-A3V, virus release was reduced approximately twofold compared to WT, an efficiency not significantly different from that of SP1-A1V and CA-G225S/SP1-A3V. Thus, the SP1-A3T but not the SP1-A3V mutation dramatically reduces particle production and impairs Pr55Gag processing, and PA-457 modestly improves both A3T virus release and Gag processing.
Morphology of PA-457-resistant mutants.
Next we performed EM analysis to examine the morphology of PA-457-resistant viruses (Fig. 9). In contrast to WT particles from untreated HeLa cells, which contain condensed conical cores (Fig. 9a), WT particles from PA-457-treated cells display aberrant, spherical, and often acentric cores with an additional electron-dense layer inside the viral membrane (23) (Fig. 9a to c). Consistent with the biochemical and replication data presented above, the PA-457-resistant viruses CA-H226Y, -L231M, and -L231F (data not shown) and SP1-A1V (Fig. 9d and e) all displayed a morphology indistinguishable from that of untreated WT viruses, showing condensed, conical cores even in the presence of PA-457.
FIG. 9.
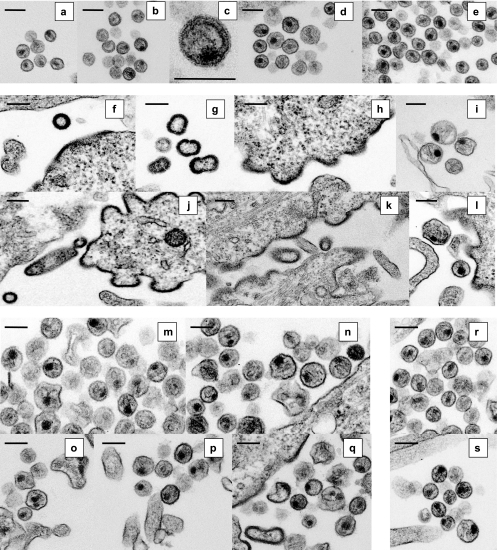
Morphology of PA-457-resistant virus particles. Thin-section transmission EM analysis of virions with the indicated mutations produced from HeLa cells cultured without PA-457 or with 5.0 μg/ml PA-457 is shown. Cells were fixed at 48 h posttransfection and were analyzed by EM. (a to c) WT pNL4-3 without (a) or with (b and c) PA-457; (d and e) SP1-A1V without (d) or with (e) PA-457; (f to i) SP1-A3V without PA-457; (j to l) SP1-A3T without PA-457; (m and n) SP1-A3V with PA-457; (o to q) SP1-A3T with PA-457; (r and s) CA-G225S/SP1-A3V without (r) or with (s) PA-457. Bars, 200 nm.
A very different phenotype was observed for the SP1-A3V and A3T mutants. In the absence of PA-457, the majority of released particles had an aberrant morphology, with an electron-dense ring resembling that of immature virions, and many were large and distorted (Fig. 9f to h, j, and k). A small number of released particles contained an aberrant condensed core (Fig. 9i and l). The abnormal morphology of these viruses is consistent with their deficient replication capacity when cultured in the absence of PA-457. Interestingly, the number of SP1-A3V and -A3T particles containing condensed cores was increased at a high concentration of PA-457. Instead of the immature-like particles produced without PA-457, the majority of released virions produced in the presence of the compound displayed some degree of core condensation, though many aberrant cores were still observed (Fig. 9m to q). The increased core condensation observed with SP1 residue 3 mutants at a high concentration of PA-457 is consistent with the enhancement in virus replication kinetics seen for these mutants in the presence of the compound (see Fig. 7).
Finally, we examined the morphology of CA-G225S/SP1-A3V virus particles. In both the absence (Fig. 9r) and presence (Fig. 9s) of PA-457, CA-G225S/SP1-A3V virions frequently contained condensed, conical cores. These EM observations are consistent with the highly fit and PA-457-resistant phenotype of CA-G225S/SP1-A3V described above.
DISCUSSION
In this study, we sought to identify the full range of replication-competent HIV-1 variants capable of conferring resistance to PA-457. Extensive selection experiments identified six single-amino-acid substitutions that independently confer PA-457 resistance: CA-H226Y, CA-L231F, CA-L231M, SP1-A1V, SP1-A3T, and SP1-A3V. The two PA-457-resistant mutants previously reported by us (SP1-A1V) (23) and others (CA-L231F) (48) were both reisolated in this study. Several of the mutants identified here were selected multiple times, with SP1-A1V arising most frequently. Upon further repetition of the selection experiments, no additional mutations were identified. Together, these results suggest that we have identified most, if not all, replication-competent NL4-3 derivatives capable of conferring resistance to PA-457. No mutations conferring resistance were selected in Gag outside the CA-SP1 boundary region or in PR, suggesting that PA-457 resistance in culture rarely, if ever, maps to residues far removed from the CA-SP1 junction.
