Abstract
Spermatogonial stem cells (SSCs) are responsible for maintaining spermatogenesis throughout life in the male by continuous production of daughter cells that differentiate into spermatozoa. However, no unique phenotypic markers to identify SSCs have been described. In this study, the SSC surface phenotype was characterized by using flow cytometric cell sorting in conjunction with a transplantation functional assay for SSCs. Highly enriched stem cell activity was found in the MHC class I (MHC-I)–Thy-1+c-kit– cell fraction of the mouse cryptorchid testis. There was little or no stem cell activity in any other fraction. The antigenic phenotype of the MHC-I–Thy-1+c-kit– SSCs was α6-integrin+CD24+αvintegrin–Sca-1–CD34–. Subsequently, testis side population (SP) cells, which are defined by a Hoechst dye efflux assay, were identified. Their surface phenotype was found to be MHC-I+Thy-1–Sca-1+, and the transplantation assay demonstrated that the testis SP and SSCs are distinct populations. In several other tissues, the SP has been shown to contain stem cells, but we found that this characteristic does not define SSCs. The identification of a surface phenotype that allows production of a highly enriched SSC population will facilitate functional and genomic studies and enable further comparison with other stem cells.
Spermatogenesis is a highly productive and tightly regulated process. Spermatogonial stem cells (SSCs) are responsible for maintaining spermatogenesis throughout life in males by continuously producing type A spermatogonia that are committed to differentiate into spermatozoa, which transmit genes to subsequent generations. This characteristic of SSCs relies on their ability to self-renew as well as to produce daughter cells that differentiate. Although normal spermatogenesis depends on strict control of the self-renewal and differentiation decision of SSCs, the mechanism of this regulation has remained elusive. Progress in understanding the mechanism involved is hampered largely because the concentration of SSCs in the testis is extremely low, presumably as few as 1 in 3,000–4,000 cells in adult mouse testis (1), and no unique phenotypic markers to identify and isolate SSCs have been described.
Stem cells of all types are defined by their biological function. Therefore, identification depends on the availability of a functional assay, which usually entails transplantation of putative stem cells and assessment of their ability to generate the differentiated tissue. Unequivocal identification of SSCs is possible by transplanting stem cell populations into the seminiferous tubules of a recipient testis, in which only a SSC can generate a colony of donor cell-derived spermatogenesis (2, 3, 4). With the transplantation assay as a functional endpoint, enrichment of murine SSCs by multiparametric separation using fluorescence-activated cell sorting (FACS) and monoclonal antibodies for specific surface markers has been developed (5). Although SSCs were enriched significantly in the side scatterlo, α6-integrin+, and c-kit–/lo or αv-integrin–/lo cell population of experimental cryptorchid testes, a single distinctive subpopulation exhibiting SSC activity was not identified. Stem cell concentration in the most enriched fraction was ≈1 in 30–40 cells. Thus, additional phenotypic characteristics of SSCs or a different strategy for effective separation is required to identify and isolate pure SSCs.
Development of a functional assay for hematopoietic stem cells (HSCs) in the early 1960s (6) greatly facilitated the study of stem cell biology in this tissue. Using FACS separation strategies and specific markers, it has been possible to isolate nearly pure HSCs from several sources (bone marrow or fetal liver). The antigenic phenotype of murine HSCs in bone marrow was demonstrated to be c-kit+Thy-1+Sca-1+Lin–/lo (7, 8). In addition, mouse HSCs can be isolated as a side population (SP) of cells on the basis of their ability to efflux rapidly a fluorescent DNA-binding dye, Hoechst 33342 (9). SP cells are identified readily by using dual-wavelength flow cytometric analysis of bone marrow cells stained with the Hoechst dye, and they represent 0.05–0.1% of nucleated bone marrow cells (10). The antigenic phenotype of the SP cells is c-kit+Sca-1+Lin–/lo, strongly suggesting that they are HSCs. Indeed, transplantation assays demonstrated that SP cells in bone marrow have long-term reconstituting ability and are HSCs (9). In addition, SP cells have been identified in several nonhematopoietic tissues (e.g., skeletal muscle, brain, and mammary gland), and studies suggest that SP cells represent stem cells in the tissues from which they were isolated (11–14). Moreover, the developmental potential of SP cells could be broader than expected, because SP cells from skeletal muscle apparently can give rise to hematopoietic cells as well as myogenic cells (11, 12). However, it is not clear whether SP cells possess complete stem cell activity, except for HSCs, because a definitive transplantation functional assay is available only for HSCs and SSCs. Although there have been a number of studies on SP cells from bone marrow and other tissues, the existence and possible function of such cells have not been determined for spermatogenic tissue.
In the present study, a combination of surface markers and FACS was used to identify SSCs as a distinctive subpopulation in adult testis. Subsequently, SP cells in the testis were identified, and their surface antigenic phenotype characterized. It was clearly established that the SP cells and SSCs are not characterized by the same surface markers, and SP cells of the testis do not generate colonies of spermatogenesis when transplanted to the testis of a recipient mouse. Therefore, testis SP cells are not SSCs. Our data show that stem cells in the testis are defined by the surface phenotype MHC class I (MHC-I)–Thy-1+c-kit–.
Materials and Methods
Donor Mice and Cell Collection. Donor testis cells were obtained from the transgenic mouse line B6.129S7-Gtrosa26 (Rosa mice; The Jackson Laboratory) that express the Escherichia coli LacZ (LacZ) gene in all cells of the seminiferous tubules of testis (15). LacZ-expressing cells can be stained blue after incubation with the substrate 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal). Experimental cryptorchid testes were produced as described (16). Single-cell suspensions from cryptorchid testes were prepared by enzymatic digestion (17) and subjected to FACS analysis (5). For details of FACS analysis, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org. Bone marrow cells were obtained from the femurs and tibias of the transgenic mouse line C57BL/6-TgN(ACTbHGFP)1Osb (The Jackson Laboratory) that express a GFP reporter gene under the control of the chicken β-actin promoter and cytomegalovirus immediate early enhancer. GFP is expressed in most cells of this mouse (18). For details of bone marrow cell transplantation and recipient analysis, see Supporting Materials and Methods.
Testis Cell Transplantation and Analysis of Recipient Testes. Donor testis cells were resuspended in 60 μl of F10-S (see Supporting Materials and Methods) and transplanted into the testes of immunologically compatible C57BL/6 × 129/SvCP F1 hybrid recipient male mice treated with busulfan (55 mg/kg) 4–6 weeks before use (2, 3). Approximately 10 μl of donor cell suspension was introduced into the seminiferous tubules of each recipient testis, resulting in 70–80% filling of the tubules (17).
Recipient testes were collected 2 mo after donor cell transplantation and analyzed by X-Gal staining (19). Appearance of individual blue-stained stretches of seminiferous tubules in recipient testes represents colonization and spermatogenesis from donor-derived stem cells. Each colony represents expansion of a single stem cell (19, 20). Colony number was counted by using a dissecting microscope. Because the number of cells that could be recovered and injected in each experiment varied, colony number was normalized to 105 cells injected. Statistical analyses were performed by using ANOVA and significant differences between means determined by using Scheffé's test.
Results
SSCs Are Enriched in the MHC-I– Cell Population of Cryptorchid Mouse Testes. About 95% of cells in wild-type mouse testes are differentiating germ cells, greatly complicating any attempt to identify and characterize the rare stem cell population. To remove these differentiating germ cells, testes for the experiments described here were obtained from cryptorchid mice, which are devoid of differentiated germ cells and have a concentration of stem cells that is 20–25 times higher than that in wild-type mice (16). The cell population in the testes of cryptorchid mice consists primarily of undifferentiated spermatogonia and somatic cells, such as Sertoli, myoid, or Leydig cells (21), and these somatic cells represent as many as 95% of total cells (1, 22). Although MHC-I molecules are expressed on almost all nucleated cells in adults, spermatogonia are believed to be an exception and to express no MHC-I molecules, at least at the protein level (23). Therefore, MHC-I molecules might be absent on the cell surface of SSCs. If somatic cells in the cryptorchid testis could be eliminated on the basis of MHC-I expression on their surface, it would leave a population of undifferentiated spermatogonia representing ≈5% of total cells in cryptorchid testis and 0.25% (5% × 5%) of total cells in wild-type testis.
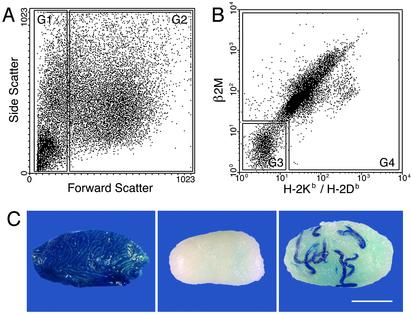
MHC-I molecules are noncovalently linked heterodimers consisting of a polymorphic MHC-I heavy chain and a nonpolymorphic β2 microglobulin (β2M) light chain. Although β2M is the common light chain, there are two groups of MHC-I heavy chain genes, highly polymorphic classical MHC-I (MHC-Ia) and less polymorphic nonclassical MHC-I (MHC-Ib). MHC-Ia is encoded by the K and D/L regions of the H-2 complex of the mouse and expressed ubiquitously. The mouse strain used in this study expresses H-2Kb and H-2Db. Expression of MHC-Ib molecules is variable in tissue distribution, and certain MHC-Ia– cells express MHC-Ib molecules (23, 24). To examine the expression of MHC-Ia and MHC-Ib on SSCs, a cell suspension from cryptorchid testes was stained with two monoclonal antibodies, one that recognizes β2M and another that recognizes MHC-Ia (H-2Kb and H-2Db), and subjected to FACS analysis. To facilitate analyses, forward scatter, which represents relative size of the objects, was used as an initial gating parameter (Fig. 1A). With this strategy, a preliminary gate (G) was established that contained predominately cellular debris (Fig. 1 A, G1). The remaining cells (Fig. 1 A, G2) could then be analyzed by using antibodies to β2M and H-2Kb/H-2Db (Fig. 1B). The gate containing cells with low or no β2M or H-2Kb/H-2Db (Fig. 1B, G3) surface antigens represented ≈14% of cells in G2 (Fig. 1A), whereas most of the remaining cells express both chains of MHC-I molecules (Fig. 1B, G4). To determine the location of SSCs, cells from each gated population (Fig. 1 A and B, G1, G3, and G4) were transplanted into the seminiferous tubules of busulfan-treated recipient mice. Testes in recipient mice were stained with X-Gal 2 mo after transplantation, and colonies of spermatogenesis in the testes were counted (Fig. 1C). Stem cell activity was detected almost exclusively in the MHC-I– cells (Fig. 1B, G3), whereas MHC-I+ cells (Fig. 1B, G4) and forward-scatterlo cells (Fig. 1 A, G1) showed almost no stem cell activity (Fig. 1 legend). Furthermore, the MHC-I– cell population contains ≈6-fold (97/16) more stem cells than unsorted control cells (P = 0.001) and produces ≈100 colonies of spermatogenesis per 105 cells transplanted (Fig. 1 legend). These results indicate that SSCs express neither MHC-Ia nor -Ib at significant levels. In the following experiments, β2M antibody was used to identify the MHC-I– cell population.
