Abstract
We report here a superfamily of “cut and paste” DNA transposons called Transib. These transposons populate the Drosophila melanogaster and Anopheles gambiae genomes, use a transposase that is not similar to any known proteins, and are characterized by 5-bp target site duplications. We found that the fly genome, which was thought to be colonized by the P element <100 years ago, harbors ≈5 million year (Myr)-old fossils of ProtoP, an ancient ancestor of the P element. We also show that Hoppel, a previously reported transposable element (TE), is a nonautonomous derivate of ProtoP. We found that the “rolling-circle” Helitron transposons identified previously in plants and worms populate also insect genomes. Our results indicate that Helitrons were horizontally transferred into the fly or/and mosquito genomes. We have also identified a most abundant TE in the fly genome, DNAREP1_DM, which is an ≈10-Myr-old footprint of a Penelope-like retrotransposon. We estimated that TEs are three times more abundant than reported previously, making up ≈22% of the whole genome. The chromosomal and age distributions of TEs in D. melanogaster are very similar to those in Arabidopsis thaliana. Both genomes contain only relatively young TEs (<20 Myr old), constituting a main component of paracentromeric regions.
Drosophila melanogaster (DM) is among the most important species shaping our knowledge of mobile or transposable elements (TEs) that populate eukaryotic and prokaryotic genomes and multiply by transferring their copies from one genomic place to another (1–4). The high variety of currently or recently active TEs and the role of DM as one of the basic genetic models were among the major factors behind successful studies of TEs. All fly TEs reported in the literature or public databases before 1999 were found experimentally. However, recent explosion of the available sequence data (5) has brought methods of computational analysis to the forefront. Computer-assisted identification of DNA patterns similar to a query sequence is fast, sensitive, and cheap. Computational analysis also permits efficient reconstruction of ancient TEs from their incomplete or mutated copies (we consider them as genomic DNA fossils). Nevertheless, there is no guarantee that any TE reconstructed from its footprints is an active element. Therefore, the final isolation of bona fide active transposable elements or verification of biological activity of reconstructed transposons must be done in a test tube.
Using approaches similar to those applied in identifying TEs in the Arabidopsis thaliana (AT) and Caenorhabditis elegans genomes (6, 7), we began a systematic computational analysis of TEs in the DM genome (DMG) in April 1999. At that time, ≈50 families of DM TEs were reported in the literature. On the basis of computational analysis, we identified and characterized ≈80 additional families of TEs harbored by the DMG. Despite stop codons, insertions, and deletions present in genomic copies of different TEs, the consensus sequences reconstructed for most of the new families are free of these mutations and contain valuable information on the evolution and function of TEs. For example, analysis of conserved protein motifs in the reconstructed proteins may be the easiest way to identify catalytic centers and other motifs involved in transpositions. Here we describe several novel TEs and an overview of DNA transposons and retrotransposons found in the DMG. