Abstract
Ultrafast IR spectroscopy is used to monitor the nonequilibrium backbone dynamics of a cyclic peptide in the amide I vibrational range with picosecond time resolution. A conformational change is induced by means of a photoswitch integrated into the peptide backbone. Although the main conformational change of the backbone is completed after only 20 ps, the subsequent equilibration in the new region of conformational space continues for times >16 ns. Relaxation and equilibration processes of the peptide backbone occur on a discrete hierarchy of time scales. Albeit possessing only a few conformational degrees of freedom compared with a protein, the peptide behaves highly nontrivially and provides insights into the complexity of fast protein folding.
Protein dynamics occurs on a large range of time scales, which can coarsely be related to various length scales of proteins: dynamics of tertiary and quaternary structure extends from milliseconds to seconds and even longer, whereas formation of secondary structure has been observed between 50 ns and a few microseconds (1–11). Nevertheless, several experiments have provided strong hints for the relevance of even faster processes from the observation of large instantaneous signals, which could not be time resolved (2, 7, 12). For example Huang et al. (7) indirectly concluded that helix nucleation might occur on a subnanosecond time scale, whereas Thompson et al. (5) estimated a “zipping time” for a 21-residue α-helix of 300 ps at 300 K. Also, molecular dynamics simulations suggest that peptides and proteins can undergo considerable structural changes within 1 ns or less (8–10, 13, 14). Hummer et al. (15) found the formation of the first α-helical turn within 0.1–1 ns in work on helix nucleation in short Ala- and Gly-based peptides. Daura and colleagues (9, 16) simulated equilibrium folding/unfolding of a β-heptapeptide at the melting point and above to obtain statistics on the populations of the folded and unfolded states. Several folding/unfolding events were observed in a 50-ns trajectory, where the actual transitions from unfolded to folded conformations could be as fast as 50–100 ps. Time-dependent 2D-IR spectroscopy has recently been applied to focus directly on single-bond dynamics in a peptide backbone, revealing that fluctuations of dihedral angles occur in <1 ps (17). These elementary conformational steps represent the lowest hierarchical level of protein dynamics. Because of a lack of appropriate techniques, however, little is known from experiment about intermediate times between about 1 ps and 10 ns. It is the aim of the present study to bridge this gap.
Conformational transitions of peptides and proteins are most commonly triggered by changing the environment of the molecule, thereby shifting the equilibrium constant of the process under investigation (see refs. 1, 3, 4, 6, 7, and 18–21 for recent reviews). Among these methods, only temperature jump experiments have been shown to allow for subnanosecond time resolution (22). Even faster triggering with a possible subpicosecond time resolution can be achieved with a molecular switch incorporated in the peptide chain (23–26). Ultrafast light-induced changes of the switching molecule (27) initiate structural dynamics of the peptide on the picosecond time scale by changing a conformational restraint (26). The peptide then starts from its old conformation to sample the conformational space now accessible under the rule of the modified potential.
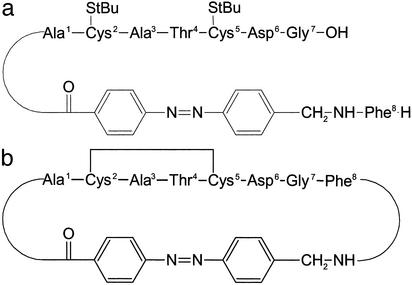
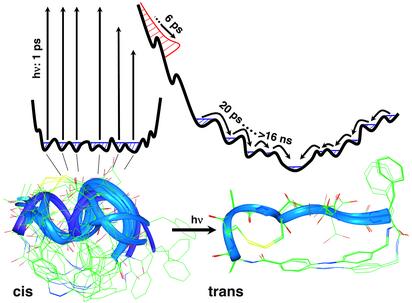
The system studied here is a cyclic octapeptide with an azobenzene photoswitch embedded in its backbone [cyclic (4-aminomethyl)phenylazobenzoic acid (cycAMPB), Fig. 1b] (25). The azobenzene unit can be reversibly switched between the cis and trans conformations by using light with different wavelengths, thereby predetermining the conformation of the backbone of the cyclic peptide. According to NMR investigations (25), the cis state of cycAMPB is a frustrated system with many close-lying conformational energy minima, leading to an ensemble of structures with different backbone geometries. The rms standard deviation (rmsd) of the nuclei positions of the backbone is 1.54 Å (Fig. 5). On switching to the trans conformation, the backbone structure is much better defined with a significantly reduced rmsd value of 0.19 Å (Fig. 5). The cis → trans transition, which is studied here, thus runs from a broad ensemble into one with a narrow distribution in conformational space. The photoreaction of this (25) and a very similar molecule (28) has been analyzed recently by probing the electronic response of the photoswitch itself by visible spectroscopy (ref. 26; J.W., S. Spörlein, B. Fonrobert, H. Satzger, C. Renner, R.B., L.M., and W.Z., unpublished observations): the cis → trans isomerization of the azobenzene unit has been found to be finished within <1 ps with a quantum efficiency of ≈50% (29). A transient signal extending up to 1 ns, with a significant contribution in the 50-ps range, has been observed that has been attributed to residual stress exerted by the peptide backbone onto the photoswitch. The time scale of these processes could be reproduced by using molecular dynamics simulations. Different concepts to photo-modulate the conformation of small peptides by using an azobenzene moiety have been reported in refs. 30 and 31.
Fig. 1.
Structure sketch of the S-tert-butylthio-protected linear precursor peptide linAMPB (a) and the disulfide-bridged cyclic peptide cycAMPB (b).
Fig. 5.
Ensemble of structures with the azobenzene unit in the cis (Lower Left) and trans conformation (Lower Right), as obtained from NMR structure analysis (25). After photoisomerizing the built-in azobenzene unit, the random distribution of cis structures is projected onto a new potential energy surface. During the driven phase (in red), the ensemble relaxes very quickly on a 6-ps time scale and entirely floods the bottom area of the potential energy surface. During the subsequent biased diffusion phase, the system equilibrates on a discrete hierarchy of time scales, extending from 20 ps to >16 ns, toward the much better defined global minimum of the trans conformation.
Here, we monitor directly the dynamics of the peptide backbone by applying ultrafast transient vibrational spectroscopy of the amide I band, rather than the indirect effect of backbone dynamics on one spectroscopic degree of freedom as in ref. 26. The amide I band originates mainly from the C O stretch vibration of the peptide units and has proven to be a sensitive probe of backbone structure (32). It is therefore expected that the amide I band monitors much more directly the complicated structural dynamics. We find that stretching of the peptide backbone is essentially completed after only 20 ps, whereas the subsequent equilibration in the new region of the conformational space continues for times >16 ns.
O stretch vibration of the peptide units and has proven to be a sensitive probe of backbone structure (32). It is therefore expected that the amide I band monitors much more directly the complicated structural dynamics. We find that stretching of the peptide backbone is essentially completed after only 20 ps, whereas the subsequent equilibration in the new region of the conformational space continues for times >16 ns.
Materials and Methods
We investigated the cyclic octapeptide formed from the thioredoxin reductase active site fragment H-Ala1-Cys2-Ala3-Thr4-Cys5-Asp6-Gly7-Phe8-OH via backbone cyclization with the photoswitchable AMPB (cycAMPB, see Fig. 1b) (25), as well as its linear precursor molecule (linAMPB, see Fig. 1a), which was used as a reference molecule. Both samples were dissolved in anhydrous DMSO with the concentration adjusted to an absorbance of ≈0.1 OD for the amide I band (≈4 mM). DMSO was used as solvent because of the good solubility of the peptides and because of its high IR transmission in the spectral range of interest. The sample was circulated through a CaF2 flow cell in a closed cycle designed for small amounts of liquid. Its Teflon spacer (thickness 100 μm) featured a small reservoir allowing for continuous irradiation and a thin channel where the flow was sufficient to completely exchange the sample between subsequent laser pulses. No indications of sample degradation during the measurements were found from visible and stationary Fourier transform IR (FTIR) spectroscopy.
The dark-adapted peptide molecules are in the trans conformation at room temperature (25). Before applying the short switch pulse, the population was shifted to cis by continuous UV irradiation of the trans π → π* band with an Ar-Ion Laser [Coherent (Palo Alto, CA) Innova 100, 363 nm, 200 mW]. Using the absorption cross sections at 363 nm and isomerization quantum yields of the cis and trans isomers, the thermal back-reaction rate (24, 25, 28), and the transmission of the cw UV laser, we estimated that ≈90% of all molecules in the closed cycle system are in the cis conformation after ≈1/2 h of irradiation. The transition from cis to trans was then initiated by a short 420-nm laser pulse (energy 8.5 μJ per pulse) obtained from a frequency-doubled 1 kHz Ti:Sapphire-laser/amplifier system (Spectra-Physics, Spitfire). To avoid unwanted nonlinear effects in the sample cell windows [such as white-light generation or the formation of color centers in the sample cell windows (23)], the pump pulses were stretched to 700 fs by guiding them through 25-cm fused silica.
The evolution of the peptide backbone after photoswitching the azobenzene unit was monitored by transient IR spectroscopy of the amide I band. IR probe pulses (center frequency 1,650 cm–1, bandwidth 240 cm–1 full width at half maximum, 100 fs) were generated using a white-light-seeded two-stage β-barium borate-optical parametric amplifier, the signal and idler pulses of which were difference frequency mixed in a AgGaS2 crystal (33). The IR beam was split into a probe and a reference pulse, which were focused into the sample with a f = 10 cm off-axis parabolic mirror, yielding a spot size of 70 μm. Both probe and reference beam were frequency dispersed in a spectrometer and imaged onto a 2 × 32 pixel HgCdTe detector array, which enables us to measure low-noise transient spectra on a single-shot basis with a resolution of 4 cm–1.
Time scans with parallel and perpendicular pump pulse polarizations were recorded simultaneously by changing the polarization of the pump beam every 400 laser shots to correlate drifts in both measurements. The pump pulses were delayed with respect to the probe pulses by means of an optical delay stage for delays up to 1.7 ns. Longer delays of up to 16 ns were created by a fixed-delay loop; therefore, no transients are available in this time range. In addition, a transient spectrum at a delay of a 1-ms spectrum was recorded by using two subsequent laser pulses of the Ti:Sapphire-amplifier with a larger focus of the 420-nm pump pulses and a reduced flow rate of the sample. Stationary cis-trans difference spectra were measured in a FTIR spectrometer (resolution 4 cm–1) after irradiation with a spectrally filtered high-power xenon lamp.