Fig. 1.
Flow cytometric analysis of cryptorchid testis cells and appearance of donor and recipient testes. (A) Gates for FACS using forward scatter. G1 is the forward-scatterlo fraction, which contains predominantly cellular debris (≈30% of objects). G2 contains the viable cells (≈70% of objects). (B) Expression pattern of MHC-I molecules in G2 of A. Cells in G3 have negative to low expression of both β2M and H-2Kb/H-2Db. G3 and G4 contain ≈14% and 86%, respectively, of cells in G2. (C) Macroscopic appearance of Rosa cryptorchid donor testis (Left), busulfan-treated testis (Center), and recipient testis (Right) transplanted with MHC-I– donor testis cells (G3) isolated by FACS. Each blue-stained stretch of cells in the recipient testis represents a colony of spermatogenesis that arises from a single stem cell (19, 20). Stain, X-Gal. (Bar = 2 mm.) The number of spermatogenic colonies generated by 105 cells transplanted to recipient testes was: G1 = 0, n = 11; G3 = 97 ± 24, n = 12; G4 = 0.4 ± 0.4, n = 12; unsorted cells = 16 ± 4.4, n = 12 (mean ± SEM).
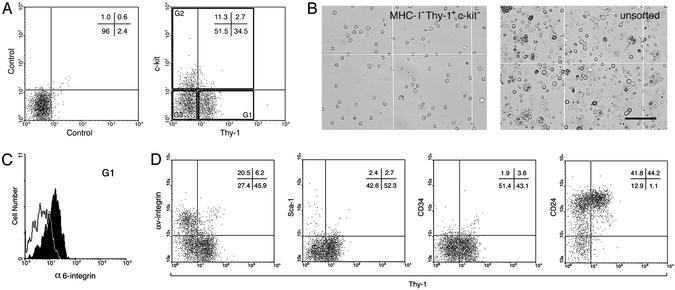
Thy-1 Is Expressed on SSCs in Cryptorchid Testes. Recent studies suggest that stem cells or primitive progenitors from various tissues in adults share several phenotypic characteristics (9–13). Thy-1 (CD90) is a glycosyl phosphatidylinositol anchored glycoprotein of the Ig superfamily, which is expressed on HSCs (7, 25, 26). In addition, Thy-1 can be detected on pluripotent stem cells in culture, such as embryonic stem (ES) cells or bone marrow-derived multipotent cells (27, 28). The c-kit tyrosine kinase receptor is expressed on HSCs, ES cells, and primordial germ cells (8, 27, 29). However, FACS analyses and transplantation studies indicate that SSC activity is enriched in c-kit– and c-kitlo testis cell populations (5). Therefore, the expression of Thy-1 and c-kit were analyzed in the MHC-I– population of cryptorchid testes by FACS (Fig. 2A). Approximately 35% of MHC-I– cells were Thy-1+c-kit–, and ≈14% (11.3 + 2.7) were c-kit+. Interestingly, c-kit+ cells were primarily Thy-1–. To determine the number of SSCs in each cell population, Thy-1+c-kit–, c-kit+, and Thy-1–c-kit– (Fig. 2 A, G1–G3, respectively) were collected by flow cytometric sorting, and each population was transplanted to seminiferous tubules of recipient mice. SSC activity was detected almost exclusively in the Thy-1+c-kit– cell population (Fig. 2 A, G1), which generated ≈340 colonies of spermatogenesis from 105 cells transplanted (Fig. 2 legend). Compared with unsorted cells from cryptorchid testes, the concentration of stem cells in the MHC-I–Thy-1+c-kit– was ≈25-fold (343/14) enriched (P < 0.001). The MHC-I–Thy-1+c-kit– cell population sorted by FACS consisted of morphologically uniform cells (Fig. 2B). Because SSCs are known to express α6-integrin, in this experiment, the cryptorchid testis cells were stained simultaneously with monoclonal antibodies for β2M, Thy-1, c-kit, and α6-integrin to assess overlap in expression patterns. The stem cell-rich population (Thy-1+c-kit–) indeed was α6-integrin+ (Fig. 2C), and these cells also had characteristic low side scatter (data not shown), consistent with previous results (5). MHC-I–c-kit+ cells expressed α6-integrin slightly, whereas MHC-I–Thy-1–c-kit– cells express no α6-integrin (data not shown). In addition, we confirmed that MHC-I–Thy-1+ cells express little αv-integrin (Fig. 2D). Taken together, these results demonstrate that the antigenic phenotype of SSCs in the mouse cryptorchid testis is restricted to the MHC-I–Thy-1+c-kit– phenotype. A similar expression profile also can be demonstrated for SSCs in wild-type and pup (5–9 days old) mouse testes (data not shown). Subsequently, we used testis cells from cryptorchid mice to examine the expression of Sca-1, CD34, and heat-stable antigen (CD24), which are antigenic markers used for identification of HSCs or neural stem cells (7, 8, 26, 31), and found that MHC-I–Thy-1+ cells express CD24 but not Sca-1 or CD34 (Fig. 2D). These results are summarized in Table 1.
Fig. 2.
Flow cytometric analysis of the MHC-I– (β2M–) cell population of cryptorchid testis cells. (A) Unstained control (Left) and staining profile of Thy-1 vs. c-kit receptor (Right) for MHC-I– cells are shown with quadrant statistics. For transplantation assay, three populations (G1–G3) were isolated by FACS. G1–G3 represent Thy-1+c-kit–, c-kit+, and Thy-1–c-kit–, respectively. The number of spermatogenic colonies generated by 105 cells transplanted into recipient testes was: G1 = 343 ± 46, n = 17; G2 = 4.1 ± 1.7, n = 18; G3 = 2.7 ± 1.3, n = 18; unsorted cells = 14 ± 2.1, n = 18 (mean ± SEM). Because colonization efficiency is ≈5% (5, 30), ≈1 in 15 cells in G1 are SSCs [105 transplanted cells/(343 colonies/5% efficiency)]. (B) MHC-I–Thy-1+c-kit– (G1) cells sorted by FACS (Left) and unsorted cryptorchid testis cells (Right) are shown. G1 cells were relatively uniform, whereas unsorted cells were highly heterogeneous. Note characteristic pseudopods on some G1 cells. (Bar = 100 μm.) (C) α6-integrin expression was analyzed. MHC-I–Thy-1+c-kit– (G1) cells were α6-integrin+. Solid line indicates unstained cells. (D) Staining profiles of Thy-1 vs. surface molecules on MHC-I– cells are shown with quadrant statistics. MHC-I–Thy-1+ cells express CD24, but not αv-integrin, Sca-1, or CD34.
Table 1. Surface antigens present on stem cells of the mouse.
| Antigen | SSC | HSC | NSC |
|---|---|---|---|
| Thy-1 | + | + | - |
| α6-integrin | + | + | ND |
| CD24 | + | + | -/lo |
| c-kit | - | + | - |
| Sca-1 | - | + | ND |
| CD34 | - | - | - |
| MHC-1 | - | + | ND |
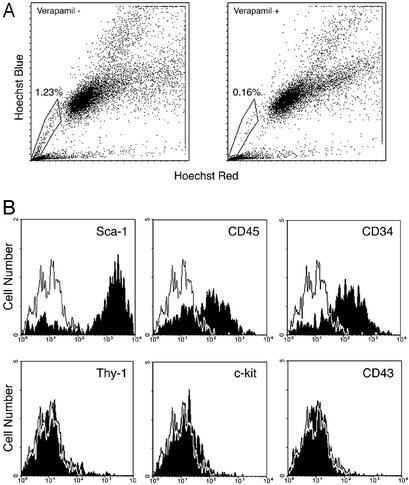
Characterization of a Testis SP Identified with Hoechst Staining. We next determined whether there is a SP in mouse testis, because it has been proposed that the SP phenotype is conserved in stem cells of different tissue origins (10–14). To identify SP cells in mouse testes, freshly isolated cell suspensions from cryptorchid testes were incubated with Hoechst 33342 for 90 min in the presence or absence of verapamil and separated by FACS. Verapamil is able to block the efflux pump for Hoechst dye exclusion and thereby to decrease the number of SP cells (9). A testis SP was identified (Fig. 3A, box), and the SP cell number was reduced from ≈1.2% to 0.2% in the presence of verapamil, indicating that the testis SP cells are verapamil-sensitive. In addition to a bone marrow SP, a skeletal muscle SP also has been characterized and shown to contain multipotent stem cells (11 12). Analyses of the antigenic phenotypes of bone marrow SP and muscle SP indicate that Sca-1 expression is a common characteristic. Therefore, we analyzed the surface phenotype of the testis SP. Consistent with results from bone marrow and muscle, testis SP cells expressed Sca-1 brightly (Fig. 3B). In addition, testis SP cells displayed very low or no expression of Thy-1, c-kit, and CD43, whereas the expression of CD45 and CD34 was heterogeneous (Fig. 3B). The antigenic profiles of testis SP, bone marrow SP, and muscle SP are summarized in Table 2.
Fig. 3.
Flow cytometric analysis of cryptorchid testis cells stained with Hoechst 33342. (A) Testis SP cells, defined by verapamil sensitivity, are indicated in the box. Shown is Hoechst 33342 staining and emission patterns of whole cryptorchid testis cells in the absence (Left) and presence (Right) of verapamil. (B) Expression of surface markers on the testis SP was analyzed. Testis SP cells express Sca-1 brightly; CD45 and CD34 heterogeneously; and Thy-1, c-kit, and CD43 at negligible levels. Solid line indicates iso-type control antibody stained cells.
