Abstract
Despite their overall accuracy, errors in macromolecular processes, such as rRNA synthesis and ribosome assembly, inevitably occur. However, whether these errors are remediated and how this might be accomplished is not known. In previous work, we showed that a double mutant strain lacking both polynucleotide phosphorylase (PNPase) and RNase R activities is inviable. In the course of examining the molecular basis for this phenotype, we found that shifting a temperature-sensitive mutant strain to 42°C led to cessation of growth and loss of cell viability. Northern analysis of RNA isolated from such cells after the temperature shift revealed that fragments of 16S and 23S rRNA accumulated to a high level, and that the amount of ribosomes and ribosomal subunits decreased due to defects in ribosome assembly. rRNA fragments were not detected at 31°C or when single mutant strains were grown at 42°C. Pulse–chase analysis showed that the rRNA fragments appeared within 5 min at 42°C, and that they accumulated before the loss of cell viability. The data are consistent with a model in which PNPase and RNase R mediate a previously unknown quality control process that normally removes defective rRNAs as soon as they are generated. In the absence of these RNases, rRNA fragments accumulate, leading to interference with ribosome maturation and ultimately to cell death.
A certain frequency of errors is associated with all cellular processes. As a consequence, incomplete or defective molecules may be produced and presumably would need to be eliminated. In the case of RNA molecules, removal would require the action of RNases. However, almost no information about such RNA quality control processes is available, particularly with regard to the stable RNAs, rRNA and tRNA.
Among the RNases present in Escherichia coli are eight distinct exoribonucleases (1). Whereas these enzymes are known to play important roles in many aspects of RNA metabolism (2, 3), their participation in RNA quality control processes has not been elucidated. It is known that PNPase and RNase II are required to remove mRNA fragments that are generated during the course of mRNA decay (4). It was also shown recently that PNPase is involved in the quality control of tRNA (5). Thus, when a defective tRNA precursor molecule is synthesized, it is polyadenylated by poly(A) polymerase and subsequently degraded by PNPase. However, removal of PNPase by mutation did not entirely eliminate the degradation of defective tRNAs, suggesting that additional RNases with similar activities, such as RNase II or RNase R, might also participate (5).
RNase R was originally identified in an E. coli strain deficient in RNase II (6), and was subsequently rediscovered in our laboratory and given the name RNase R, due to its activity on ribosomal RNA (7). RNase R has been purified to homogeneity and been shown to be a 3′ to 5′ exoribonuclease (8). The rnr gene encoding RNase R was identified and shown to be identical to the vacB gene required for virulence in Shigella and enteroinvasive E. coli (9). Efforts to construct mutant strains deficient in RNase R, alone or in combination with other exoribonucleases, showed that RNase R- single mutant strains are essentially normal, as are multiple mutant strains lacking RNases R and T, RNases R and PH, or RNases R, II, D, and BN (9). However, a mutant strain lacking both RNase R and PNPase activities could not be made (9), suggesting that such cells are inviable and that the two exoribonucleases might overlap in some important function that had not yet been identified.
In this paper, we examine the molecular basis for this phenotype. We constructed a conditional-lethal mutant strain that carries both a temperature-sensitive allele of PNPase (10), and a deletion-interruption allele of RNase R (9), and have used this strain to examine the physiological consequences of removal of both RNases. The data confirm our previous suggestion that cells lacking both PNPase and RNase R are inviable. Most importantly, we show that, in the absence of these RNases, fragments of rRNA accumulate to high levels, ribosome maturation and assembly become defective, and cells ultimately die. The data suggest that an essential function of PNPase and RNase R is the maintenance of quality control of rRNA metabolism.
