Abstract
Disease-related amyloidogenic propensity has been unexpectedly found in proteins driven to adopt a monomeric uncomplexed state at high concentrations under near-physiological conditions. This situation occasionally arises in new health treatments, such as kidney dialysis. Assuming that under such conditions a partial retention of native structure takes place, this work identifies a structural characteristic indicating amyloidogenic propensity: a high density of backbone hydrogen bonds exposed to water attack in monomeric structure. On this basis, we propose a diagnostic tool based on the identification of hydrogen bonds with a paucity of intramolecular dehydration or “wrapping.” We use this predictor to identify potentially pathogenic mutations that foster amyloidogenic propensity in human prions. Such mutations either enhance the intramolecular dehydration of β-sheet hydrogen bonds, thus stabilizing the nucleus for rearrangement into the scrapie fold, or contribute to the destabilization of the cellular form by introducing additional underwrapped hydrogen bonds. Our predictions are consistent with known disease-related mutations and lead to a cogent explanation of the pathogenic nature of specific mutations affecting the cellular prion protein structural wrapping. On the other hand, a different wrapping of a very similar fold, mouse doppel, induces a dramatically different level of amyloidogenic propensity, suggesting that the packing within the fold, and not the fold itself, contains the signal for aggregation.
Keywords: amyloidosis, prions, protein structure, structural wrapping, hydrogen bonds
Amyloid fibril formation is associated with illnesses such as Alzheimer's disease and spongiform encephalopathies (1–8) and apparently with a nucleation-induced process resulting in a tightly packed regular aggregate of protein molecules. Unexpectedly, a hazardous fibrillogenic propensity has been discovered in which some proteins, e.g., β2-microglobulin in kidney dialysis (1, 8), reach high enough concentrations to form fibrils. Apparently, an overexpression coupled with a scarcity of binding partners may induce certain proteins to rearrange and aggregate (1–8, 9). Under physiological conditions where some structural features of the native protein are retained (8), we expect to infer amyloidogenic propensity from a structural analysis (9–14). Thus, we address the following question: Are there any kinds of structural defects in the monomeric protein that are removed as the protein rearranges and aggregates? We shall focus on abnormal levels of exposure of native backbone hydrogen bonds to water attack in monomeric structures (9–11). A profusion and localization of such packing defects on the protein surface is here shown to mark a tendency for aggregation.
This approach is suggested by the link between amyloidogenic propensity and structural instability (6, 7) and its association with specific structural motifs like amphiphilic helix (15), alternating hydrophobic pattern (16), edgy β-strand (17), discordant α-helix (18), etc. However, a general diagnosis stemming from examination of native monomeric structure remains a challenge.
Here we propose and benchmark a tool for such a diagnosis. Our approach involves the identification of deficiencies in the native packing of backbone hydrogen bonds, often partially overcome upon complexation (9, 10), and regions of the protein surface with high densities of defects that may act as aggregation nuclei (19–21).
The stability of native structure for soluble proteins is contingent on the intramolecular protection of backbone hydrogen bonds from water attack (9, 10). Here we argue that deficiencies in this protection on the protein surface can be correlated with amyloidogenic propensity if the protein becomes overexpressed with respect to the concentration of its binding partners. To detect such deficiencies we examine the intramolecular environments around native backbone hydrogen bonds, focusing on residues with nonpolar groups clustered in their vicinity. Backbone hydrogen bonds are stable and become structural determinants when water in the surrounding environment is excluded or highly structured (9–11, 22). Thus, the proximity of “wrapping” hydrophobic groups stabilizes such hydrogen bonds by destabilizing the nonbonded state, i.e., by hindering the solvation of the unpaired amides and carbonyls. Here we focus on backbone hydrogen bonds insufficiently shielded from water attack, or “underwrapped” hydrogen bonds (UWHBs), and show that a concentration of such defects might yield an aggregation-inducing nucleus.
Methods
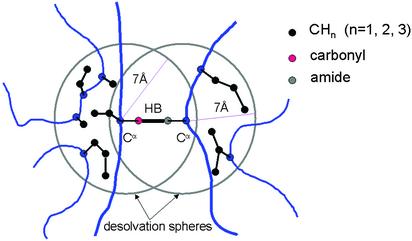
We define underwrapped in relation to an “average” native environment. The extent of hydrogen-bond desolvation is here operationally defined by the number of hydrophobic residues with at least two nonpolar carbonaceous groups (CHn, n = 1, 2, 3) whose β-carbon is contained in a specific desolvation domain. This domain consists of two intersecting R = 7-Å-radius spheres centered at the α-carbons of the residues paired by the hydrogen bond (Fig. 1) (9–11). The desolvators wrapping the hydrogen bond strengthen the electrostatic energy contribution by decreasing the charge screening of the proton donor and acceptor (10, 22). The desolvation statistics taken across all hydrogen bonds in a native structure of course highly depend on the choice of R, but the results are robust within the range 6.4 ≤ R ≤ 7.5 Å in the sense that the hydrogen bonds identified as underwrapped are invariably the same in at least 96% of the Protein Data Bank (PDB) entries sampled.
Fig. 1.
The desolvation domain of a backbone hydrogen bond represented as two intersecting 7-Å spheres centered at the α-carbons of the paired residues. The extent of desolvation or hydrophobic wrapping is determined by the number of hydrophobic residues (marked in black) whose β-carbon lies within the desolvation domain of the hydrogen bond.
The desolvation radius represents a typical cutoff distance to evaluate interactions between residues in spatial proximity. An amide-carbonyl hydrogen bond is defined by an N—O (heavy-atom) distance within the range 2.6–3.4 Å (typical extreme bond lengths) and a 60° latitude in the N—H—O angle.
Alternative (9) and even more complicated ways of measuring the extent of hydrogen bond desolvation are possible (23), and they yield equivalent results. For example, computing the number of hydrophobic carbonaceous side-chain groups within the desolvation spheres produces different statistics (9) but identifies the same UWHBs and yields the same relative ordering of PDB proteins in terms of decreasing average wrapping.
The average extent of hydrogen-bond desolvation, denoted ρ, over all backbone hydrogen bonds of a monomeric structure is readily accessible from PDB structural data. To obtain meaningful statistics, we examined a nonredundant sample of 2,811 PDB structures. The average ρ over the entire sample is 6.61 and its dispersion across backbone H bonds of a structure averaged over all sampled structures is σ = 1.46. These statistics dictate a way to define UWHBs as those at the extreme of the distribution, containing three or fewer wrapping residues in their desolvation domains.
Results
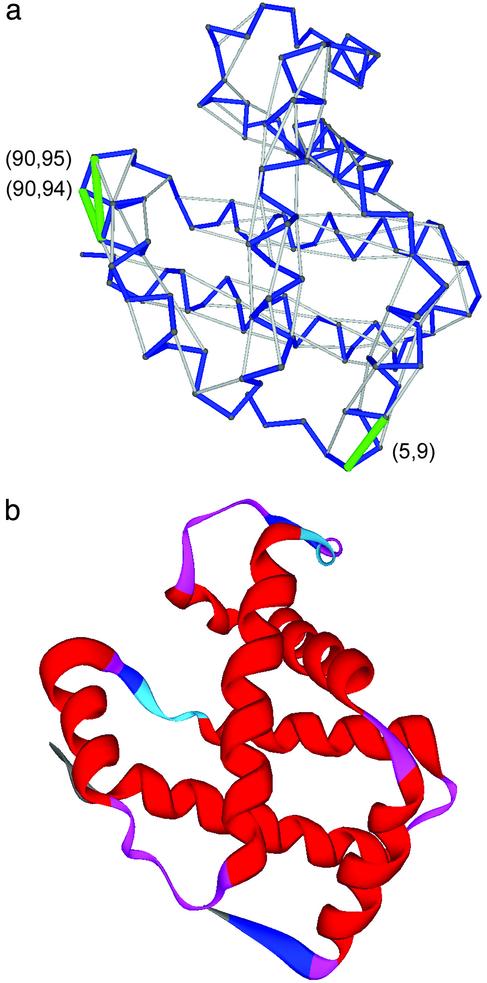
Structure Wrapping and Molecular Disease. Figs. 2 and 3 display the UWHBs for Hb β-subunit (pdb. 1bz0, chain B) and human cellular prion protein (pdb. 1qm0) (12–14). Within the natural interactive context of the Hb subunit, the UWHBs signal crucial binding regions (24): UWHBs (90, 94), (90, 95) are associated with the β-FG corner involved in the quaternary α1β2 interface; UWHB (5, 9) is adjacent to Glu-6 which in sickle cell anemia mutates to Val-6 and is located at the Val-6-(Phe-85, Leu-88) interface in the deoxyHbS fiber.
Fig. 2.
(a) Pattern of UWHBs, represented as green segments joining paired α-carbons, for human Hb β-subunit. The virtual-bond chain backbone is shown schematically as a blue polygonal joining α-carbons. The sufficiently wrapped hydrogen bonds are indicated as gray segments joining the α-carbons of the paired residues. (b) Ribbon display for human Hb.
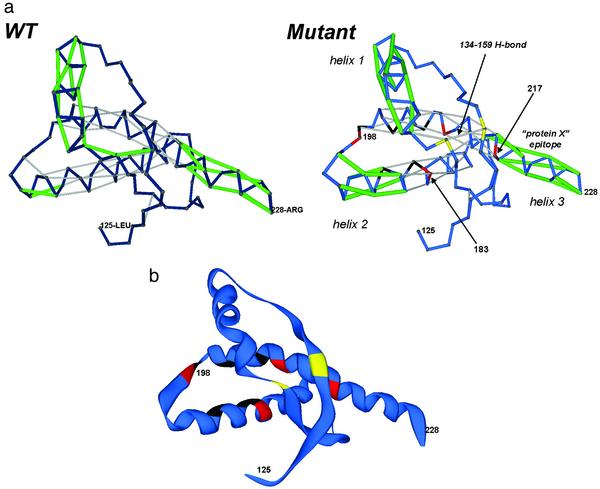
Fig. 3.
(a) UWHB patterns for the wild-type (WT) and mutant (Gln -217→ Val, Phe-198→ Ser) human cellular prion protein PrPC. In the wild type, the (134, 159) β-sheet hydrogen bond is underwrapped, and thus, the nucleus for scrapie conversion is highly unstable. Notice how and why the β-sheet becomes stable in the mutant because of the extra wrapping provided by Val-217. The four sites for pathogenic mutation that affect the wrapping of backbone hydrogen bonds are marked in red. The hydrogen-bonded pairs of residues whose desolvation levels and decrease due to pathogenic mutation Phe-198→ Ser are marked in black, and those for which the extent of desolvation increases are marked in yellow. (b) Ribbon representation of the mutant PrPC structure following the color convention given in Fig. 2b.
In the cellular prion protein (Fig. 3), 55% of the hydrogen bonds are UWHBs, indicating that parts of the structure, i.e., α-helix 1 with a high concentration of UWHBs, must be vulnerable to water attack, thus prone to rearrangement. The highest concentration of defects occurs in helix 1 (residues 143–156), where 100% of the hydrogen bonds are UWHBs. This observation is consistent with current research, which identifies helix 1 as undergoing an α-helix-to-β-strand transition (12–14). Furthermore, helix 3 (residues 199–228) contains a significant concentration of UWHBs at the C terminus, a region assumed to define the epitope for protein X binding (12). The remaining UWHBs occur at the helix–loop junctures and bestow flexibility required for rearrangement.
Our structural analysis identifies the site mutations that would improve the packing of the β-sheet hydrogen bonds (129,163) and (134,159), thereby stabilizing the part of the cellular prion protein that is believed to nucleate the cellular-to-scrapie transition (Fig. 3). Such mutations should be expected to be pathogenic, and thus it is worth contrasting them with those assumed to trigger the Creutzfeld–Jakob or Gerstmann–Sträussler–Scheinker syndrome (14). Although the (129, 163) hydrogen bond is well desolvated locally (ρ = 5) by Tyr-128, Tyr-162, Tyr-163, Arg-164, and Leu-130, the (134, 159) hydrogen bond, with ρ = 3, is underwrapped (only protected locally by Val-161 and Arg-136), and thus its stability is very sensitive to mutations that alter the large-scale context preventing water attack. This finding leads us to postulate that a factor that triggers the prion disease is the stabilization of the (134, 159) β-sheet hydrogen bond by mutations that foster its intramolecular desolvation beyond wild-type levels.