Table 2. Surface antigens present on Hoechst-stained SP cells.
| Antigen | Testis SP | BM SP | SM SP |
|---|---|---|---|
| Sca-1 | + | + | + |
| Thy-1 | - | -/IO | ND |
| c-kit | - | + | - |
| CD34 | +/- | -/IO | ND |
| CD43 | - | + | - |
| CD45 | +/- | + | +/- |
| MHC-I | + | + | ND |
The expression of CD45, a unique marker for hematopoietic cells, in testis SP cells prompted us to determine whether the testis SP arises from hematopoietic cells circulating in the blood vessels of the testis. Therefore, bone marrow cells from mice expressing GFP in hematopoietic cells were transplanted into the jugular vein of newborn W54/WV mice. The genetic defect in the c-kit receptor of W54/WV mice allows donor HSCs to colonize host bone marrow without irradiation of the recipient mouse (34). Eight to nine months after bone marrow transplantation, recipient mice were killed, and the testes were analyzed for the presence of donor-derived hematopoietic cells in testis SP cells (Fig. 5, which is published as supporting information on the PNAS web site). FACS analysis showed that 50–60% of CD45+ cells in the spleen and bone marrow were GFP+ cells (Fig. 5A), indicating that donor bone marrow cells were engrafted successfully in the W recipient mice. Testis cells from the same recipients were stained with Hoechst dye followed by CD45 staining, and FACS analysis indicated that the percentage (1.2%) of the cells in the SP of W testes was similar to that in cryptorchid testes (Fig. 5B). However, GFP+ cells were negligible (<5%) in the testis SP of W recipients, whereas >70% of bone marrow SP cells in the same recipient mouse were GFP+ (Fig. 5C). Because the testis SP in GFP mice showed a strong GFP signal (data not shown), failure to detect donor cells in the recipient testis SP is not due to the absence of GFP expression in the SP cells. These results indicate that there is little, if any, contribution of bone marrow cells to testis SP cells, even though the latter express CD45.
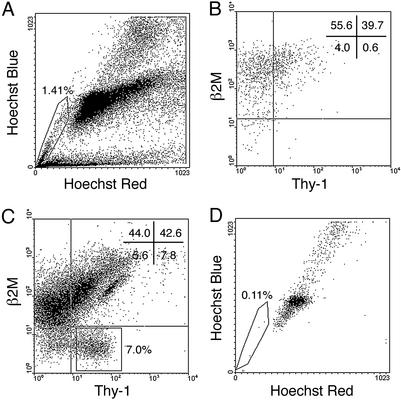
SSCs Are Not Enriched in the Testis SP. SSCs are MHC-I–Thy-1+Sca-1–, whereas testis SP cells are Thy-1–Sca-1+ and, therefore, distinct cell populations (Tables 1 and 2). FACS analysis, indeed, demonstrated unambiguously that there was no overlap between MHC-I–Thy-1+ cells and testis SP cells (Fig. 4). Testis SP cells expressed β2M, indicating that they are MHC-I+. Although MHC-I+ cells showed little stem cell activity (Fig. 1 legend), there was a possibility that SP cells are rare in the large number of MHC-I+ cells and, therefore, SSC activity was not detected in the transplantation assay results. To address directly the question whether the testis SP has SSC potential, flow cytometric-sorted SP cells and MHC-I–Thy-1+ cells from testis cells treated with Hoechst dye were transplanted into seminiferous tubules of busulfan-treated mice. Two months after transplantation, experimental testes were stained with X-Gal and colonies of spermatogenesis were counted. In this experiment, the number of spermatogenic colonies was reduced by toxicity of the Hoechst dye. A deleterious effect of Hoechst dye also has been noted for hematopoietic and neuronal cells (14, 35). Unsorted testis cells generated fewer colonies (compare values in Fig. 4 legend with those in Figs. 1 and 2 legends), and the viability of unsorted cells was only 65 ± 1.2% (mean ± SEM, n = 3) in Hoechst experiments compared with 80 ± 1.3% (mean ± SEM, n = 5) in other experiments. However, the viability of sorted testis SP cells was constantly high, 96 ± 0.3% (mean ± SEM, n = 3), presumably due to their dye efflux ability. Nonetheless, transplantation results demonstrated that the testis SP does not have stem cell activity, whereas MHC-I–Thy-1+ cells generated numerous colonies of spermatogenesis (Fig. 4 legend), as observed in previous experiments. Clearly, the testis SP does not contain SSCs.
Fig. 4.
Flow cytometric analysis of cryptorchid testis cells stained with Hoechst 33342 followed by antibody staining for β2M and Thy-1. (A) Hoechst staining pattern of cryptorchid testis cells. Testis SP cells are enclosed in box. The testis SP disappeared in the presence of verapamil (data not shown). (B) β2M vs. Thy-1 expression profile of the testis SP gated in A, with quadrant statistics. There are few β2M–Thy-1+ cells in the testis SP cell fraction. (C) Staining profile of Thy-1 vs. β2M for cryptorchid testis cells. β2M–Thy-1+ cells (≈7%) are gated in the rectangle. (D) Hoechst emission profile of β2M–Thy-1+ cells gated in C is shown. SP cells and β2M–Thy-1+SP– cells were sorted simultaneously for transplantation assay. The number of spermatogenic colonies generated by 105 cells transplanted to recipients was: SP in A = 0, n = 18; β2M–Thy-1+ in C = 76 ± 15; unsorted cells = 4.7 ± 0.7, n = 18 (mean ± SEM).
Discussion
In this study, we describe properties of SSCs with respect to surface phenotype and Hoechst dye efflux activity, both of which are used commonly to identify and characterize stem cells from various tissues. A transplantation functional assay for SSCs detected highly enriched stem cell activity in the MHC-I–Thy-1+c-kit– cell fraction of the mouse cryptorchid testis. This cell population does not contain Sca-1+, CD34+, or αv-integrin+ cells but does express CD24 uniformly. In addition, the MHC-I–Thy-1+c-kit– cells are side scatterlo and α6-integrin+, which have been demonstrated to be SSC characteristics in previous studies (5). Because there is little stem cell activity in any other fraction (MHC-I+, MHC-I–Thy-1–c-kit–, or MHC-I–c-kit+), the results suggest that MHC-I–Thy-1+c-kit– cells are the only SSCs in the testis. In addition to identifying a unique testis cell population (MHC-I–Thy-1+c-kit–) that contained essentially all of the SSCs, we found that the population had a stem cell concentration of ≈1 in 15 (Fig. 2 legend).
Mouse bone marrow cells stained with Hoechst dye and separated by FACS on the basis of fluorescence in both far red and blue emission channels generate a group of cells designated SP, which represents 0.05–0.1% of the bone marrow cells (9, 10). Because it was shown that SP cells are enriched for HSC activity in various species (10), the protocol was established as a new method to identify HSCs. Visualization of the SP with flow cytometric analysis is based on the efflux of Hoechst dye and does not require cell surface staining with antibodies, which makes the method versatile (9, 10). In addition to identifying HSCs, recent studies suggested that the technique could be used to identify putative stem cells from various tissue sources, including skeletal muscle, brain, and mammary gland (11–14). These findings suggest a potential universal method to identify stem cell populations with the SP protocol. Therefore, whether Hoechst staining of testis cells identifies a SP, and if so, whether the testis SP contains SSCs are very important questions in stem cell biology. SSCs represent a particularly valuable cell population in which to test whether the SP universally contains stem cells, because of the unequivocal identification of function provided by the transplantation assay. In this study, Hoechst staining of a testis cell suspension clearly identified a SP, which represents ≈1.2–1.4% of cryptorchid testis cells and 0.1–0.2% of normal wild-type testis cells (unpublished data). The surface phenotype of the testis SP is Sca-1+Thy-1–c-kit–CD34+/– CD43–CD45+/–MHC-I+. This antigenic profile is similar to that found for the SP in bone marrow or skeletal muscle (Table 2). However, although >70% of testis SP cells express a hematopoietic marker, CD45, bone marrow transplantation experiments clearly demonstrated that testis SP cells are not contaminating cells from circulating blood cells (Fig. 5), suggesting that the testis SP comprises resident cells in this tissue. Moreover, FACS analysis indicated that the surface phenotype of the testis SP does not overlap MHC-I–Thy-1+c-kit– SSCs. In agreement with the phenotypic difference of these two populations, the functional assay for SSCs demonstrated that the testis SP does not contain SSCs. Taken together, our data clearly demonstrate that the antigenic phenotype of SSCs in the mouse testis is Thy-1+α6-integrin+CD24+MHC-I–c-kit–αv-integrin–Sca-1–CD34–, and that SSCs are not found in the testis SP.
Although Hoechst staining demonstrated a distinct difference between SSCs and HSCs, surface molecule similarities do exist between SSCs and certain other stem cells. First, we found that SSCs express few, if any, MHC-I antigens. The major function of MHC-I is in regulating immunological self and non-self recognition by presenting peptides to cytotoxic T cells. Although MHC-I molecules are expressed on virtually all nucleated somatic cells, the expression is very limited in certain types of stem cells, primitive cells in the embryo, and spermatogonia (23, 24, 28). In the mouse, undifferentiated embryonic stem cells and teratocarcinomas, which are pluripotent stem cells, do not express MHC-I antigens (36, 37). Because these pluripotent cells are reported to give rise to germ cells after transplantation into blastocysts, our result suggests that the MHC-I negative phenotype may be a common characteristic among stem cells that have the potential to take part in generation of the germ line.
A second characteristic similarity of SSCs and other stem cells is expression of Thy-1. Although Thy-1 is a differentiation marker for thymocytes, T cells, and some neuronal cells, expression of Thy-1 on HSCs was reported more than two decades ago (25). Because Thy-1 is found on HSCs in many species (7, 25, 26), it appears to be a highly conserved HSC marker. Expression of Thy-1 on HSCs is considerably lower than that on T cells; therefore, the phenotype is designated Thy-1lo (7). Studies in mouse suggest that Thy-1 expression decreases with commitment and differentiation of HSCs (32). However, despite a long history of research on its conserved characteristics in molecular structure and expression pattern, the function of Thy-1 on HSCs is unclear. In the cryptorchid testis, removal of MHC-I+ cells is required to visualize Thy-1+ SSCs in FACS analysis, because a large number of autofluorescent cells and large cells obscure the Thy-1+ subpopulation. The intensity of Thy-1 expression on SSCs is only ≈10% of that on T cells in FACS analysis (unpublished data), indicating that the expression on SSCs is relatively low and similar to HSCs. In addition, Thy-1+ cells are primarily c-kit–, and in the MHC-I– cells there is a reciprocal relationship between Thy-1 and c-kit expression. Because c-kit is acquired as spermatogonia differentiate, perhaps down-regulation of Thy-1 and up-regulation of c-kit expression occur during SSC differentiation. Because SSCs are slow-dividing cells, Thy-1 may have an important role in transducing a growth inhibitory signal as has been suggested for neuronal Thy-1 (38).
One of the ultimate goals of stem cell biology is to understand mechanisms that control properties of stem cells, such as self-renewal and multilineage differentiation. To achieve this goal, it is necessary to identify and isolate stem cells, which has been facilitated by functional assay systems to identify stem cell activity and FACS cell fractionation. With these techniques, molecular determinants representing phenotypic characteristics of stem cells such as the surface antigens Sca-1 and Thy-1 or metabolic pump molecules such as Mdr1a/1b or Bcrp1 have been identified. Subsequently, knockout mice for each of these genes were generated to examine the effect on stem cell systems in mutant animals (39–42). Surprisingly, none of these molecules appeared essential for stem cell-dependent self-renewing systems. For example, Bcrp1 null animals showed no SP in Hoechst-stained bone marrow, but they have a normal number of functional c-kit+Sca-1+Lin– HSCs (42). These findings suggest that molecular markers used to identify stem cells are not necessarily essential regulators of functional activity, and Sca-1, Thy-1, Mdr1a/1b, and Bcrp1 fall into this nonessential category. Recent studies comparing gene expression profiles among HSCs, neural stem cells, and embryonic stem cells identified a number of genes expressed selectively on all three stem cells (43, 44). Among these molecules may be key regulators of stem cell function, which are also critical for SSCs. Because HSC and SSC transplantations are the most definitive assays of stem cell activity, these two systems represent a powerful resource to understand stem cell biology. Thus, the identification of SSCs by surface phenotype, which allows production of greatly enriched stem cell populations, will make possible sophisticated functional and genomic studies on these important cells, which provide for continuity between generations.
Supplementary Material
Acknowledgments
We thank Drs. R. Behringer, K. Orwig, and E. Sandgren for critical evaluation of the manuscript and helpful comments. We thank C. Freeman and R. Naroznowski for assistance with animal maintenance and experimentation, C. Brensinger for statistical analysis, W. Murphy for flow cytometry, and J. Hayden for photography. This work was supported by National Institutes of Health Institute of Child Health and Human Development Grant 36504, the Commonwealth and General Assembly of Pennsylvania, and the Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation.
Abbreviations: FACS, fluorescence-activated cell sorting; G, gate; HSC, hematopoietic stem cell; MHC-I, MHC class I; SP, side population; SSC, spermatogonial stem cell; X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside; β2M, β2 microglobulin.
References
- 1.Tegelenbosch, R. A. & de Rooij, D. G. (1993) Mutat. Res. 290, 193–200. [DOI] [PubMed] [Google Scholar]
- 2.Brinster, R. L. & Zimmermann, J. W. (1994) Proc. Natl. Acad. Sci. USA 91, 11298–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinster, R. L. & Avarbock, M. R. (1994) Proc. Natl. Acad. Sci. USA 91, 11303–11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinster, R. L. (2002) Science 296, 2174–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinohara, T., Orwig, K. E., Avarbock, M. R. & Brinster, R. L. (2000) Proc. Natl. Acad. Sci. USA 97, 8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Till, J. E. & McCulloch, E. A. (1961) Radiat. Res. 14, 213–222. [PubMed] [Google Scholar]
- 7.Spangrude, G. J., Heimfeld, S. & Weissman, I. L. (1988) Science 241, 58–62. [DOI] [PubMed] [Google Scholar]
- 8.Randall, T. D. & Weissman, I. L. (1998) Stem Cells 16, 38–48. [DOI] [PubMed] [Google Scholar]
- 9.Goodell, M. A., Brose, K., Paradis, G., Conner, A. S. & Mulligan, R. C. (1996) J. Exp. Med. 183, 1797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodell, M. A., Rosenzweig, M., Kim, H., Marks, D. F., DeMaria, M., Paradis, G., Grupp, S. A., Sieff, C. A., Mulligan, R. C. & Johnson, R. P. (1997) Nat. Med. 3, 1337–1345. [DOI] [PubMed] [Google Scholar]
- 11.Gussoni, E., Soneoka, Y., Strickland, C. D., Buzney, E. A., Khan, M. K., Flint, A. F., Kunkel, L. M. & Mulligan, R. C. (1999) Nature 401, 390–394. [DOI] [PubMed] [Google Scholar]
- 12.Asakura, A., Seale, P., Girgis-Gabardo, A. & Rudnicki, M. A. (2002) J. Cell Biol. 159, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welm, B. E., Tepera, S. B., Venezia, T., Graubert, T. A., Rosen, J. M. & Goodell, M. A. (2002) Dev. Biol. 245, 42–56. [DOI] [PubMed] [Google Scholar]
- 14.Murayama, A., Matsuzaki, Y., Kawaguchi, A., Shimazaki, T. & Okano, H. (2002) J. Neurosci. Res. 69, 837–847. [DOI] [PubMed] [Google Scholar]
- 15.Nagano, M. & Brinster, R. L. (1998) Acta Pathol. Microbiol. Immunol. Scand. 106, 47–55. [DOI] [PubMed] [Google Scholar]
- 16.Shinohara, T., Avarbock, M. R. & Brinster, R. L. (2000) Dev. Biol. 220, 401–411. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa, T., Arechaga, J. M., Avarbock, M. R. & Brinster, R. L. (1997) Int. J. Dev. Biol. 41, 111–122. [PubMed] [Google Scholar]
- 18.Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. (1997) FEBS Lett. 407, 313–319. [DOI] [PubMed] [Google Scholar]
- 19.Nagano, M., Avarbock, M. R. & Brinster, R. L. (1999) Biol. Reprod. 60, 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobrinski, I., Ogawa, T., Avarbock, M. R. & Brinster, R. L. (1999) Mol. Reprod. Dev. 53, 142–148. [DOI] [PubMed] [Google Scholar]
- 21.Nishimune, Y., Aizawa, S. & Komatsu, T. (1978) Fertil. Steril. 29, 95–102. [DOI] [PubMed] [Google Scholar]
- 22.Vergouwen, R. P., Huiskamp, R., Bas, R. J., Roepers-Gajadien, H. L., Davids, J. A. & de Rooij, D. G. (1993) J. Reprod. Fertil. 99, 479–485. [DOI] [PubMed] [Google Scholar]
- 23.Klein, J. (1986) in Natural History of Major Histocompatibility Complex (Wiley, New York), pp. 152–175.