Materials and Methods
Bacterial Strains and Plasmid. The strains used in this study are derivatives of E. coli K-12. Strain CA244I- (rna zbe-279::Tn10) is an RNase I- derivative (7) of strain CA244 (11), and was considered the wild type for the studies described here. Strain SK6639 (pnp-200 Camr) encoding a temperature-sensitive PNPase, and strain SK5662 carrying a Tn10 near argG6 were obtained from S. R. Kushner (University of Georgia, Athens). They were used to construct CA244PNPts by using cotransduction of pnp-200 (10) with argG. Transductants containing a ts PNPase were identified by assaying extracts from cells that had been incubated at 42°C for 1 h for PNPase synthetic activity. CA244PNPtsR- (pnp-200 rnr::kan Camr) was constructed by transducing CA244PNPts by using P1 phage grown on CAN20-12ER- (9). Strains CA 244R- (9), CA 244PNPts, and CA244PNPtsR- were made RNase I- (rna zbe-279::Tn10) via P1-mediated transduction. Plasmid pKK3535 containing the ribosomal RNA operon rrnB was a kind gift from A. E. Dahlberg (Brown University, Providence, RI) (12).
Growth. Cells were grown in YT medium (13) at the temperature indicated. Antibiotics were included as needed at the following concentrations: 25 μg/ml for kanamycin (Kan), 34 μg/ml for chloramphenicol (Cam), and 10 μg/ml for tetracycline (Tet). To make solid media, 1.5% agar was added. Low-phosphate YT medium was made by using a procedure adapted from the protocol of Nelson and Kornberg for making low-phosphate SNB (supplemented nutrient broth; ref. 14). Strains used in these studies were unaffected by growth in low-phosphate YT medium.
Materials. PCR Master Mix and Prime-a-Gene Labeling System were products of Promega. Restriction endonucleases and T4 polynucleotide kinase were from New England Biolabs. [α-32P]dATP [3,000 Ci/mmol (1 Ci = 37 GBq)], [γ-32P]ATP (6,000 Ci/mmol), [32P]orthophosphate, and [3H]uridine (35–50 Ci/mmol) were from Perkin–Elmer. The DNA oligonucleotides used were as follows: 23S-2, 5′-GTA CCG GTT AGC TCA ACG CAT CGC-3′, complementary to residues 2857–2880 of 23S rRNA; 23S-4, 5′-CCT TCA TCG CCT CTG ACT GCC A-3′, complementary to residues 34–55 of 23S rRNA; 16S-5′-ZC, 5′-CCT GTT ACC GTT CGA CTT GC-3′, complementary to residues 57–76 of 16S rRNA; 16S-2, 5′-CAT GAC TGG GGT GAA GTC GTA AC-3′, corresponding to residues 1479–1501 of 16S rRNA; and 16S-5, 5′-GGA GGT GAT CCA ACC GCA GG-3′, complementary to residues 1520–1539 of 16S rRNA. All other chemicals were reagent grade.
RNA Preparation. Total RNA was prepared from E. coli cells by using the hot phenol method (15).
Northern Hybridization Analysis. To examine 16S and 23S rRNAs, 1 μg of total RNA was resolved in a 1.2% agarose gel (16) in TAE buffer (17) and transferred to a nylon membrane by downward capillary transfer for 2 h by using 5× SSC/10 mM NaOH as the transfer solution. Probes corresponding to full-length 23S and 16S rRNA were labeled by random primer extension (as per the manufacturer's instructions) by using a DNA fragment from each rRNA gene. For 16S rRNA, the 1.5-kb BstEII-BclI fragment derived from plasmid pKK3535 was used; for 23S rRNA, the fragment used was a 2.8-kb BsrFI fragment cleaved from a 3.4-kb PCR product amplified from plasmid pKK3535 by using DNA oligonucleotides 16S-2 and 23S-2 as the primers. DNA oligonucleotide probes were labeled at their 5′ ends by T4 polynucleotide kinase. To examine small RNA species, including 5S rRNA and tRNA, 1 μg of total RNA was resolved on a 6% polyacrylamide-8.3 M urea gel and transferred to a nylon membrane by electroblotting. Membranes were analyzed by either autoradiography or PhosphorImager (Molecular Dynamics).
Primer Extension. DNA oligonucleotide primers were labeled at their 5′ ends with 32P. Primer extension analysis was carried out as described (18).
Site-Directed RNase H Cleavage Analysis. This technique can be used to examine the end of large RNA molecules at single nucleotide resolution. Analysis was carried out as described (19). The reaction products were resolved on a 6% polyacrylamide-8.3 M urea gel and analyzed by Northern hybridization.
Ribosome Analysis. Cell extracts (S30) were prepared and analyzed in 5–20% sucrose gradients as described (20).
Pulse–Chase Labeling of Cells. Cells were grown in either YT medium (for [3H]uridine incorporation) or low-phosphate YT medium (for 32Pi incorporation) and pulse labeled at the indicated times for a period of 5 min followed by a chase with rifampicin. Samples were taken at various times and treated as described (20).
Gel Analysis of Pulse-Labeled RNA. Total RNA was dissolved in RNA loading buffer and directly loaded on a 4% polyacrylamide-8.3 M urea gel. Electrophoresis was stopped after the xylene cyanol FF dye had migrated 21 cm. The gel was stained by ethidium bromide to visualize total RNA, and then fixed with a solution containing 10% acetic acid and 10% methanol, dried, and analyzed with a PhosphorImager to visualize the pulse-labeled RNA.
Results
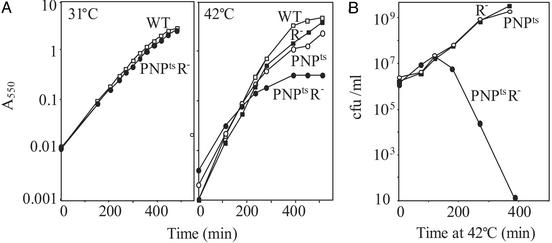
Growth of a PNPtsR– Mutant Strain at Elevated Temperature. In previous work, based on our inability to construct a PNP-R- double mutant, we suggested that such strains are not viable (9). To examine this point in more detail, growth of a PNPtsR- strain was examined on a temperature-shift from a permissive temperature of 31°C to a nonpermissive temperature of 42°C. Initially, growth was monitored by A550 measurements (Fig. 1A). At 31°C, double mutant cells grew normally. However, when shifted to 42°C, growth ceased after a number of generations. In contrast, PNPts and R- single mutant strains grew essentially like the wild-type strain. Further examination revealed that the double mutant strain rapidly lost viability after 2 h at 42°C despite the fact that the A550 of the culture continued to increase for several more hours (Fig. 1B). In fact, as shown in Fig. 1B, essentially all cells were dead at the time when the A550 of the PNPtsR- strain reached a plateau (Fig. 1 A). On the other hand, the number of viable cells in cultures of PNPts and R- single mutant strains increased steadily, and was roughly proportional to the A550 values (Fig. 1). These data show that the absence of both PNPase and RNase R activities is lethal to E. coli cells.
Fig. 1.
Growth of the PNPtsR- strain. Cultures grown overnight with shaking in liquid YT medium at 31°C were diluted into fresh YT medium and incubated with shaking at 31°C. For growth at 42°C, the cultures were incubated at 31°C for 1 h before shifting to 42°C. (A) Growth was monitored by measuring A550.(B) Viable cells were monitored by plating at 31°C on YT medium with antibiotics, as needed, and determining the number of colonies.
At various times after shifting a mutant culture to 42°C, cells were fixed, stained with 4′,6-diamidino-2-phenylindole (DAPI) and examined by phase-contrast microscopy. Cells were essentially normal until 1 h after the shift, but became more and more elongated with multiple nucleoids thereafter (data not shown). Single mutant cells displayed a morphology similar to that of the wild type and contained at most two nucleoids. The data suggest that cells lacking PNPase and RNase R activity are defective in cell division. The molecular basis for this phenotype is not known, and is not explored further here. However, the observation that PNPtsR- cells become elongated and contain multiple nucleoids may help explain the growth experiments (Fig. 1) in which the A550 value of the culture continued to increase even though the number of viable cells was decreasing.
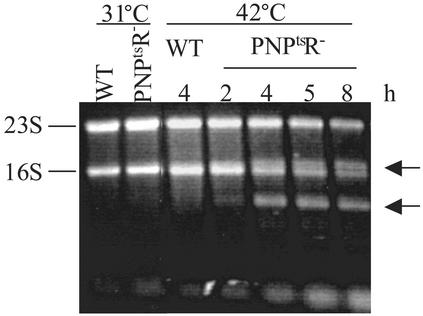
Analysis of RNA Metabolism in PNPtsR– Cells. The inviability of strains lacking PNPase and RNase R indicated that these two enzymes participate in some important cell function(s), and that at least one has to be present to retain cell viability. Inasmuch as these enzymes are RNases, it seemed possible that some RNA molecule that ordinarily would be degraded or processed might accumulate in their absence. To examine this point, total RNA was isolated (20) from the PNPtsR- strain at various times after it had been shifted to 42°C, and was analyzed on an agarose gel to determine whether any additional RNA molecules could be seen. As shown in Fig. 2, several RNA bands (noted by arrows) that were not present in wild-type cells appeared within 2 h after the temperature shift. By 4 h, these bands were present at levels comparable to that of the rRNAs, and they remained at these levels at later times. Concomitant with the appearance of these new bands, there was a decrease in the amounts of the mature rRNAs, suggesting that the newly appearing molecules might be related to 16S and 23S rRNA.
Fig. 2.
Analysis of total RNA. Cultures were grown as in Fig. 1, and portions were taken at the indicated times and temperatures. Total RNA was prepared, resolved in a 1.2% agarose gel, and stained with ethidium bromide. The positions of 23S and 16S rRNA are indicated on the left. Arrows indicate the major additional bands appearing in the PNPtsR- mutant cells at 42°C. Numbers above the lanes indicate the hours at 42°C. Cultures at 31°C were in log phase.
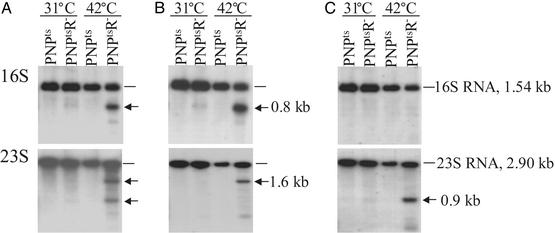
To examine this finding in more detail, Northern analysis was carried out by using full-length probes complementary to 16S, 23S, and 5S rRNAs. As shown in Fig. 3A, the shorter molecules that accumulated after 4 h were, in fact, fragments of 16S and 23S rRNA. As above, essentially no fragments were observed when the double mutant was grown at 31°C or with a PNPts single mutant at either temperature. Fragments of 5S rRNA were not observed in this analysis, even at 42°C (data not shown). More detailed examination revealed that the fragments derived from 16S rRNA also could be detected by a short oligonucleotide complementary to the 5′ end of 16S rRNA (Fig. 3B), but not by a probe complementary to the 3′ end (Fig. 3C). Consequently, these data indicate that the 16S rRNA fragments are shortened at their 3′ ends. In contrast, one of the major 23S rRNA bands (the 1.6-kb band) was detected by an oligonucleotide complementary to the 5′-end of 23S rRNA (Fig. 3B), whereas the other major band (the 0.9-kb band) was detected by an oligonucleotide complementary to the 3′ end of 23S rRNA (Fig. 3C). Thus, the 23S rRNA fragments seem to be shortened at either end.
Fig. 3.
Northern analysis of RNA. Cultures were grown as in Fig. 1 with incubation either at 31°Ctoan A550 of 0.1–0.2 or at 42°C for 4 h. Total RNA was prepared and purified as described in Materials and Methods.(A) Full-length probes. (B) Oligonucleotide probes complementary to 5′ ends of rRNAs. (C) Oligonucleotide probes complementary to 3′ ends of rRNAs. Arrows indicate positions of major new bands.
Analysis of Full-Length 23S and 16S rRNAs. In addition to the rRNA fragments, apparently full-length 23S and 16S rRNAs also were present in the double mutant at 42°C. Thus, it was of interest to determine whether these species had mature 5′ and 3′ termini or whether they also might be partially degraded, because this result could not be ascertained from the Northern experiments. To determine the 5′ termini of the rRNAs, primer extension analysis was performed by using primers complementary to sequences close to the 5′ ends of 23S or 16S rRNA. Based on these experiments, the apparently full-length 23S and 16S RNA molecules carried normal 5′ termini (data not shown). To examine the 3′ termini, chimeric oligonucleotide-directed RNase H cleavage was used (19). These analyses indicated that both the 23S and 16S rRNA species in the double mutant at 42°C have normal 3′ termini as well (data not shown). Thus, from these data, we conclude that both full-length rRNAs and rRNA fragments are present in the PNPtsR- mutant at 42°C. The absence of 3′ extended forms of either 16S or 23S RNA in the double mutant at 42°C also indicates that removal of PNPase and RNase R activities does not affect 3′ maturation of these RNA species.