This study extends the determinants of PA-457 resistance and hence its molecular target beyond the residues that immediately flank the CA/SP1 cleavage site. Residues to which PA-457 resistance was previously mapped, CA-L231 and SP1-A1, flank the scissile bond between CA and SP1, defined as the P1 and P1′ positions, respectively, of the PR recognition site. While SP1-A3 also lies within the PR recognition site (at the P3′ position), CA-H226 lies upstream of those residues involved in PR substrate recognition (39). All of the amino acid positions to which PA-457 resistance maps are, however, located in the putative α-helical domain that spans the CA-SP1 boundary and is proposed to promote Gag multimerization (3, 24, 25, 31).
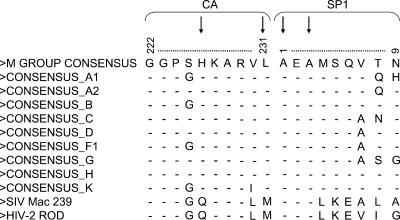
The identification of a PA-457-resistant isolate with a Leu-to-Met change at CA residue 231 (CA-L231M) is noteworthy because HIV-2 and simian immunodeficiency virus SIVmac both encode a Met at this position (Fig. 10). CA residue 226, at which a His-to-Tyr change in HIV-1 confers PA-457 resistance, is also not conserved between HIV-1 and HIV-2 or SIVmac. This observation is significant because both HIV-2 and SIVmac are naturally resistant to the action of PA-457 (23, 48). Replacement of the HIV-1 P2 and P1 residues (CA-V230 and -L231) with the corresponding SIVmac amino acids also results in PA-457 resistance, and conversely, introduction of HIV-1 CA-SP1 residues into the SIVmac CA-SP1 region renders SIVmac sensitive to PA-457 (46). Although several residues at the CA/SP1 junction are not conserved between HIV-1, HIV-2, and SIVmac, this study demonstrates that the L231M or H226Y substitution alone is sufficient to confer PA-457 resistance.
FIG. 10.
Amino acid positions to which PA-457 resistance maps are highly conserved between HIV-1 isolates. An amino acid sequence alignment of the CA-SP1 boundary region of Gag is shown. The alignment was constructed from the 2004 Los Alamos HIV-1 sequence database group M consensus sequences and SIVmac239. Arrows indicate amino acid positions to which PA-457 resistance maps (22; http://hiv-web.lanl.gov/content/hivdb/CONSENSUS/M_GROUP/Consensus.html).
The comprehensive selection for PA-457-resistant isolates conducted in this study enables us to anticipate the potential emergence of PA-457 resistance in vivo. This is significant, as PA-457 is currently undergoing clinical trials and the emergence of resistant isolates could have major implications for the treatment of HIV-1-infected patients. This study shows that PA-457 resistance can arise as independent single-amino-acid substitutions at a number of positions within the CA/SP1 boundary region and that the majority of these mutations (CA-H226Y, CA-L231M, CA-L231F, and SP1-A1V) do not impose a significant replication cost to the virus in culture. In contrast, the two mutations at the third SP1 residue, SP1-A3V and SP1-A3T, significantly impaired virus replication. Second-site mutations were readily acquired by SP1-A3V, which, as demonstrated for CA-G225S, could compensate for the replication defect imposed by SP1-A3V. The amino acid positions to which the second-site mutations map are not conserved within HIV-1 isolates, and at CA residue 225 Gly and Ser occur interchangeably (Fig. 10). These data could be interpreted as predicting that resistance to PA-457 will arise relatively easily in vivo. However, all of the residues to which PA-457 resistance maps are highly conserved among HIV-1 isolates (Fig. 10), implying that there may be a fitness cost associated with changes at these positions in vivo. Furthermore, in a phase IIa, 10-day monotherapy clinical study with HIV-1-infected patients, none of the resistance-conferring mutations described here were detected. The absence of resistance-conferring mutations is consistent with the lack of viral load rebound during the dosing period (unpublished data). While these initial clinical results are encouraging with respect to resistance, longer-term studies will be needed to determine the frequency and rate with which resistance to PA-457 emerges in patients on combination therapy. The results presented here will provide valuable guidance in evaluating patient-derived sequences for potential PA-457 resistance.
Characterization of the PA-457-resistant viral isolates described here demonstrated that the SP1 residue 3 mutants differed significantly from those bearing changes in CA or in SP1 residue 1. Replacement of SP1-A3 with Val or Thr imposed a moderate to severe virus replication defect. This defect correlated with aberrant virion morphology; in the absence of PA-457, the majority of released SP1-A3V and -A3T particles displayed an immature morphology. In the case of SP1-A3T, virus particle production was also reduced 20-fold, and detectable virus replication was not observed for up to 1 month in culture. SP1-A3T Pr55Gag processing was also impaired, suggesting a defect in membrane binding (33). In this regard, it is noteworthy that other studies have shown that mutations elsewhere in the CA-SP1 assembly domain disrupt the binding of Gag to membrane (14, 15, 25).
Both the SP1-A3V and A3T mutants displayed some level of PA-457 dependence. At a high drug concentration (i) replication of SP1-A3V was enhanced and occurred readily in the absence of second-site mutations, whereas in the absence of PA-457 or at a low drug concentration secondary mutations compensating for the A3V replication defect were frequently acquired; (ii) virus particle maturation was enhanced; (iii) SP1-A3T virus particle production and Gag processing were improved; and (iv) the replication of the double mutant CA-G225S/SP1-A3T was facilitated. Although high concentrations of PA-457 improved the defective phenotypes associated with the SP1-A3V and -A3T mutations, it should be noted that viral replication and core condensation were not fully restored to WT levels. This study provides the first description of HIV-1 isolates that are both PA-457 resistant and partially PA-457 dependent, although other examples of HIV-1 isolates that are both resistant to and dependent on other drugs have been reported (2, 6, 28).
The distinct phenotypes associated with the mutants described in this study imply the existence of multiple mechanisms by which HIV-1 can acquire resistance to PA-457. Although the mechanism of action of PA-457 is not fully elucidated, recent data suggest that PA-457 inhibits maturation by associating with a binding site formed during oligomerization of Gag during particle assembly (38, 47). The double mutant CA-V230L/L231M, which confers PA-457-resistance, significantly reduced PA-457 incorporation into immature particles (47), indicating that mutations in the CA-SP1 boundary region can confer PA-457 resistance by preventing PA-457 binding to Gag. While this may be a feasible resistance mechanism for some of the mutants isolated in this study, the PA-457 enhancement effect observed for SP1-A3V and -A3T suggests that these mutations do not block PA-457 binding but rather alter the consequences of binding. The SP1-A3V and -A3T mutations could induce PA-457 to bind in an alternative conformation or orientation or could cause the compound to interact with a different binding site. In either case, the effect of PA-457 binding to these mutants stimulated rather than disrupted virion maturation.
An alternative mechanism by which mutations in the CA-SP1 boundary region could confer resistance to PA-457 might be by significantly altering the rate of PR-mediated processing at the CA-SP1 cleavage site. For example, a marked increase in the rate of CA-SP1 cleavage could render processing at that site insensitive to PA-457 binding. Amino acid substitutions in HIV-1 Gag processing sites have been shown to inhibit or enhance the rate of proteolytic processing (references 26 and 35 and references therein). To examine this possible mechanism of resistance, we measured the extent of CA-SP1 processing for each of our PA-457-resistant mutants. We observed a variable rate of CA-SP1 cleavage within the set of six resistant mutants, indicating that, in general, there is no correlation between the extent of CA-SP1 processing and PA-457 resistance. However, the SP1-A1V mutation did cause a modest increase in the degree of CA-SP1 cleavage both in cells (Fig. 3) and in an in vitro assembly system (Fig. 4) (38), suggesting that the increased CA-SP1 processing rate may contribute to resistance for this mutant.
This study provides significant and novel insights into the emergence of resistance to PA-457. The results presented here will inform both the analysis of PA-457 resistance in patients and the development of additional compounds that, like PA-457, block HIV-1 maturation by disrupting specific steps in the Gag processing pathway.