Experimental Results and Assignments
Steady-State Spectroscopy. The absorption spectra of the cyclic peptide cycAMPB in the trans configuration, as well as those of its linear precursor linAMPB, show a broad amide I band centered at 1,675 cm–1 with only little structure (Figs. 2 and 3 Top). Because the peptide chain contains 8 amino acids and the C O group of the azobenzene moiety, in total nine amide I vibrators, which are not spectrally resolved, lie underneath this band. The band at 1,720 cm–1 originates from the carboxyl groups of the aspartic acid and (in the case of linAMPB) the C-terminal group. The difference spectra between the trans and cis conformations obtained under steady-state illumination in a FTIR spectrometer, on the other hand, reveal a very different response for the two samples (Figs. 2 and 3 Bottom). In these difference spectra, negative signals originate from the depleted start (cis) conformation and positive signals from the generated (trans) conformation. The linear precursor molecule linAMPB exhibits only a small response on cis → trans isomerization of the azo switch. In the open chain-like form of the molecule, the conformation of the AMPB chromophore does not impose a severe restraint on the backbone structure of the linear peptide. In contrast, the difference spectrum of cycAMPB reveals a blue shift of the amide I band, because the backbone is forced to adopt a new conformation.
O group of the azobenzene moiety, in total nine amide I vibrators, which are not spectrally resolved, lie underneath this band. The band at 1,720 cm–1 originates from the carboxyl groups of the aspartic acid and (in the case of linAMPB) the C-terminal group. The difference spectra between the trans and cis conformations obtained under steady-state illumination in a FTIR spectrometer, on the other hand, reveal a very different response for the two samples (Figs. 2 and 3 Bottom). In these difference spectra, negative signals originate from the depleted start (cis) conformation and positive signals from the generated (trans) conformation. The linear precursor molecule linAMPB exhibits only a small response on cis → trans isomerization of the azo switch. In the open chain-like form of the molecule, the conformation of the AMPB chromophore does not impose a severe restraint on the backbone structure of the linear peptide. In contrast, the difference spectrum of cycAMPB reveals a blue shift of the amide I band, because the backbone is forced to adopt a new conformation.
Fig. 2.
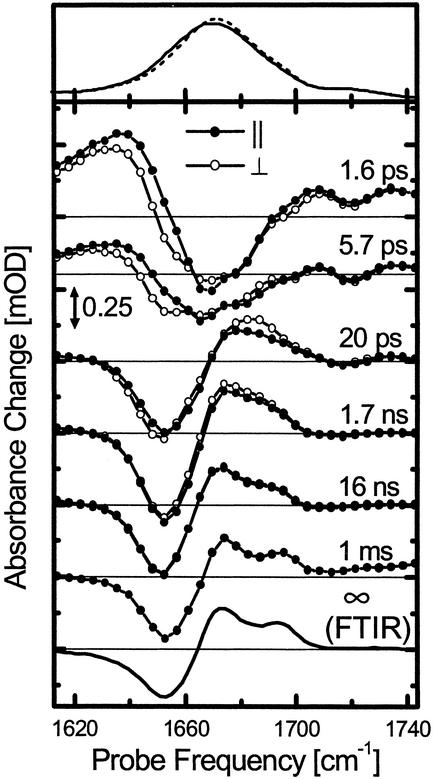
Absorption spectra of the cis (Top, solid line) and trans conformer (Top, dashed line), transient difference spectra at selected delay times (Middle) and stationary cis-trans difference spectrum measured in the FTIR spectrometer (Bottom) of the cyclic peptide cycAMPB. The FTIR difference and absorption spectra have been scaled to facilitate direct comparison with the time-resolved spectra. The transient responses with both parallel (filled circles) and perpendicular (open circles) polarization of pump and probe pulses are shown (both have been measured simultaneously and have not been scaled with respect to each other; see Materials and Methods). The scale of the time-resolved spectra is the same as in Fig. 3.
Fig. 3.
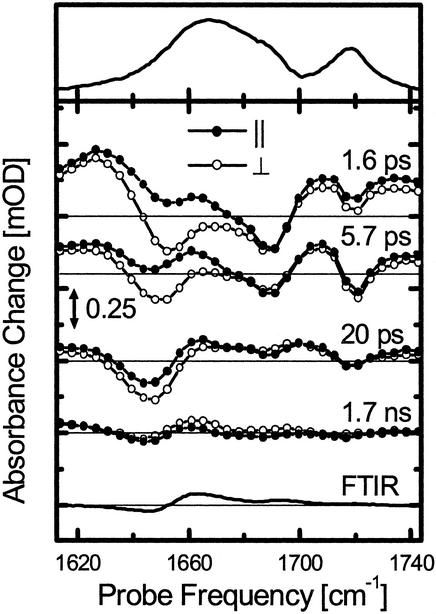
Absorption spectrum (Top), transient difference spectra at selected delay times (Middle), and stationary cis-trans difference spectrum measured in the FTIR spectrometer (Bottom) of the linear precursor peptide linAMPB. The responses with both parallel (filled circles) and perpendicular (open circles) polarization of pump and probe pulses are shown (both have been measured simultaneously and have not been scaled with respect to each other; see Materials and Methods). The FTIR difference and absorption spectra have been scaled to facilitate direct comparison with the time-resolved spectra. The scale of the time-resolved spectra is the same as in Fig. 2.
It is well established that the amide I band is a sensitive probe of protein secondary structure and is, for example, commonly used to distinguish between α-helical and β-sheet structure motives in large proteins (34). The structure sensitivity is attributed mainly to the distance and angle dependence of the coupling between individual amide I vibrators (32, 35). This coupling was originally introduced by Krimm and Bandekar (32) as transition dipole coupling. However, because the size of the peptide unit is about the same as the distance between adjacent groups, the dipole approximation is crude, and more sophisticated approaches using ab initio quantum-chemistry calculations have been developed recently (36, 37). Applying the model outlined in ref. 35 to the cis and trans structures of cycAMPB derived from NMR spectroscopy (25), the experimentally observed blue shift can be reproduced. Hence, we conclude that the blue shift directly reflects the stretching of the peptide backbone on cis→trans isomerization of the azobenzene unit.
Transient Spectroscopy. The time evolution of both AMPB peptides on their way from the start to the end conformation is investigated by transient IR spectroscopy (Figs. 2, 3, 4). Two main phases can clearly be distinguished from the experimental data.
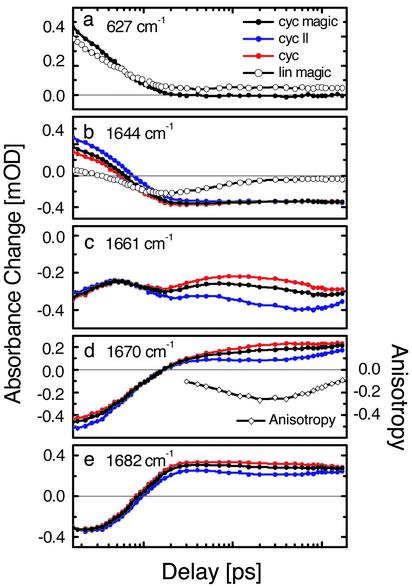
Fig. 4.
Polarization-dependent (parallel, perpendicular, and magic angle) transient absorbance change of the cycAMPB and linAMPB-peptide at selected probe frequencies. An example of the anisotropy is shown for 1,670 cm–1.
Fast Phase, 0–20 ps (Driven and Cooling Phase). Immediately after photoswitching of the azobenzene unit from cis to trans, a transient red shift of the amide I band of the peptide is observed (Fig. 2, 1.6 ps), which decays on a 4-ps time scale and vanishes within 14 ps (Fig. 4a). Test measurements on the linear precursor molecule linAMPB, as well as on a compound containing only one amino acid attached to the photoswitch (data not shown), revealed an almost identical red shift at early times (Fig. 4a). In view of this universal response, we attribute this red shift to a heating, or nonthermal vibrational excitation, of low-frequency modes that are anharmonically coupled to the amide I vibrations. A similar response has been observed for pure azobenzene and has been analyzed in detail in ref. 38.
A somewhat slower process leads to the transient blue shift observed after 20 ps (Fig. 2). We used a global exponential fit to separate this process from the heat signal and obtained a time constant of 6 ps for the formation of the blue-shifted signature. The intensity and peak positions in the difference spectrum reached after 20 ps are almost equivalent to that observed in the 1-ms difference spectrum. Because the blue shift of the amide I band can be directly related to the change of backbone structure (see above), we conclude that stretching of the peptide conformation, in a coarse sense, is completed after only 20 ps.
The linear reference molecule linAMPB shows a response comparable to that of cycAMPB up to ≈20 ps (Figs. 3 and 4 a and b). In particular, the bleach signal at 1,644 cm–1 ≈20 ps (Fig. 4b) indicates a significant change of the linear molecule backbone structure, even though the backbone is not closed to a loop. Apparently, friction fixes the open end of the linear peptide on a short time scale and thus allows the flipping photoswitch to indeed perturb the conformation of the peptide backbone. In contrast to cycAMPB, however, this bleach decays within ≈100 ps (Fig. 4b). As expected, and as confirmed by the steady-state FTIR difference spectra, the conformation of the azoswitch has no significant long-lasting influence on the backbone structure of the linear molecule. Apparently, conformational diffusion of the backbone quickly reestablishes its initial structure.
Interestingly, the early 1.6-ps spectrum of the linear peptide linAMPB is more strongly modulated than the corresponding cycAMPB spectrum. It shows the bleach and heating of two bands at 1,650 cm–1 and 1,690 cm–1 (Fig. 3), which we tentatively assign to the peptide units directly attached to the azobenzene unit, because these are expected to receive most of the dissipated energy during isomerization. In contrast, a broad response throughout the whole amide I band is observed in the cyclic peptide, indicating that essentially all units of the cyclic peptide are perturbed instantaneously by pulling apart both ends of the peptide chain.