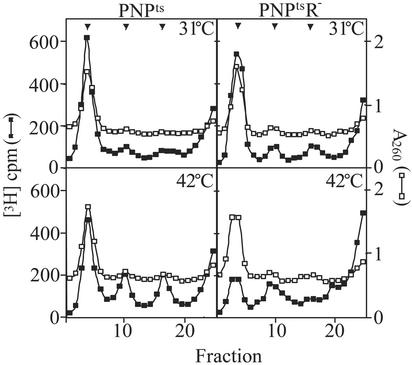
Analysis of Ribosomes in the PNPtsR– Mutant Strain. The presence of full-length 16S and 23S rRNA in PNPtsR- mutant cells that had been incubated at 42°C for 4 h was unexpected, given that these cells were rapidly losing viability (Fig. 1B), and raised the question of the status of ribosomes in these cells. To answer this question, extracts were prepared in a buffer containing 10 mM Mg2+ to help preserve 70S ribosomes, and sedimentation analysis was performed (Fig. 4). After 4 h at 42°C, the level of 70S ribosomes in PNPtsR- cells was substantially reduced, compared with that of the same cells at 31°C or of the PNPts single mutant strain at either temperature. These data show that ribosomes are affected in the double mutant at 42°C, and they suggest either that preexisting ribosomes are being degraded or that new ribosomes cannot be assembled, or both (see below).
Fig. 4.
Sedimentation analysis of ribosomes. Cultures were grown as described in Fig. 1 except that 5 μCi/ml [3H]uridine was added to the fresh YT medium at 42°C. 3H-labeled S30 extracts (≈20,000 cpm) were layered on 11-ml 5–20% sucrose gradients and centrifuged at 18,500 rpm in an SW41 rotor (Beckman) for 15 h at 4°C. Fractions were collected from the bottom, and 3H counts were determined by scintillation counting. An excess of unlabeled, wild-type extract was present in each gradient to determine the positions of the 70S, 50S, and 30S peaks (shown by arrows) by absorption at 260 nm. Total cpm in all of the samples were normalized to the same value to correct for loading and recovery variations.
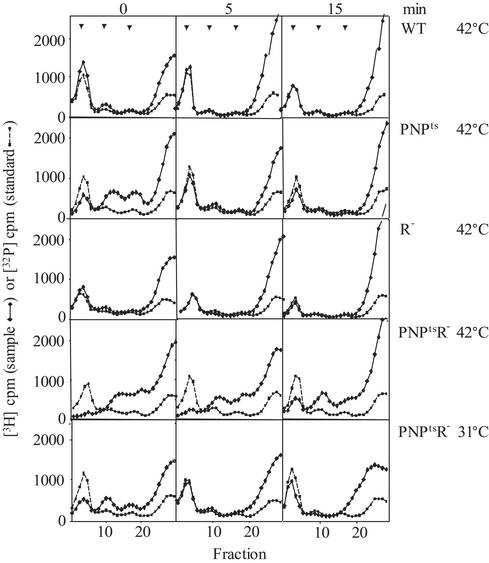
Pulse–Chase Analysis of Ribosomes and RNA. To help distinguish between ribosome breakdown and defective ribosome assembly, and also to attempt to identify the primary lesion that leads to the mutant phenotype when PNPase and RNase R activities are absent, pulse–chase analysis of ribosomes and ribosomal RNA was carried out. Shown in Fig. 5 are ribosome profiles of cells labeled for 5 min with [3H]uridine and chased with rifampicin for 0, 5, or 15 min. Cells were incubated at 42°C for 3 h before the pulse for the experiments at elevated temperature. Under these conditions, relatively little [3H]uridine was found in 70S ribosomes of the PNPts R- strain, even after a 15-min chase period. Rather, particles sedimenting at a size smaller than 50S and 30S subunits accumulated (compare with 32P standard in gradient), and these were poorly chased into 70S ribosomes. In contrast, the wild-type and single mutant strains, and the double mutant at 31°C all synthesized 70S ribosomes. Synthesis was extremely rapid in the wild-type and R- strains, but somewhat slower in PNPts cells and in PNPtsR- cells at 31°C. However, even in the latter strains, label was efficiently chased into 70S ribosomes by 5 min. The data indicate that, after 3 h at 42°C, PNPtsR- cells are defective in ribosome assembly.