In the wild type, the only nonadjacent residue in the desolvation domain of hydrogen bond (134,159) is Val-210, thus conferring marginal stability at best. We predict that four sites can be mutated to increase significantly the extent of desolvation of this hydrogen bond without disrupting the underlying helical structure of the cellular prion protein: Thr-183, Val-210, Met-213, and Gln-217. Thus, the known pathogenic mutations Thr-183→Ala, Val-210→Ile and Gln-217→Val would all increase the desolvation level of the (134, 159) hydrogen bond, consolidating the nucleus for the cellular-to-scrapie transition. In addition, we predict that substitution of Met-213 with a hydrophobic residue, such as Val, Leu, or Ile, should be equally pathogenic, in so far as it enhances the stability of the β-sheet nucleus. Conversely, substitutions that destabilize the (134, 159) hydrogen bond, such as Val-161–Ala should act as suppressors of the prion disease. This last prediction now awaits experimental confirmation. It is likely to have passed unnoticed because it is not pathogenic, or merely regarded as part of the prion polymorphism.
On the other hand, the known pathogenic substitution Phe-198→Ser causes a drastic reduction in the desolvation of hydrogen bonds (188, 184), (199, 203), (202, 206), (203, 207), which become underwrapped in the mutant (Fig. 3). The net consequence of this destabilization is that the helix 2–helix 3 junction becomes f lexible, without directly affecting the β-sheet nucleus. Thus, within our scheme this mutation should be conducive to the transition to the scrapie form. The Phe-198–Ser mutation would leave helix 3 with 60% of UWHBs; thus, its stability would be severely compromised, diminishing the pathogenic potential of this mutation unless the presumed binding partner, protein X, associates at the C terminus, as speculated (12), and provides intermolecular desolvation of UWHBs in that region.
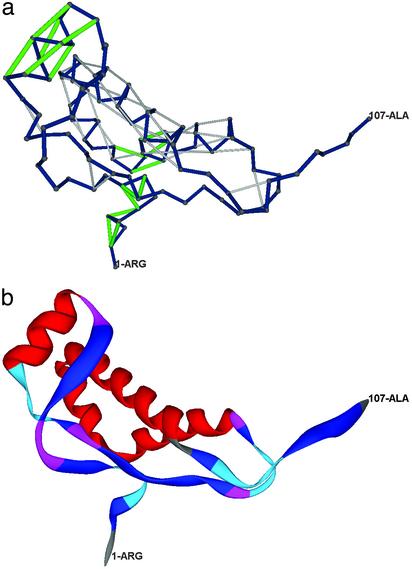
To verify that the structural wrapping and not the fold per se is the signal for amyloidogenic propensity, we compared the wrapping of the cellular prion protein with that of the folding analogue doppel protein (24), not known to undergo conversion into a scrapie-like form. The secondary structure wrapping for doppel (pdb. 1I17) is presented in Fig. 4. Although the folding topologies are very similar, major differences exist in the wrapping of doppel and prion. As in the prion, helix 1 is severely underwrapped in the doppel protein (Fig. 4b). However, the rest of the doppel helices are well wrapped at the helix–loop junctions and helix 3 lacks entirely the underwrapped C-terminal region presumed to serve as epitope for protein X binding. Furthermore, the purported scrapienucleating region, consisting in the small β-sheet is more unstable than in the prion: Of its four backbone hydrogen bonds, two of them, Gly-8–Tyr-41 and Ala-9–Tyr-41, are severely underwrapped (ρ < 3) and the N terminus region is also severely exposed. Thus, it is unlikely that this region, with its inherent instability can serve as a nucleating element for condensation of a rearranged helix 1 region. An unstable scrapie-inducing nucleus together with the lack of a protein X epitope make the doppel an unlikely candidate for scrapie-like condensation, as recent work confirms (25).