Slow Phase, >20 ps (Biased Diffusion Phase). Although stretching of the peptide backbone is essentially finished after 20 ps in cycAMPB, it is clear from the further evolution of the transient spectra that the system is not yet in equilibrium (Figs. 2 and 4 c–e). Most prominent is the formation of a shoulder at the blue side of the product band (1,695 cm–1) after 16 ns, the onset of which is visible already after 1.7 ns. As this shoulder develops into a well separated band in the 1-ms spectrum, we conclude that equilibration of the system is not completed after 16 ns. On the 1-ms time scale, however, the transient and FTIR difference spectra are identical, indicating that the system is now entirely relaxed.
An additional measure of internal motion of the peptide backbone is obtained from the time evolution of the polarization dependence of the transient signals. The negative part of the signal at 20 ps is only weakly polarization dependent, in agreement with the broad distribution of the orientations of the C O groups in the various cis structures with respect to the electronic transition dipole of the azobenzene unit (Fig. 5). In contrast, the anisotropy α = (ΔA∥–ΔA⊥)/(ΔA∥ + 2ΔA⊥) of the product band at 1,670 cm–1 is initially close to 0 but decreases until 200 ps to α ≈ –0.2 (Fig. 4d). As the anisotropy measures the angle ϕ between pumped and probed transition through α = 1/5(3cosϕ – 1), the negative anisotropy is consistent with the perpendicular orientation of most amide I transition dipoles to the electronic transition dipole of the azobenzene unit, in agreement with the NMR structure for the trans conformation (Fig. 5) (25). In other words, the alignment of the peptide units still changes between 20 and 200 ps. However, it should be noted that the strong overlap of cis and product trans band might overstate the anisotropies. On a longer time scale, rotational diffusion of the entire molecule blurs the effect of internal motion and diminishes the anisotropy after ≈200 ps. The time scale of that anisotropy decay matches the rotational correlation time of 1.5 ns obtained from NMR relaxation experiments (25).
O groups in the various cis structures with respect to the electronic transition dipole of the azobenzene unit (Fig. 5). In contrast, the anisotropy α = (ΔA∥–ΔA⊥)/(ΔA∥ + 2ΔA⊥) of the product band at 1,670 cm–1 is initially close to 0 but decreases until 200 ps to α ≈ –0.2 (Fig. 4d). As the anisotropy measures the angle ϕ between pumped and probed transition through α = 1/5(3cosϕ – 1), the negative anisotropy is consistent with the perpendicular orientation of most amide I transition dipoles to the electronic transition dipole of the azobenzene unit, in agreement with the NMR structure for the trans conformation (Fig. 5) (25). In other words, the alignment of the peptide units still changes between 20 and 200 ps. However, it should be noted that the strong overlap of cis and product trans band might overstate the anisotropies. On a longer time scale, rotational diffusion of the entire molecule blurs the effect of internal motion and diminishes the anisotropy after ≈200 ps. The time scale of that anisotropy decay matches the rotational correlation time of 1.5 ns obtained from NMR relaxation experiments (25).
The dynamics of the equilibration process is governed by a complex hierarchy of time scales. This becomes directly evident from the transient at the quasiisosbestic point at 1,661 cm–1 (Fig. 4c), which is very sensitive to minor shifts of substates of the amide I band induced by the underlying conformational dynamics. These frequency shifts might originate directly from rearrangements of the individual C O groups and/or from the opening and closing of intramolecular hydrogen bonds as a result of such rearrangements (intermolecular hydrogen bonds to the solvent should play a minor role in DMSO). At least five phases can be distinguished during the first 1.7 ns of equilibration. To illustrate the complexity of the response, we attempted a global exponential fit of the cycAMPB data at all probe frequencies and for delay times up to 1.7 ns (the time range for which we were able to measure transients; see Materials and Methods). A reasonable fit is obtained when including five time constants of 2.2 ps, 6.2 ps, 17 ps, 430 ps, and 1 ns for the magic angle signal, whereas somewhat different values are obtained when fitting the data with either parallel or perpendicular polarization of pump and probe pulse. A singular value decomposition of the same data sets, on the other hand, suggests the presence of at least six linearly independent dynamical contributions between 0 and 1.7 ns. The further evolution of the signal between 1.7 and 16 ns, which is not included in this analysis, requires even more kinetic components for a complete temporal description of the conformational dynamics.
O groups and/or from the opening and closing of intramolecular hydrogen bonds as a result of such rearrangements (intermolecular hydrogen bonds to the solvent should play a minor role in DMSO). At least five phases can be distinguished during the first 1.7 ns of equilibration. To illustrate the complexity of the response, we attempted a global exponential fit of the cycAMPB data at all probe frequencies and for delay times up to 1.7 ns (the time range for which we were able to measure transients; see Materials and Methods). A reasonable fit is obtained when including five time constants of 2.2 ps, 6.2 ps, 17 ps, 430 ps, and 1 ns for the magic angle signal, whereas somewhat different values are obtained when fitting the data with either parallel or perpendicular polarization of pump and probe pulse. A singular value decomposition of the same data sets, on the other hand, suggests the presence of at least six linearly independent dynamical contributions between 0 and 1.7 ns. The further evolution of the signal between 1.7 and 16 ns, which is not included in this analysis, requires even more kinetic components for a complete temporal description of the conformational dynamics.
We cannot rule out that part of the complexity of the response is due to the 50% of molecules that do not undergo cis → trans isomerization but relax back to the original cis-conformation after electronic excitation of the azo-compound (i.e., the 50% quantum efficiency of the cis → trans isomerization). However, we expect that the <1-ps lifetime of the electronically excited state of the azoswitch is too short to significantly deform the cis structures so that the excess energy is not converted into directional motion but into heat, which is dissipated within 10 ps.
The dynamics might also be influenced by the use of the relatively slow solvent DMSO. Indeed, experiments have shown that the solvation response of DMSO contains a small (10%) component with a 10-ps time constant (39), significantly slower than water. However, solvation dynamics is still fast enough to not perturb the dynamics of the equilibration process (>20 ps), but it might slow down slightly the driven phase.
Discussion
Combining all experimental observations, the course of the conformational transition can be sketched as shown in Fig. 5. The reaction starts from the cis state of cycAMPB, which is a frustrated system with many close-lying conformational energy minima. The cis → trans isomerization transfers the peptide to a modified potential surface by changing the conformational restraint. The area where the molecular ensemble is launched on the modified surface is determined by the region of the conformation space occupied by the equilibrium ensemble before the arrival of the switch pulse. Starting from the broad distribution of cis structures, the conformational distribution on the modified surface directly after switching is expected to be similarly inhomogeneous with many possible downhill pathways. Furthermore, a good fraction of huge excess energy pumped into the photoswitch (420 nm ≙ 290 kJ/mol!) is certainly expected to remain in the molecule as heat and/or strain within the early phase. Hence, the potential energy surface in the vicinity of the well defined global minimum of the trans conformation might be entirely flooded even if the downhill pathways were homogeneous. In any case, we expect to find a broad distribution of structures after the initial driven phase. In other words, the peptide is forced into an almost stretched configuration at the end of the driven phase but the backbone is not yet in the most favorable conformation and the dihedral angles have yet to adjust to the elongated structure. The initial phase may be imagined as an essentially barrierless process on the steep part of the energy surface, driven by a large amount of excess energy. This is by far the fastest large-scale peptide backbone motion ever observed directly, which exceeds previous results (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11) by two to three orders of magnitudes. The subsequent much slower phase at the bottom of the funnel is diffusion controlled. The potential energy landscape is still biased in this region because the rest of strain is not yet released, but solvent driven conformational fluctuations will now be necessary to escape from traps on the way to the final state.
It is instructive to compare the present results with related experiments by Hochstrasser and coworkers (23), who have investigated a cyclic peptide closed by a disulfide bridge. After breaking the disulfide bridge using a short UV pulse, the liberated molecules either underwent geminate recombination or diffused apart and allowed the peptides to change conformation. However, no significant shift of the amide I band, which would be indicative for a conformational change of the backbone, has been detected during the first 2 ns. In our case, a notable fraction of the energy pumped into the azobenzene unit is converted into a directional force, driving the peptide backbone far away from the new equilibrium conformation within an extremely short time. The subsequent relaxation of the nonequilibrium ensemble hence includes very fast processes.
One would expect a two-state kinetics if the peptides reached the trans state directly by a smooth and steep funnel. In contrast, the observed conformational transition is surprisingly complex. This becomes particularly clear at the quasiisosbestic point at 1,661 cm–1 (Fig. 4c), which emphasizes that relaxation and equilibration of the backbone occurs on multiple time scales. It is important to note that we observe discrete processes with alternating signs of the change of the absorption signal near the quasiisosbestic point, so that the signal oscillates on the logarithmic time scale (used in Fig. 4c). Logarithmic oscillations have been suggested by Jortner and coworkers (40) to be indicative for a discrete hierarchy of relaxation processes on a rugged energy landscape. The more traditional picture of protein dynamics starts from the assumption of a continuum of time scales, leading to a nonexponential but monotonic decay, which commonly is modeled by stretched exponentials or power laws (23, 41–44). The latter response is frequently observed in much larger systems, in particular at low temperatures, and emphasizes the glass-like behavior of proteins (1–4, 5–7). In view of the small size of the peptide studied here, its complex response seems surprising. However, we apply a sharp trigger on a small system and directly probe the backbone dynamics with high time resolution. The amide I band, consisting of in total nine spectroscopic states, can sense very directly even minor rearrangements of the C O groups. Hence, we are able to monitor details of the backbone dynamics that are obscured in larger systems due to averaging over too many spectroscopic states or when using a more indirect, i.e., electronic probe.
O groups. Hence, we are able to monitor details of the backbone dynamics that are obscured in larger systems due to averaging over too many spectroscopic states or when using a more indirect, i.e., electronic probe.