Fig. 5.
Sedimentation analysis of pulse–chased ribosomes. Cultures were grown as described in Fig. 1. Cells at 31°Corat42°C for 4 h were pulse-labeled with 5 μCi/ml [3H]uridine for 5 min and chased with 400 μg/ml rifampcin for the times indicated at the top of the figure. For size standards, wild-type cells were grown in low-phosphate YT medium with 32Pi at 37°C. 3H- and 32P-labeled S30 extracts (≈20,000 cpm) were analyzed as in Fig. 4. The temperature of the pulse–chase and the cells used are indicated on the right. Arrows indicate positions of 70S, 50S, and 30S ribosomes.
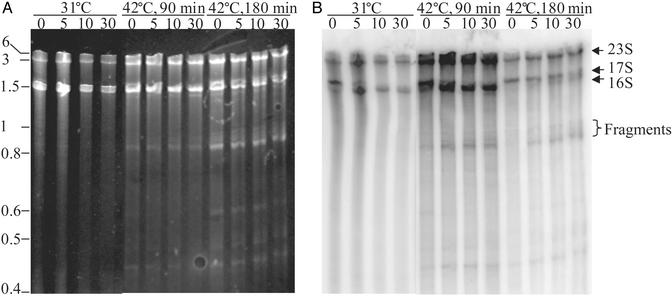
Pulse–chase analysis was also carried out on rRNA to determine when the defect in ribosome metabolism was initiated and when rRNA fragments appeared. Briefly, PNPtsR- cells were grown in low-phosphate YT medium at 31°C, transferred to 42°C for either 90 or 180 min, and then pulse-labeled with 32Pi for 5 min. Rifampicin was then added to stop further transcription initiation, and samples were taken at the indicated times for RNA analysis (Fig. 6). At 31°C, 17S rRNA precursor, which is a major product after the pulse, was rapidly converted to 16S mature rRNA. Likewise, when the pulse was carried out 90 min after the shift to 42°C, the same rapid conversion of 17S precursor RNA to the mature form occurred. However, in these cells, additional RNA bands, 0.8–1 kb in size, were observed as soon as 5 min after the chase. These bands were not observed when the pulse–chase was carried out in double mutant cells growing at 31°C (Fig. 6), or in either of the single mutant strains or in wild-type cells, even 180 min after the temperature shift to 42°C (data not shown). In contrast, in PNPtsR- cells 180 min after the shift (Fig. 6), not only were the smaller bands present, but conversion of the 17S RNA precursor to 16S rRNA also was slowed dramatically. Moreover, overall incorporation of 32Pi was decreased dramatically, as might be expected for cells that were rapidly losing viability at this time.
Fig. 6.
Pulse–chase analysis of RNA from PNPtsR- strain. Cultures were grown as in Fig. 1 except that the dilution was into low-phosphate YT medium. Cells were incubated for 1 h at 31°C before a portion of the culture was shifted to 42°C. Samples grown at 31°C or 90 and 180 min after shifting to 42°C were concentrated to an A550 ≈ 1 for pulse labeling. Cells were labeled with 40 μCi/ml 32Pi for 5 min, and chased with 400 μg/ml rifampcin for the times indicated above each lane in min. Portions of the cultures (500 μl) were taken and transferred to 1 ml of cold 80% ethanol/1% diethylpyrocarbonate. Cells were collected by centrifugation, and total RNA was isolated as described in Materials and Methods. Two micrograms of RNA were loaded on a 4% polyacrylamide–8.3 M urea gel for analysis. (A) Ethidium bromide staining of the gel. (B) PhosphorImager analysis of the gel. The positions of mature and precursor rRNAs are indicated on the right. Positions of size standards are shown on the left.
