Abstract
RNA interference (RNAi) is an evolutionarily conserved mechanism in plant and animal cells that directs the degradation of messenger RNAs homologous to short double-stranded RNAs termed small interfering RNA (siRNA). The ability of siRNA to direct gene silencing in mammalian cells has raised the possibility that siRNA might be used to investigate gene function in a high throughput fashion or to modulate gene expression in human diseases. The specificity of siRNA-mediated silencing, a critical consideration in these applications, has not been addressed on a genomewide scale. Here we show that siRNA-induced gene silencing of transient or stably expressed mRNA is highly gene-specific and does not produce secondary effects detectable by genomewide expression profiling. A test for transitive RNAi, extension of the RNAi effect to sequences 5′ of the target region that has been observed in Caenorhabditis elegans, was unable to detect this phenomenon in human cells.
RNA interference (RNAi) is an evolutionarily conserved mechanism of gene silencing that is thought to inhibit the replication and expression of selfish DNA elements and viruses (reviewed in refs. 1 and 2). In Caenorhabditis elegans, RNAi was first observed as a silencing of endogenous genes homologous to injected double-stranded RNA (dsRNA) (3). Studies over the last several years have demonstrated that RNAi is mediated by the generation of 21- to 23-nt dsRNA molecules, termed small interfering RNA (siRNA). dsRNA molecules are processed by the RNaseIII-like enzyme Dicer to generate siRNAs, and the siRNAs direct the recognition and subsequent degradation of homologous mRNAs by a multiprotein complex. The enzymatic machinery for generating siRNA also appears to be used for the production of a second class of endogenously encoded, small RNA molecules termed microRNAs (miRNAs). miRNA are processed from endogenous transcripts that form hairpin structures, and miRNAs are thought to mediate the translational control of other genes by binding to the 3′ ends of their messenger RNAs in animals (1, 2).
Because RNAi can, in principle, allow the silencing of a gene given its sequence, RNAi has become a popular research tool to annotate gene function. In particular, once the genome of an organism has been sequenced, it may be feasible to design RNAi experiments to target every gene in the genome and screen for specific phenotypes (4) (5). In mammalian cells, the utility of RNAi had been limited by the innate immune response triggered by dsRNA; long dsRNAs induce the IFN response, which leads to the inhibition of protein translation by the PKR pathway, and activation of RNase L (6). These responses inhibit gene expression generally and significantly alter the cell physiology. The recent demonstration that synthetic siRNAs can trigger sequence-specific RNAi in mammalian cells has stimulated interest in using siRNAs to annotate gene function in human cells and as therapeutic agents (7, 8). However, although siRNAs are too short to induce the IFN response, the specificity of the gene silencing induced by siRNAs in mammalian cells has not been systematically examined. For instance, we do not know whether siRNAs can trigger additional antiviral mechanisms that significantly alter cell physiology, nor how the introduction of exogenous siRNAs might impact the processing and regulation of endogenous miRNAs and the genes that miRNAs control.
The cardinal features of RNAi in C. elegans are the potency and persistence of gene silencing. RNAi can be successfully induced in C. elegans with a few molecules of the trigger dsRNA per cell, and the silencing effect is propagated to the progeny of the treated animals (3). These results suggested the presence of amplification mechanisms in RNAi. Recently, “degradative PCR” was proposed as a mechanism underlying amplification in RNAi in Drosophila embryos and C. elegans (9) (10). In this model, the antisense strand of siRNA hybridizes to the target mRNA and primes an RNA-dependent RNA polymerase reaction to generate double stranded RNA 5′ of sense sequence. The newly synthesized dsRNA are then subject to Dicer digestion and generate many secondary siRNAs from the extended regions that can target additional mRNA molecules for degradation. This model explains the catalytic efficiency and potency of RNAi, but it presents potential problems for the specificity of RNAi as a research tool. A consequence of the generation of the secondary siRNAs is the spreading of the RNAi specificity to sequences 5′ to the original target sequence in the mRNA. This phenomenon, termed transitive RNAi, has been demonstrated in vivo in C. elegans (10). Transitive RNAi poses the possibility of silencing of a significant number of unintended genes within the genome with each siRNA experiments, and it is a greater concern with human cells because of the increased complexity of domain structures in the human proteome. Thus, an understanding of the specificity of siRNA-mediated gene silencing in human cells will be essential for the appropriate design and interpretation of RNAi experiments and RNAi-based therapeutic strategies (11, 12).