Protein folding is an inherently very difficult problem. Microscopic all-atom molecular dynamics simulation, albeit being a powerful tool, encounters great difficulties to reach the time scales of protein folding with present computer power (8). Therefore, simplifying models have been proposed, which try to describe protein folding with kinetic models using a more macroscopic description of the molecule (4–6). Depending on whether one is interested in secondary or tertiary structure, macroscopic states could be, for example, either native or nonnative conformations of each peptide unit (4, 5, 45) or contacts between whole α-helical strands, which themselves are treated as building blocks (46). The number of degrees of freedom needed to describe the state of the system is thereby reduced dramatically. It is hoped that dynamics of the system can be understood without having to consider a microscopic level. However, these approaches intrinsically assume a separation of time scales of the fast internal motion within each macroscopic state and the much slower kinetic transitions between them. The peptide studied here is too small to form any distinct secondary structure. Nevertheless, the dynamics of this small fragment shows a series of processes overlapping in time, which extend well into the »10-ns range, much longer than individual steps in the formation of, for example, an α-helix, which have been proposed to occur within <1 ns (5–7, 9, 14). In other words, equilibration of a small peptide fragment can be considerably slower than kinetic transitions in simplified, macroscopic protein folding models (4–6, 45, 46). In view of this result, it seems questionable whether a separation of time scales is possible that would allow the discussion of protein dynamics on certain length scales without having to consider a microscopic atomic level (43).
Conclusion
We have studied the backbone response of a small cyclic peptide after cis→trans isomerization of a built-in photoswitch applying ultrafast vibrational spectroscopy. The experiments show that important conformational changes of peptide fragments can be extremely fast (i.e., a few picoseconds) when a sufficiently strong force is driving them. The subsequent relaxation of the peptide ensemble far away from equilibrium is governed by a discrete hierarchy of time scales, extending from ≈20 ps to >16 ns. This finding indicates that the potential energy surface, on which the trans ensemble equilibrates, is rugged and contains hierarchical energy barriers (40). The system studied here is small enough to allow resolving many of the entangled processes involved in its conformational transition directly by ultrafast vibrational spectroscopy all the way from the start to the very end. Furthermore, the system is still within the scope of thorough all-atom molecular dynamics simulation (26), which can be connected directly to the experiment and at the same time have the capability to reveal a microscopic picture of the underlying processes. Nevertheless, albeit possessing only a few conformational degrees of freedom compared with a protein, the peptide behaves highly nontrivially and provides insights into the complexity of protein dynamics on short time scales and small length scales.
Acknowledgments
The work was partially supported by the Swiss Science Foundation and the Deutsche Forschungsgemeinschaft (SFB 533 A8 and B8).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FTIR, Fourier transform IR; AMPB, (4-aminomethyl)phenylazobenzoic acid; cycAMPB, cyclized peptide with AMPB; linAMPB, linear precursor molecule of cycAMPB.
References
- 1.Ballew, R. M., Sabelko, J. & Gruebele, M. (1996) Nat. Struct. Biol. 3, 923–926. [DOI] [PubMed] [Google Scholar]
- 2.Williams, S., Causgrove, T. P., Gilmanshin, R., Fang, K. S., Callender, R. H., Woodruff, W. H. & Dyer, R. B. (1996) Biochemistry 35, 691–697. [DOI] [PubMed] [Google Scholar]
- 3.Gilmanshin, R., Williams, S., Callender, R. H., Woodruff, W. H. & Dyer, R. B. (1997) Proc. Natl. Acad. Sci. USA 94, 3709–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muñoz, V., Thompson, P. A., Hofrichter, J. & Eaton, W. A. (1997) Nature 390, 196–199. [DOI] [PubMed] [Google Scholar]
- 5.Thompson, P. A., Muñoz, V., Jas, G. S., Henry, E. R., Eaton, W. A. & Hofrichter, J. (2000) J. Phys. Chem. B 104, 378–389. [Google Scholar]
- 6.Werner, J. H., Dyer, R. B., Fesinmeyer, R. M. & Andersen, N. H. (2002) J. Phys. Chem. B 106, 487–494. [Google Scholar]
- 7.Huang, C.-Y., Getahun, Z., Zhu, Y., Klemke, J. W., DeGrado, W. F. & Gai, F. (2002) Proc. Natl. Acad. Sci. USA 99, 2788–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, Y. & Kollman, P. A. (1998) Science 282, 740–744. [DOI] [PubMed] [Google Scholar]
- 9.Daura, X., Jaun, B., Seebach, D., van Gunsteren, W. F. & Mark, A. E. (1998) J. Mol. Biol. 280, 925–932. [DOI] [PubMed] [Google Scholar]
- 10.Zhou, Y. & Karplus, M. (1999) Nature 401, 400–402. [DOI] [PubMed] [Google Scholar]
- 11.Bieri, O., Wirz, J., Hellrung, B., Schutkowski, M., Drewello, M. & Kiefhaber, T. (1999) Proc. Natl. Acad. Sci. USA 96, 9597–9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leeson, D. T., Gai, F., Rodriguez, H. M., Gregoret, L. M. & Dyer, R. B. (2000) Proc. Natl. Acad. Sci. USA 97, 2527–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazaridis, T. & Karplus, M. (1997) Science 278, 1928–1930. [DOI] [PubMed] [Google Scholar]
- 14.Hummer, G., Garcia, A. E. & Garde, S. (2000) Phys. Rev. Lett. 85, 2637–2640. [DOI] [PubMed] [Google Scholar]
- 15.Hummer, G., Garcia, A. E. & Garde, S. (2001) Proteins 42, 77–84. [PubMed] [Google Scholar]
- 16.Daura, X., van Gunsteren, W. F. & Mark, A. E. (1999) Proteins 34, 269–280. [DOI] [PubMed] [Google Scholar]
- 17.Woutersen, S., Mu, Y., Stock, G. & Hamm, P. (2001) Proc. Natl. Acad. Sci. USA 98, 11254–11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volk, M. (2001) Eur. J. Org. Chem. 2605–2621.