The 0.8–1 kb RNA products observed in these pulse–chase experiments migrate the same as the 0.8- and 0.9-kb rRNA fragments that accumulated in the steady-state measurements (Figs. 2 and 3), when run side by side (data not shown). Moreover, from the stained gel (Fig. 6B), it is evident that multiple bands are present in this region. These bands were not well resolved in Figs. 2 and 3. Their appearance, within 90 min of the temperature shift, and before the onset of the 17S to 16S rRNA processing defect and before loss of cell viability, suggests that accumulation of these RNA products might be a major contributor to the phenotype of the double mutant strain.
Discussion
Construction of a temperature-sensitive double mutant strain enabled us to demonstrate conclusively that E. coli cells lacking both PNPase and RNase R activity are not viable. Most importantly, the availability of this strain made it possible to examine what overlapping function of PNPase and RNase R might be responsible for this loss of viability. One function revealed by these studies is that, in the absence of the two RNase activities, large amounts of 23S and 16S rRNA fragments accumulate. Pulse–chase analysis indicates that these RNA fragments appear rapidly, within 5 min of the pulse, and that they already are present in cells that have not yet ceased growth at 42°C (i.e., at 90 min after the temperature shift). However, the origin of these abnormal products is not yet known (see below). Whatever their source, the present work strongly suggests that PNPase and RNase R are the enzymes responsible for degrading the fragments, and that when both activities are absent, fragments accumulate. In fact, the fragments are substrates for PNPase and RNase R, but not of RNase II, in vitro (unpublished observation). The data also suggest that the presence of these fragments is deleterious to the cell. It might be expected that rRNA fragments would compete with nascent rRNA transcripts for the pool of available ribosomal proteins, and thereby interfere with ribosome assembly. This result, in turn, would lead to the production of additional rRNA fragments as the unassembled rRNAs are ultimately degraded. Moreover, there may be other cellular processes with which the rRNA fragments interfere. As shown, PNPtsR- cells ultimately lose viability; however, further work will be needed to prove conclusively that it is the accumulation of rRNA fragments that is directly responsible for the cell death, and not some other toxic product or the absence of some essential component.
Nevertheless, the presence in cells of rRNA fragments, and their removal by PNPase and RNase R, draw attention to the concept of RNA quality control, particularly as it relates to metabolism of the stable RNAs, rRNA, and tRNA. Until recently, this had been a neglected area. However, in recent work (5), our laboratory showed that defective precursor tRNA molecules are degraded by a mechanism that involves poly(A) polymerase and PNPase. Extension of that work has revealed that RNase R also may participate in the process (M.P.D., S. Reimers, and C. Kim, unpublished observations). Thus, it is of particular interest that PNPase and RNase R also are required for the quality control of rRNA metabolism described here. Taken together, all of these observations suggest that cells may contain dedicated machinery for the degradation of defective, stable RNA molecules. Ribosomal and tRNAs are known to be highly stable under normal growth conditions (21, 22). Yet, under starvation conditions, these molecules become destabilized, and are degraded (22). It would be of interest to examine whether PNPase and RNase R might be involved in this process as well.
How the initial rRNA fragments arise is not yet clear, but they apparently are normal by-products of rRNA metabolism. Presumably, they have not been observed previously because they are rapidly degraded by PNPase and RNase R. It is only when the two enzymes are eliminated that fragments accumulate. It has been estimated that rapidly growing cells contain about 70,000 ribosomes, and that they are synthesized continually (23). Ribosome biogenesis is a complex process that includes transcription and processing of rRNAs and of mRNAs for ribosomal proteins, translation of ribosomal proteins, and assembly of the 30S and 50S ribosomal subunits. Although cells have evolved mechanisms to enhance the accuracy of these processes, mistakes such as misincorporation, premature termination of transcription, and incomplete assembly can still occur. It is known that a fraction of newly transcribed rRNAs is degraded. This fraction is high when the growth rate is extremely low (≈70% degraded when the doubling time is 10 h) and is low in fast-growing cells (≈10% degraded when the doubling time is ≈40 min) (24, 25). It is also known that certain single-base substitutions in the leader sequence of rRNA operons can affect processing and ribosome assembly (26, 27). Presumably, the portion of rRNA molecules that cannot be properly assembled into ribosomes is subject to degradation. Consequently, there are multiple, potential mechanisms that could lead to the accumulation of the rRNA fragments that are observed in this work. Whereas further studies on the origin of the fragments will be of considerable interest, the work presented here demonstrates their existence and helps to clarify the enzymatic machinery by which they are eliminated.