Acknowledgments
We thank Wei-Shau Hu and members of the Freed lab for critical reviews of the manuscript. The HIV-Ig was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; by the Intramural AIDS Targeted Antiviral Program; and also by the University of Oklahoma Health Sciences Center. This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Abdurahman, S., S. Hoglund, L. Goobar-Larsson, and A. Vahlne. 2004. Selected amino acid substitutions in the C-terminal region of human immunodeficiency virus type 1 capsid protein affect virus assembly and release. J Gen. Virol. 85:2903-2913. [DOI] [PubMed] [Google Scholar]
- 2.Aberham, C., S. Weber, and W. Phares. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamson, C. S., and I. M. Jones. 2004. The molecular basis of HIV capsid assembly—five years of progress. Rev. Med. Virol. 14:107-121. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 78:12428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson-Viitanen, S., J. Manfredi, P. Viitanen, D. E. Tribe, R. Tritch, C. A. Hutchison III, D. D. Loeb, and R. Swanstrom. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retroviruses 5:577-591. [DOI] [PubMed] [Google Scholar]
- 8.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 9.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujioka, T., Y. Kashiwada, R. E. Kilkuskie, L. M. Cosentino, L. M. Ballas, J. B. Jiang, W. P. Janzen, I. S. Chen, and K. H. Lee. 1994. Anti-AIDS agents, 11-betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. J. Nat. Prod. 57:243-247. [DOI] [PubMed] [Google Scholar]
- 12.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 13.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 14.Guo, X., and C. Liang. 2005. Opposing effects of the M368A point mutation and deletion of the SP1 region on membrane binding of human immunodeficiency virus type 1 Gag. Virology 335:232-241. [DOI] [PubMed] [Google Scholar]
- 15.Guo, X., A. Roldan, J. Hu, M. A. Wainberg, and C. Liang. 2005. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J. Virol. 79:1803-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi, A., K. Nagashima, and E. O. Freed. 2006. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, Gag-Gag interactions, Gag-membrane binding, and virion maturation. J. Virol. 80:7939-7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanamoto, T., Y. Kashiwada, K. Kanbara, K. Gotoh, M. Yoshimori, T. Goto, K. Sano, and H. Nakashima. 2001. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrob. Agents Chemother. 45:1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan, A. H., J. A. Zack, M. Knigge, D. A. Paul, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in abberant virus assembly and the formation of noninfectious particles. J. Virol. 67:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krausslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krausslich, H. G., H. Schneider, G. Zybarth, C. A. Carter, and E. Wimmer. 1988. Processing of in vitro-synthesized Gag precursor proteins of human immunodeficiency virus (HIV) type 1 by HIV proteinase generated in Escherichia coli. J. Virol. 62:4393-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber. 2005. HIV sequence compendium. LA-UR 06-0680. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 23.Li, F., R. Goila-Gaur, K. Salzwedel, N. R. Kilgore, M. Reddick, C. Matallana, A. Castillo, D. Zoumplis, D. E. Martin, J. M. Orenstein, G. P. Allaway, E. O. Freed, and C. T. Wild. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. USA 100:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, C., J. Hu, R. S. Russell, A. Roldan, L. Kleiman, and M. A. Wainberg. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis, J. M., I. T. Weber, J. Tozser, G. M. Clore, and A. M. Gronenborn. 2000. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 49:111-146. [DOI] [PubMed] [Google Scholar]
- 27.Melamed, D., M. Mark-Danieli, M. Kenan-Eichler, O. Kraus, A. Castiel, N. Laham, T. Pupko, F. Glaser, N. Ben-Tal, and E. Bacharach. 2004. The conserved carboxy terminus of the capsid domain of human immunodeficiency virus type 1 Gag protein is important for virion assembly and release. J. Virol. 78:9675-9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menzo, S., A. Monachetti, C. Balotta, S. Corvasce, S. Rusconi, S. Paolucci, F. Baldanti, P. Bagnarelli, and M. Clementi. 2003. Processivity and drug-dependence of HIV-1 protease: determinants of viral fitness in variants resistant to protease inhibitors. AIDS 17:663-671. [DOI] [PubMed] [Google Scholar]
- 29.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 62:3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morellet, N., S. Druillennec, C. Lenoir, S. Bouaziz, and B. P. Roques. 2005. Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci. 14:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman, J. L., E. W. Butcher, D. T. Patel, Y. Mikhaylenko, and M. F. Summers. 2004. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 13:2101-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono, A., A. A. Waheed, A. Joshi, and E. O. Freed. 2005. Association of human immunodeficiency virus type 1 Gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 79:14131-14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettit, S. C., G. J. Henderson, C. A. Schiffer, and R. Swanstrom. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 76:10226-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakalian, M., S. S. Dittmer, A. D. Gandy, N. D. Rapp, A. Zabransky, and E. Hunter. 2002. The Mason-Pfizer monkey virus internal scaffold domain enables in vitro assembly of human immunodeficiency virus type 1 Gag. J. Virol. 76:10811-10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakalian, M., C. P. McMurtrey, F. J. Deeg, C. W. Maloy, F. Li, C. T. Wild, and K. Salzwedel. 2006. 3-O-(3′,3′-Dimethysuccinyl)betulinic acid inhibits maturation of the human immunodeficiency virus type 1 Gag precursor assembled in vitro. J. Virol. 80:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 40.Tang, C., E. Loeliger, P. Luncsford, I. Kinde, D. Beckett, and M. F. Summers. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 101:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tritch, R. J., Y. E. Cheng, F. H. Yin, and S. Erickson-Viitanen. 1991. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 65:922-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 43.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 85:9580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. P. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta. Crystallogr. D 55:85-92. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, J., C. H. Chen, and C. Aiken. 2004. The sequence of the CA-SP1 junction accounts for the differential sensitivity of HIV-1 and SIV to the small molecule maturation inhibitor 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid. Retrovirology 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, J., L. Huang, D. L. Hachey, C. H. Chen, and C. Aiken. 2005. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J. Biol. Chem. 280:42149-42155. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, J., X. Yuan, D. Dismuke, B. M. Forshey, C. Lundquist, K. H. Lee, C. Aiken, and C. H. Chen. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]