In this report, we evaluated the properties of siRNA-mediated gene silencing on a genomic scale. We assessed the specificity of siRNAs in human cells by using global gene expression profiling. Because the activity of the genome as a whole reflects the interplay of all of the cell's metabolic and regulatory pathways, the global gene expression pattern is a highly sensitive indicator of alterations in the cell's physiology. Finally, we formally tested for transitive RNAi activity in human cells by using an experimental design closely similar to one that elicited the phenomenon in C. elegans.
Materials and Methods
Cells and Reagents. Human embryonic kidney (HEK) 293 cells (American Tissue Culture Collection) and 293-derived Phoenix amphotropic packaging cell line (G. Nolan, Stanford University, Stanford, CA) are obtained from the indicated sources. Single stranded dTdT RNA oligonucleotides (Dharmacon) were annealed to generate siRNAs: E1 [begins at 435th nucleotide (nt), sequences in Fig. 1], E2 (begins at 441th nt, sequences in Fig. 1) and E3 (begins at 2nd nt, 5′-UGGUGAGCAAGGGCGAGGAdTdT-3′ and 5′-UCCUCGCCCUUGCUCACCAdTdT-3′) that target the indicated coding sequences of eGFP. Stable GFP-expressing Phoenix cells were produced by transient transfection of pMIGR (gift of W. Pear, University of Pennsylvania, Philadelphia) into amphotropic Phoenix cells and followed by two rounds of fluorescence-activated cell sorting (FACS) selection of GFP+ cells. The resultant cells were >95% GFP+ and remained so subsequently without additional selection. Constructs: eGFP-N3, dsRED, yellow fluorescent protein (YFP)-actin, and pSEAP2-control (CLONTECH), and pGL3 luciferase (Promega) were obtained from indicated sources. The XhoI–BamHI actin fragment from YFP-actin was released by restriction digestion and cloned into eGFP-N3 and pGL3-control to generate ActinS-GFP, ActinAS-GFP, and Luciferase-actin constructs.
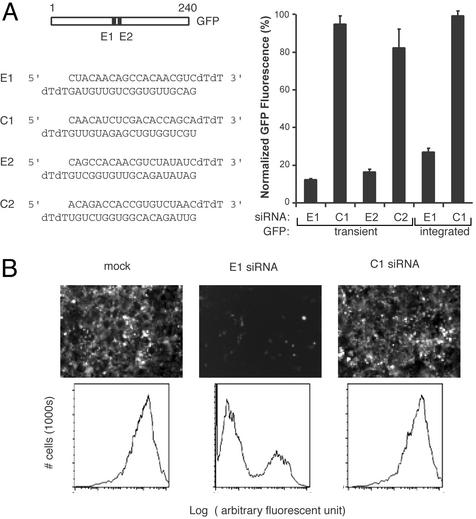
Fig. 1.
Silencing of a model gene by siRNAs. (A) Silencing of transiently expressed and integrated GFP gene by siRNAs. Sequences of the siRNAs used are indicated on the left. For silencing of transiently transfected GFP, 0.3 μgof pGFP was transfected with 1 μg of pSEAP2-control and 12 picomoles of the indicated siRNA in HEK293 cells. For silencing of an integrated GFP gene, HEK293-derived Phoenix cells expressing GFP after retroviral transduction (Materials and Methods) were transfected with the 12 picomoles of the indicated siRNA and 1 μg of pSEAP2-control. GFP expression was determined by FACS 48 h (transient GFP target) or 72 h (integrated GFP target) after transfection. The mean fluorescence intensity was normalized for transfection efficiency by the alkaline phosphatase activity of pSEAP2-control (Materials and Methods). The experiments were done in triplicate, and the means (± standard deviation) of GFP fluorescence intensity relative to mock transfected cells (no siRNA) are shown. (B) Fluorescence photomicroscopy and FACS plots of cells stably expressing GFP and transfected with the indicated siRNAs.
siRNA Experiments. Expression constructs and siRNAs were transfected by using Lipofectamine 2000 (Invitrogen) as described (7). GFP expression was assayed by either FACS or fluorescence microscopy 48–72 h after transfection. Transfection efficiency was normalized by dividing GFP or luciferase fluorescent units with the secreted placental alkaline phosphatase activity generated from cotransfected pSEAP2-control plasmid.
Microarray Procedures and Statistical Methods. Messenger RNA was purified with oligo(dT) beads by using Fastrack (Invitrogen) following the manufacturer's instructions. A reference mRNA standard prepared by pooling RNA from 11 cell lines was used in all experiments. The mRNAs were used to synthesize fluorescent cDNA probes by reverse transcription with oligo(dT) primers and Cy3 or Cy5 dUTPs. The fluorescent probes were competitively hybridized without fragmentation on cDNA arrays as described (13, 14). The gene expression data from three sets of siRNA experiments were derived from 27 microarrays and were analyzed separately in three data sets. In each data set, genes were considered well measured if the reference channel had >1.5-fold of signal intensity over background and was present for >80% of data set. The three sets of genes were each analyzed by multiclass comparison by using significance analysis of microarrays (SAM) (15), and the predicted false discovery rate for the top 10 SAM-selected genes was calculated. The top 10 genes from each data set were collated, and the expression data for this set of 30 genes from each data set were retrieved and grouped by hierarchical clustering (16).
Results
Global View of Gene Silencing by siRNA. To evaluate the specificity of siRNA, we used a target gene that has no normal role or known physiological effects in the cell, so that its presence or absence would not otherwise perturb the transcriptome. We chose the enhanced GFP of Aequoria victoria as a model target because the protein level is easily monitored, it is an exogenous protein that has no normal function in human cells, and it is relatively nontoxic and known to be well tolerated in normal development. Transient transfection of HEK293 cells with GFP and the two siRNAs directed toward GFP sequences (termed E1 and E2) suppressed the level of GFP activity by >80%, but cotransfection of GFP with scrambled siRNAs matched for nucleotide content (termed C1 and C2, respectively) did not affect GFP activity compared with mock-transfected cells, which were not exposed to siRNA (Fig. 1). C1 and C2 did not have significant homology to any human gene or ESTs in the nonredundant and EST database when analyzed with BLAST program in NCBI. The transfection efficiency was >80% as judged by GFP fluorescence. To address the specificity of RNAi against an integrated and nuclear gene, we established a population of cells stably expressing a GFP gene that was introduced by retroviral transduction (Materials and Methods). Transfection of these stable GFP-expressing cells with the E1 siRNA silenced GFP expression by >70%, but GFP expression was unaffected by mock or C1 transfection (Fig. 1 B and C).
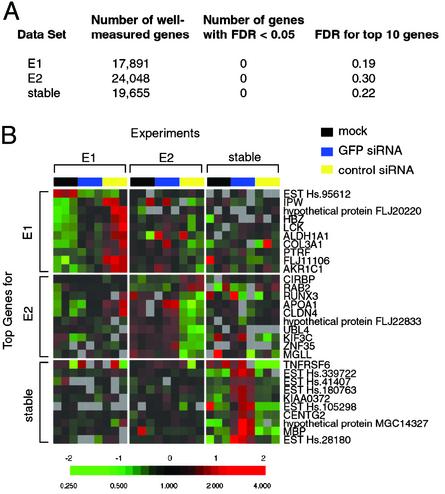
The global gene expression patterns of cells after mock transfection, silencing of transiently expressed or stably expressed GFP by E1 or E2 siRNA, and control silencing by C1 or C2 siRNA were determined by using human cDNA microarrays. The microarrays contained ≈43,000 elements, corresponding to ≈36,000 genes based on Unigene data. Because even small differences in cell passage or media metabolism can lead to differences in global gene expression pattern, control and siRNA experiments were always performed in parallel in sets of three and in triplicate as described above. To search for gene expression responses associated with RNA interference, we performed a statistical test (SAM) to identify genes whose expression varied accordingly in response to the experimental manipulations we tested (15). SAM is a permutation-based technique that permits the estimation of a false discovery rate (FDR) for set of genes identified (15). The FDR is analogous to P value in standard statistical tests, but the FDR can accommodate the effects of nonnormal distribution in the data and multiple testing (15). For each of the three sets of gene expression data, none of ≈20,000 well measured mRNAs was consistently affected by the siRNA treatments, with a FDR <0.05 (Fig. 2A). The 10 genes that showed the most consistent changes in expression with the experimental manipulations had estimated FDRs that ranged from 0.19 to 0.30 in the three experiments. Fig. 2B displays the top 10 genes identified by SAM in all three data sets. We note that the genes that showed the largest apparent responses in the three sets of experiments did not overlap, and the magnitude of the changes in expression of any of these genes was small (mostly <2-fold). Moreover, these small variations in gene expression did not consistently distinguish the siRNA-silenced samples from the mock treated samples (Fig. 2B). Among all of the genes that showed variation in expression in the experiments identifying either transiently or stably GFP, none showed a consistent response pattern. Thus, we believe that the small observed variations are likely to be caused by experimental noise, rather than resulting from the siRNA treatment. Collectively, we interpret these results to indicate that no consistent “off-target” gene expression perturbation is associated with the process of siRNA-mediated gene silencing. To the detectable limits of our cDNA array method, siRNA-mediated gene silencing in the tested cells appears to be highly sequence-specific.
Fig. 2.
Global gene expression changes associated with RNAi. (A) Summary of gene expression data. Global gene expression patterns in three siRNA experiments were analyzed; in each set, the gene expression of cells that were mock transfected (no siRNA), transfected with GFP siRNA, or cognate control siRNA were determined in parallel in triplicate. Data sets: E1, HEK293 cells with transiently expressed GFP target treated with E1, C1, or no siRNA; E2, HEK293 cells with transiently expressed GFP target treated with E2, C2, or no siRNA; stable, Phoenix cells stably expressing an integrated GFP gene treated with E1, C1, or no siRNA. Genes that had signal intensity >1.5-fold of the local spot element background in the reference channel and were present for >80% of the data set were considered well measured. The number of well measured genes are shown on the second column; these genes were analyzed in the multiclass comparison by using SAM (15). The number of genes that had an estimated FDR of <0.05 and the FDR of the top 10 performing genes for each data set are shown on the right two columns. (B) Minimal gene expression changes associated with siRNA-mediated RNAi. The 10 genes with the most consistent changes in expression in response to the experimental manipulation, in each of the three siRNA experiments, were collated into a nonredundant gene list. The expression changes of this group of genes in all experiments are displayed in matrix format (16). The expression ratios were mean-centered within each data set, and the gene expression changes are indicated by the color scale as indicated below.
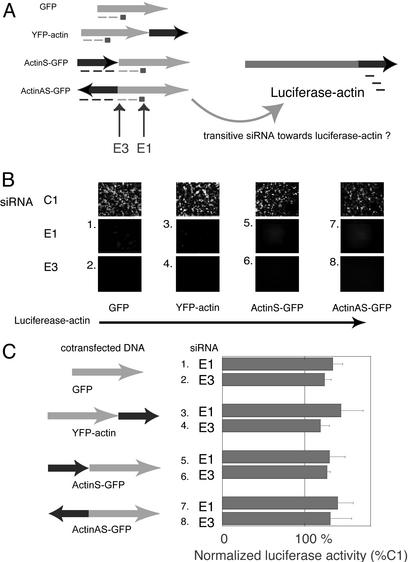
Evaluation of Transitive RNAi in Human Cells. Although siRNAs appear to be highly sequence-specific, the extension of RNAi-mediated silencing to sequences 5′ to the mRNA sequence complementary to the siRNA could generate secondary siRNAs that could potentially target other mRNAs with sequence similarity. Such a phenomenon, termed “transitive RNAi” has been shown to occur in C. elegans (10). To test for the occurrence of transitive RNAi in human cells, we cotransfected into HEK 293 cells two sets of reporter genes (GFP/YFP and luciferase) with sequence overlap engineered by fusing a sequence for the actin gene to both sets of constructs (Fig. 3A). We used siRNA to target the first reporter genes (GFP or YFP, which contain the same cognate sequence) and verified the RNA silencing by monitoring the fluorescence of transfected HEK293 cells. If transitive RNAi were active in 293 cells, silencing of GFP/YFP-actin fusion mRNA should generate secondary siRNAs targeting the actin sequences and thereby initiate the silencing of the second reporter gene, luciferase-actin, resulting in diminished luciferase activity. Because transitive RNAi in C. elegans demonstrated polarity (that is, diminished effect with greater distance), we designed a siRNA E3 that target the first 21 nucleotides of GFP coding regions (Fig. 3A) and <20 nt 3′ of actin sequences in both actin-GFP fusion proteins. We tested the transitive effects of silencing GFP expressed alone or in the form of fusion transcripts with actin fused at either the 3′ end of YFP-actin, or at the 5′ end of GFP in both orientations (ActinS-GFP, ActinAS-GFP) with E1 and E3 compared with control siRNA C1. Fluorescent microscopy confirmed that siRNA-mediated RNA silencing of the primary target gene was achieved for all pairs of different fluorescent proteins (Fig. 3B). In all experiments, the luciferase activity in the cells silenced by GFP siRNA (E1 or E3) was not lower than that in cells treated with control siRNA (C1) (Fig. 3C). These results indicate that transitive RNAi, at least on the scale demonstratable in C. elegans, does not occur during siRNA-mediated silencing in 293 cells. This result may be related to the relatively inefficient silencing mediated by siRNA in mammalian cells compared with that seen in C. elegans.
Fig. 3.