Fig. 4.
(a) UWHB pattern for the doppel protein (24), following the repre- sentational convention for Fig. 2b. (b) Ribbon representation of the doppel protein.
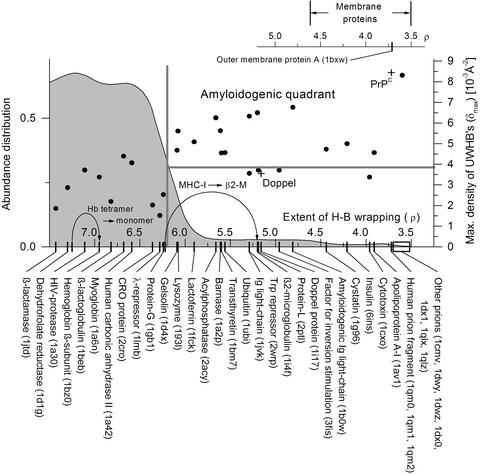
Toward a Structural Diagnosis. The distribution of proteins according to their average extent of hydrogen bond wrapping and their spatial concentration of structural defects is shown in Fig. 5 (see also ref. 23). The sample of 2,811 PDB proteins is large enough to define a reliable abundance distribution with an inflection point at ρ = 6.20. The integration of the distribution over a ρ-interval gives the fraction of proteins whose ρ lies within that range. Of the 2,811 proteins examined, 2,572 have ρ > 6.20, and none of them is known to yield amyloid aggregation under physiological conditions entailing partial retention of structure. Strikingly, relatively few disease-related amyloidogenic proteins are known in the sparsely populated, underwrapped 3.5 < ρ < 6.20 range, with the cellular prion proteins located at the extreme of the spectrum (3.53 < ρ < 3.72).
Fig. 5.
Abundance distribution of soluble proteins grouped according to their average extent of H-bond wrapping ρ. The abundance density distribution (gray area) was obtained from a large nonredundant PDB sample (see text). The number of isolated soluble proteins with extent of wrapping lower than the inflection point (ρ < 6.2) constitutes ≈10% of the sample. Representative proteins are displayed on the abscissas. The change in ρ upon dissociation of a complex is indicated by an arrow in two illustrative cases: the β-subunit of Hb taken from complexed to isolated form, and the β2-microglobulin within the MHC-I complex and taken in isolation. For comparison the ρ-value range of membrane proteins is displayed. The ordinate on the right gives the maximum density, ρmax, of underwrapped H bonds for representative proteins. The human prion (PrPC) and the mouse doppel protein, both sharing a similar fold but with very different wrapping, are marked by crosses (+). Disease-related amyloidogenic proteins are singled out by the conditions ρ < 6.2, ρmax > 3.8, and thus correspond to a sector indicated. The thresholds are indicated by gray lines.
This analysis reveals that extensive exposure of H bonds to water attack might be a necessary but not a sufficient condition (see below) to imply a propensity for organized aggregation. At most, 10% of PDB proteins may possess this propensity, although the critical region 3.5 < ρ < 6.20 is so sparsely populated that no reliable statistics can be drawn from our sample in this regard. Approximately 60% of the proteins in the critical region 3.5 < ρ < 6.20, which are not known to be amyloidogenic, are neurotoxins or toxins whose H bonds are significantly undershielded from water attack, while their structures are stabilized mostly by disulfide bridges. It remains to be seen whether their toxicity is linked to their overexposed surface hydrogen bonds.
The range of H-bond wrapping 3.5 < ρ < 4.6 of 20 sampled PDB membrane proteins has been included in Fig. 5 for comparison. As expected, such proteins do not have the stringent H-bond packing requirements of soluble proteins for their H bonds at the lipid interface. Thus, this comparison becomes suggestive in terms of elucidating the driving factor for aggregation in soluble proteins: Although the UWHB constitutes a structural defect in a soluble protein because of its vulnerability to water attack, it is not a structural defect in a membrane protein. The exposure of the polar amide and carbonyl of the unbound state to a nonpolar phase is thermodynamically unfavorable (22). The virtually identical ρ value for human prion and outer-membrane protein A (Fig. 5) is revealing in this regard.