- 24.Kubota, H. & Reid, L. M. (2000) Proc. Natl. Acad. Sci. USA 97, 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldschneider, I., Gordon, L. K. & Morris, R. J. (1978) J. Exp. Med. 148, 1351–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baum, C. M., Weissman, I. L., Tsukamoto, A. S., Buckle, A. M. & Peault, B. (1992) Proc. Natl. Acad. Sci. USA 89, 2804–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling, V. & Neben, S. (1997) J. Cell. Physiol. 171, 104–115. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, Y., Jahagirdar, B. N., Reinhardt, R. L., Schwartz, R. E., Keene, C. D., Ortiz-Gonzalez, X. R., Reyes, M., Lenvik, T., Lund, T., Blackstad, M., et al. (2002) Nature 418, 41–49. [DOI] [PubMed] [Google Scholar]
- 29.Anderson, R., Fassler, R., Georges-Labouesse, E., Hynes, R. O., Bader, B. L., Kreidberg, J. A., Schaible, K., Heasman, J. & Wylie, C. (1999) Development (Cambridge, U.K.) 126, 1655–1664. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa, T., Ohmura, M., Yumura, Y., Sawada, H. & Kubota, Y. (2003) Biol. Reprod. 68, 316–322. [DOI] [PubMed] [Google Scholar]
- 31.Rietze, R. L., Valcanis, H., Brooker, G. F., Thomas, T., Voss, A. K. & Bartlett, P. F. (2001) Nature 412, 736–739. [DOI] [PubMed] [Google Scholar]
- 32.Kondo, M., Weissman, I. L. & Akashi, K. (1997) Cell 91, 661–672. [DOI] [PubMed] [Google Scholar]
- 33.Wagers, A. J., Allsopp, R. C. & Weissman, I. L. (2002) Exp. Hematol. (Charlottesville, VA) 30, 176–185. [DOI] [PubMed] [Google Scholar]
- 34.Capel, B., Hawley, R., Covarrubias, L., Hawley, T. & Mintz, B. (1989) Proc. Natl. Acad. Sci. USA 86, 4564–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, C. Z., Li, M., de Graaf, D., Monti, S., Gottgens, B., Sanchez, M. J., Lander, E. S., Golub, T. R., Green, A. R. & Lodish, H. F. (2002) Proc. Natl. Acad. Sci. USA 99, 15468–15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croce, C. M., Linnenbach, A., Huebner, K., Parnes, J. R., Margulies, D. H., Appella, E. & Seidman, J. G. (1981) Proc. Natl. Acad. Sci. USA 78, 5754–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian, L., Catt, J. W., O'Neill, C. & King, N. J. (1997) Biol. Reprod. 57, 561–568. [DOI] [PubMed] [Google Scholar]
- 38.Tiveron, M. C., Barboni, E., Pliego Rivero, F. B., Gormley, A. M., Seeley, P. J., Grosveld, F. & Morris, R. (1992) Nature 355, 745–748. [DOI] [PubMed] [Google Scholar]
- 39.Nosten-Bertrand, M., Errington, M. L., Murphy, K. P., Tokugawa, Y., Barboni, E., Kozlova, E., Michalovich, D., Morris, R. G., Silver, J., Stewart, C. L., et al. (1996) Nature 379, 826–829. [DOI] [PubMed] [Google Scholar]
- 40.Schinkel, A. H., Mayer, U., Wagenaar, E., Mol, C. A., van Deemter, L., Smit, J. J., van der Valk, M. A., Voordouw, A. C., Spits, H., van Tellingen, O., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanford, W. L., Haque, S., Alexander, R., Liu, X., Latour, A. M., Snodgrass, H. R., Koller, B. H. & Flood, P. M. (1997) J. Exp. Med. 186, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, S., Morris, J. J., Barnes, Y., Lan, L., Schuetz, J. D. & Sorrentino, B. P. (2002) Proc. Natl. Acad. Sci. USA 99, 12339–12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. (2002) Science 298, 597–600. [DOI] [PubMed] [Google Scholar]
- 44.Ivanova, N. B., Dimos, J. T., Schaniel, C., Hackney, J. A., Moore, K. A. & Lemischka, I. R. (2002) Science 298, 601–604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.