Test of transitive RNAi in HEK293 cells. (A) Experimental strategy for transitive RNAi. The square indicates the original trigger siRNA, and the dashed lines indicate secondary siRNAs. The regions of GFP targeted by E1 and E3 are indicated by arrows. (B) Effect of siRNAs (E1 and E3) on expression of GFP fusion constructs. HEK293 cells were transfected with the indicated constructs and siRNAs and photographed by fluorescence microscopy 48 h after transfection. (C) Effect of siRNAs on luciferase-actin expression. The luciferase activity in cells transfected with the indicated constructs and siRNA (E1 or E3) were compared with those of corresponding constructs and control siRNA (C1); the values shown are the means of relative activity (E1/C1 or E3/C1) ± standard deviation of triplicate experiments.
Discussion
By using DNA microarrays to profile global gene expression, we have demonstrated that siRNA-mediated gene silencing has exquisite sequence specificity for the target mRNA and does not induce detectable secondary changes in the global gene expression pattern. We tested for transitive RNAi by using paired, highly expressed transcripts with overlapping sequence identity, conditions that easily afforded detection of transitive RNAi in C. elegans (10). The lack of robust transitive RNAi in human cells is consistent with published reports of selective targeting of splicing isoforms using siRNA (17 18), the lack of an obvious RNA-dependent RNA polymerase in the human genome, and the dispensability of priming activity of siRNAs for RNAi in Drosophila and mammalian cells (19–22). Although it will be important to examine the possibility that different mammalian cell types might respond to siRNAs differently, these results provide further impetus for using siRNA-mediated RNAi as a research and therapeutic tool. The high specificity observed in these experiments, if confirmed for additional cell types and target genes, should increase the confidence with which phenotypes observed with siRNA-mediated silencing can be ascribed to the targeted genes. The results are also encouraging for the prospects that siRNA-based therapeutic agents could have useful molecular specificity. Because the process of siRNA-mediated silencing does not appear, in general, to produce nonspecific gene expression changes, global changes of gene expression patterns may provide an assay with which to study and annotate the function of unknown genes, especially based on comparisons to gene expression patterns of mutants in known pathways (23).
Acknowledgments
We thank Pate Skene, David Botstein, and members of the Brown laboratory for insightful discussions. We acknowledge C. Bondre for technical assistance. We are also indebted to Mike Fero and staffs at Stanford Functional Genomics Facility and Stanford Microarray Database for their helps and advice. This work was supported by grants from the National Cancer Institute. P.O.B. is an investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAi, RNA interference; dsRNA, double-stranded RNA; siRNA, small interfering RNA; miRNA, microRNA; YFP, yellow fluorescent protein; FACS, fluorescence-activated cell sorting; SAM, significance analysis of microarrays.
See commentary on page 6289.
References
- 1.Hannon, G. J. (2002) Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 2.Zamore, P. D. (2002) Science 296, 1265–1269. [DOI] [PubMed] [Google Scholar]
- 3.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 4.Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M. & Ahringer, J. (2000) Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 5.Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S. J., Copley, R. R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. (2000) Nature 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 6.Bass, B. L. (2001) Nature 411, 428–429. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 8.Carmichael, G. G. (2002) Nature 418, 379–380. [DOI] [PubMed] [Google Scholar]
- 9.Lipardi, C., Wei, Q. & Paterson, B. M. (2001) Cell 107, 297–307. [DOI] [PubMed] [Google Scholar]
- 10.Sijen, T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., Plasterk, R. H. & Fire, A. (2001) Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- 11.Gitlin, L., Karelsky, S. & Andino, R. (2002) Nature 418, 430–434. [DOI] [PubMed] [Google Scholar]
- 12.Novina, C. D., Murray, M. F., Dykxhoorn, D. M., Beresford, P. J., Riess, J., Lee, S. K., Collman, R. G., Lieberman, J., Shankar, P. & Sharp, P. A. (2002) Nat. Med. 8, 681–686. [DOI] [PubMed] [Google Scholar]
- 13.Perou, C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747–752. [DOI] [PubMed] [Google Scholar]
- 14.Sherlock, G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29, 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kisielow, M., Kleiner, S., Nagasawa, M., Faisal, A. & Nagamine, Y. (2002) Biochem. J. 363, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celotto, A. M. & Graveley, B. R. (2002) RNA 8, 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu, Y.-L. & Rana, T. M. (2002) Mol. Cell 10, 549–561. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz, D. S., Hutvagner, G., Haley, B. & Zamore, P. D. (2002) Mol. Cell 10, 537–548. [DOI] [PubMed] [Google Scholar]
- 21.Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. (2002) Cell 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 22.Roignant, J. Y., Carre, C., Mugat, B., Szymczak, D., Lepesant, J. A. & Antoniewski, C. (2003) RNA 9, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109–126. [DOI] [PubMed] [Google Scholar]