Furthermore, all known amyloidogenic proteins that occur naturally in complexed form have sufficient H-bond wrapping within their respective complexes (ρ value near 6.2). Their amyloidogenic propensity appears only under conditions in which the protein is dissociated from the complex (compare Fig. 5). This finding is corroborated by the following computation. If an intramolecular hydrogen bond is underwrapped within the isolated protein molecule but located at an interface upon complexation, then to determine its extent of wrapping within the complex, we take into account the additional residues in the binding partner that lie within the desolvation domain of the intramolecular H bond. Thus, the uncomplexed or monomeric β2-microglobulin (pdb. 1i4f) (21) has ρ = 5.2, putting it in the purported amyloidogenic region. However, upon complexation within the MHC-I, its ρ increases to 6.22.
The underwrapping of H bonds in the isolated protein might be a necessary condition but it is unlikely to be sufficient to determine amyloidogenic propensity. For example, protein L (2ptl) is not known to possess this property under the conditions specified, and nevertheless its ρ = 5.06 places it within the range of overall insufficient wrapping. A similar situation arises with trp-repressor (pdb. 2wrp, ρ = 5.29) and the factor for inversion stimulation (pdb. 3fis, ρ = 4.96), as shown in Fig. 5. Most neurotoxins, e.g., pdb. 1cxo, ρ = 3.96, fall in this category as well.
In general, the existence of short fragments endowed with fibrillogenic potential (19, 20, 21, 26, 27, 28, 29) suggests a localization or concentration of the amyloid-related structural defects. Thus, we introduce a complementary parameter, the maximum density, δmax, of UWHBs on the protein surface. It reveals the highest concentration of structural defects within each protein and is obtained by sampling regions of the protein surface and computing the number of UWHBs within the regions, together with their solvent-exposed areas. The δmax values for representative proteins are given in Fig. 5 (right ordinate). A threshold δmax = 3.8/(103 Å2) distinguishes the known disease-related amyloidogenic proteins from other proteins with a low extent of surface hydrogen bond wrapping. Again, with δmax = 8.4/(103 Å2), prions possess the highest concentration of UWHBs. All known amyloidogenic proteins found in the sampled PDB and only such proteins lie in the quadrant defined by ρ < 6.20, δmax > 3.8/(103 Å2).
On the basis of this combined analysis we predict that the following proteins probably possess a marked and hitherto undetected amyloidogenic propensity: angiogenin [pdb. 1b1e, pdb. 2ang ρ = 4.31, δmax = 3.9/(103 Å2)]; plasminogen [pdb. 1b2i, ρ = 4.08, δmax = 3.9/(103 Å2)]; anti-oncogene A [pdb. 1a1u, ρ = 3.53, δmax = 4.0/(103 Å2)]; meizothrombin [pdb. 1a0h, ρ = 3.7, δmax = 3.8/(103 Å2)]; RADR zinc finger peptide [pdb. 1a1k, ρ = 3.3, δmax = 4.4/(103Å2)]; and rubredoxin [pdb. 1b20, ρ = 3.82, δmax = 4.6/(103 Å2)]. These proteins have been selected from our nonredundant PDB sample by imposing the stringent filtering conditions: ρ < 4.5, δmax > 3.8/(103 Å2); thus, they all lie within the amyloidogenic quadrant defined in Fig. 5.
At this point we may ask whether the regions of high concentration of structural defects assessed by the indicator introduced in this work are invariably correlated with the primary sites for protein–protein association (9). A systematic study of the compensatory role of complexation for proteins is reported in Table 1: Invariably, the density δint of intramolecular UWHBs at the binding interface is significantly larger than the average density δ of UWHBs of the binding partners. This result reveals that UWHBs might signal binding sites because their desolvation domains can be “filled” (leading to ρ ≈ 6.2 or larger) upon binding. This increase in the extent of hydrogen-bond desolvation arises as a desolvating residue of a binding partner pene- trates the domain of a hydrogen bond in the other partner upon association. Strikingly, in all cases in which complexation of amyloidogenic proteins represents a means of compensating for the structural defects of the isolated molecule, we have found that δint ≈ δmax. This result validates our approach and reveals the potential hazard in the uncomplexed or out-of-context protein that usually requires a binding partner to dehydrate its hydrogen bonds adequately.