- 19.Gruebele, M. (1999) Annu. Rev. Phys. Chem. 50, 485–516. [DOI] [PubMed] [Google Scholar]
- 20.Callender, R. H., Dyer, R. B., Gilmanshin, R. & Woodruff, W. H. (1998) Annu. Rev. Phys. Chem. 49, 173–202. [DOI] [PubMed] [Google Scholar]
- 21.Eaton, W. A., Muñoz, V., Hagen, S. J., Jas, G. S., Lapidus, L. J., Henry, E. R. & Hofrichter, J. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 327–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips, C. M., Mizutani, Y. & Hochstrasser, R. M. (1995) Proc. Natl. Acad. Sci. USA 92, 7292–7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volk, M., Kholodenko, Y., Lu, H. S. M., Gooding, E. A., De Grado, W. F. & Hochstrasser, R. M. (1997) J. Phys. Chem. B 101, 8607–8616. [Google Scholar]
- 24.Behrendt, R., Renner, C., Schenk, M., Wang, F. Q., Wachtveitl, J., Oesterhelt, D. & Moroder, L. (1999) Angew. Chem. Int. Ed. 38, 2771–2774. [DOI] [PubMed] [Google Scholar]
- 25.Renner, C., Cramer, J., Behrendt, R. & Moroder, L. (2000) Biopolymers 54, 501–514. [DOI] [PubMed] [Google Scholar]
- 26.Spörlein, S., Carstens, H., Renner, H. S. C., Behrendt, R., Moroder, L., Tavan, P., Zinth, W. & Wachtveitl, J. (2002) Proc. Natl. Acad. Sci. USA 99, 7998–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nägele, T., Hoche, R., Zinth, W. & Wachtveitl, J. (1997) Chem. Phys. Lett. 252, 489–495. [Google Scholar]
- 28.Renner, C., Behrendt, R., Spörlein, S., Wachtveitl, J. & Moroder, L. (2000) Biopolymers 54, 489–500. [DOI] [PubMed] [Google Scholar]
- 29.Wachtveitl, J., Spörlein, S., Fonrobert, B., Renner, C., Behrendt, R., Moroder, L. & Zinth, W. (2003) in Ultrafast Processes in Spectroscopy XII, eds. Califano, S., Foggi, P. & Righini, R. (Olschki, Florence, Italy), in press.
- 30.Ulysse, L., Cubillos, J. & Chmielewski, J. (1995) J. Am. Chem. Soc. 117, 8466–8467. [Google Scholar]
- 31.Kumita, J. R., Smart, O. S. & Wooley, G. A. (2000) Proc. Natl. Acad. Sci. USA 97, 3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krimm, S. & Bandekar, J. (1986) Adv. Protein Chem. 38, 181–364. [DOI] [PubMed] [Google Scholar]
- 33.Hamm, P., Kaindl, R. A. & Stenger, J. (2000) Opt. Lett. 25, 1798–1800. [DOI] [PubMed] [Google Scholar]
- 34.Surewicz, W. K. & Mantsch, H. H. (1993) Biochemistry 32, 389–394. [DOI] [PubMed] [Google Scholar]
- 35.Torii, H. & Tasumi, M. (1992) J. Chem. Phys. 96, 3379–3387. [Google Scholar]
- 36.Torii, H. & Tasumi, M. (1998) J. Raman Spectrosc. 29, 81–86. [Google Scholar]
- 37.Hamm, P. & Woutersen, S. (2002) Bull. Chem. Soc. Jpn. 75, 985–988. [Google Scholar]
- 38.Hamm, P., Ohline, S. M. & Zinth, W. (1997) J. Chem. Phys. 106, 519–529. [Google Scholar]
- 39.Horng, M. L., Gardecki, J. A., Papazyan, A. & Maroncelli, M. (1995) J. Phys. Chem. 99, 17311–17337. [Google Scholar]
- 40.Metzler, R., Klafter, J. & Jortner, J. (1999) Proc. Natl. Acad. Sci. USA 96, 11085–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. (1991) Science 254, 1598–1603. [DOI] [PubMed] [Google Scholar]
- 42.Sabelko, J., Ervin, J. & Gruebele, M. (1999) Proc. Natl. Acad. Sci. USA 96, 6031–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karplus, M. (2000) J. Phys. Chem. B 104, 11–27. [Google Scholar]
- 44.Metzler, R., Klafter, J., Jortner, J. & Volk, M. (1998) Chem. Phys. Lett. 293, 477–484. [Google Scholar]
- 45.Young, W. S. & Brooks, C. L., III (1996) J. Mol. Biol. 259, 560–572. [DOI] [PubMed] [Google Scholar]
- 46.Karplus, M. & Weaver, D. L. (1993) Protein Sci. 3, 650–668. [DOI] [PMC free article] [PubMed] [Google Scholar]