Acknowledgments
This work was supported by Grant GM16317 from the National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Deutscher, M. P. & Li, Z. (2001) Prog. Nucleic Acid Res. Mol. Biol. 66, 67–105. [DOI] [PubMed] [Google Scholar]
- 2.Li, Z. & Deutscher, M. P. (1996) Cell 86, 503–512. [DOI] [PubMed] [Google Scholar]
- 3.Li, Z., Pandit, S. & Deutscher, M. P. (1998) Proc. Natl. Acad. Sci. USA 95, 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donovan, W. P. & Kushner, S. R. (1986) Proc. Natl. Acad. Sci. USA 83, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, Z., Reimers, S., Pandit, S. & Deutscher, M. P. (2002) EMBO J. 21, 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasai, T., Gupta, R. S. & Schlessinger, D. (1977) J. Biol. Chem. 252, 8950–8956. [PubMed] [Google Scholar]
- 7.Deutscher, M. P., Marlor, C. W. & Zaniewski, R. (1984) Proc. Natl. Acad. Sci. USA 81, 4290–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Z.-F. & Deutscher, M. P. (2002) J. Biol. Chem. 277, 21624–21629. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Z.-F., Zuo, Y., Li, Z., Rudd, K. E. & Deutscher, M. P. (1998) J. Biol. Chem. 273, 14077–14080. [DOI] [PubMed] [Google Scholar]
- 10.Yancey, S. D. & Kushner, S. R. (1990) Biochimie 72, 835–843. [DOI] [PubMed] [Google Scholar]
- 11.Brenner, S. & Beckwith, J. R. (1965) J. Mol. Biol. 13, 629–637. [Google Scholar]
- 12.Brosius, J., Dull, T. J., Sleeter, D. D. & Noller, H. F. (1981) J. Mol. Biol. 148, 107–127. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 413–415.
- 14.Nelson, D. L. & Kornberg, A. (1970) J. Biol. Chem. 245, 1137–1145. [PubMed] [Google Scholar]
- 15.Huang, S. & Deutscher, M. P. (1992) J. Biol. Chem. 267, 25609–25613. [PubMed] [Google Scholar]
- 16.Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B. & Alexander, J. S. (1997) Biochem. Biophys. Res. Commun. 238, 277–279. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 18.Li, Z. & Deutscher, M. P. (1995) Proc. Natl. Acad. Sci. USA 92, 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Z., Pandit, S. & Deutscher, M. P. (1999) RNA 5, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Z., Pandit, S. & Deutscher, M. P. (1999) EMBO J. 18, 2878–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidhardt, F. C. (1964) Prog. Nucleic Acid Res. Mol. Biol. 3, 145–181. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Hamida, F. & Schlessinger, D. (1966) Biochim. Biophys. Acta 119, 183–191. [DOI] [PubMed] [Google Scholar]
- 23.Gourse, R. L., Gaal, T., Bartlett, M. S., Appleman, J. A. & Ross, W. (1996) Annu. Rev. Microbiol. 50, 645–677. [DOI] [PubMed] [Google Scholar]
- 24.Norris, T. E. & Koch, A. L. (1972) J. Mol. Biol. 64, 633–649. [DOI] [PubMed] [Google Scholar]
- 25.Gausing, K. (1977) J. Mol. Biol. 115, 335–354. [DOI] [PubMed] [Google Scholar]
- 26.Theiben, G., Thelen, L. & Wagner, R. (1993) J. Mol. Biol. 233, 203–218. [DOI] [PubMed] [Google Scholar]
- 27.Schäferkordt, J. & Wagner, R. (2001) Nucleic Acids Res. 29, 3394–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]