Table 1. Complexes identified by their PDB accession codes.
| Complex name, PDB code | Yint | Y | δ 10-3 [Å-2] | δint 10-3 [Å-2] | δmax 10-3 [Å-2] |
|---|---|---|---|---|---|
| HLA antigen A-2 + β2-microglobulin, 1i4f | 6 | 36 | 1.58 | 6.50 | 6.50 |
| Ig-light chain dimer, 1jvk | 8 | 26 | 1.78 | 6.33 | 6.33 |
| Transthyretin dimer, 1bm7 | 5 | 14 | 1.01 | 4.55 | 4.55 |
| Insulin dimer, 6ins | 5 | 22 | 2.80 | 5.00 | 5.00 |
| HIV-1 protease dimer + inhibitor, 1a30 | 7 | 12 | 1.87 | 3.51 | 3.70 |
| Simian immunodeficiency virus protease dimer, 1siv | 4 | 14 | 1.06 | 2.65 | 3.35 |
| Chey complex, 1fqw | 4 | 10 | 1.02 | 3.07 | 3.30 |
| Antitrypsin polymers, 1d5s | 14 | 22 | 1.01 | 2.76 | 3.50 |
| Bombyxin, 1bon | 4 | 5 | 0.60 | 3.02 | 3.25 |
| FcγRIII receptor + Ig, 1e4k, B, C | 7 | 22 | 0.97 | 3.08 | 3.50 |
| Colicin + ligand, 1bxi | 6 | 12 | 0.92 | 3.27 | 3.60 |
| Colicin + ligand, 1emv | 5 | 11 | 0.86 | 3.30 | 3.66 |
| Serpin + ligand, 1as4 | 14 | 31 | 1.40 | 2.02 | 2.90 |
| Troponin heterodimer, 1pon | 6 | 10 | 1.34 | 3.22 | 3.50 |
| MHC, antigen + receptor, 1im9, A—D | 3 | 22 | 0.84 | 2.22 | 2.85 |
Complexes are described by the following quantities: Y, number of intramolecular UWHBs in the individual binding partners; Yint, number of intramolecular UWHBs at the binding interface; δ, average density of UWHBs in the binding partners; δint, density of intramolecular UWHBs at the binding interface satisified on complexation; δmax, maximum density of intramolecular UWHBs from the binding partners.
Much of the current research on amyloidosis is focused on severe denaturation conditions (28, 29). Such endeavors are outside the scope of this work, which defines and benchmarks a disease-related signal encoded in the native fold: a high concentration of insufficiently desolvated backbone hydrogen bonds within a poorly wrapped structure. Our assay of struc- tural anomaly is a step toward a proteomics-based diagnosis of molecular disease and here has been shown to be instrumental in assessing the pathogenic potential of specific site mutations. Furthermore, this study might invite a paradigmatic shift because we have demonstrated that the fold itself is not indicative of a pathogenic propensity, rather, the signal is enshrined in the wrapping of the fold.
Acknowledgments
A.F. acknowledges the support of Osaka University, which provided a visiting professorship for the summer of 2002, where this work originated. R.S.B. acknowledges the support of the National Science Foundation.
Abbreviations: UWHB, underwrapped hydrogen bond; PDB, Protein Data Bank.
References
- 1.Gejyo, F., Yamada, T., Odani, S., Nakagawa, Y., Arakawa, M. & Kunitomo, T. (1985) Biochem. Biophys. Res. Commun. 129, 701–706. [DOI] [PubMed] [Google Scholar]
- 2.Shtilerman, M. D., Ding, T. T. & Lansbury, P. T., Jr. (2002) Biochemistry 41, 3855–3860. [DOI] [PubMed] [Google Scholar]
- 3.Rochet, J. C. & Lansbury, P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60–68. [DOI] [PubMed] [Google Scholar]
- 4.Dobson, C. M. (1999) Trends. Biochem. Sci. 24, 329–332. [DOI] [PubMed] [Google Scholar]
- 5.Koo, E. H., Lansbury, P. T., Jr. & Kelly, J. W. (1999) Proc. Natl. Acad. Sci. USA 96, 9989–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth, D. R., Sunde, M., Bellotti, V., Robinson, C. V., Hutchinson, W. L., Fraser, P. E., Hawkins, P. N., Dobson, C. M., Radford, S. E., Blake, C. C. & Pepys, M. B. (1997) Nature 385, 787–793. [DOI] [PubMed] [Google Scholar]
- 7.Souillac, P. O., Uversky, V. N., Millett, I. S., Khurana, R., Doniach, S. & Fink, A. L. (2002) J. Biol. Chem. 277, 12657–12665. [DOI] [PubMed] [Google Scholar]
- 8.Verdone, G., Corazza, A., Viglino, P., Pettirossi, F., Giorgetti, S., Mangione, P., Andreola, A., Stoppini, M., Bellotti, V. & Esposito, G. (2002) Protein Sci. 11, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández, A. & Berry, R. S. (2002) Biophys. J. 83, 2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández, A. & Scheraga, H. A. (2003) Proc. Natl. Acad. Sci. USA 100, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández, A., Sosnick, T. R. & Colubri, A. (2002) J. Mol. Biol. 321, 659–675. [DOI] [PubMed] [Google Scholar]
- 12.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahn, R., Liu, A., Luhrs, T., Riek, R., von Schroetter, C., Lopez Garcia, F., Billeter, M., Calzolai, L., Wider, G. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riek, R., Wider, G., Billeter, M., Hornemann, S., Glockshuber, R. & Wüthrich, K. (1998) Proc. Natl. Acad. Sci. USA 95, 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi, Y., Yamashita, T., Ueno, A. & Mihara H. (2000) Tetrahedron 56, 7011–7018. [Google Scholar]
- 16.Broome, B. M. & Hecht, M. H. (2000) J. Mol. Biol. 296, 961–968. [DOI] [PubMed] [Google Scholar]
- 17.Richardson, J. S. & Richardson, D. C. (2002) Proc. Natl. Acad. Sci. USA 99, 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallberg, Y., Gustafsson, M., Persson, B., Thyberg, J. & Johansson, J. (2001) J. Biol. Chem. 276, 12945–12950. [DOI] [PubMed] [Google Scholar]
- 19.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol. 9, 332–336. [DOI] [PubMed] [Google Scholar]
- 20.McParland, V. J., Kalverda, A. P., Homans, S. W. & Radford, S. E. (2002) Nat. Struct. Biol. 9, 326–331. [DOI] [PubMed] [Google Scholar]
- 21.Kozhukh, G. V., Hagihara, Y., Kawakami, T., Hasegawa, K., Naiki, H. & Goto, Y. (2002) J. Biol. Chem. 277, 1310–1315. [DOI] [PubMed] [Google Scholar]
- 22.Roseman, M. A. (1998) J. Mol. Biol. 201, 621–623. [DOI] [PubMed] [Google Scholar]
- 23.Fernández, A. & Berry, R. S. (2003) Proc. Natl. Acad. Sci. USA 100, 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voet, D. & Voet, J. G. (1990) Biochemistry (Wiley, New York).
- 25.Ho, H., Moore, R., Cohen, F., Westaway, D., Prusiner, S. B., Wright, P. E. & Dyson, H. J. (2001) Proc. Natl. Acad. Sci. USA 98, 2352–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azriel, R. & Gazit, E. (2001) J. Biol. Chem. 276, 34156–34161. [DOI] [PubMed] [Google Scholar]
- 27.DePace, A. H., Santoso, A., Hillner, P. & Weissman, J. S. (1998) Cell 93, 1241–1252. [DOI] [PubMed] [Google Scholar]
- 28.Fandrich, M., Fletcher, M. A. & Dobson, C. M. (2001) Nature 410, 165–166. [DOI] [PubMed] [Google Scholar]
- 29.Guijarro, J. I., Sunde, M., Jones, J. A., Campbell, I. D. & Dobson, C. M. (1998) Proc. Natl. Acad. Sci. USA, 95, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]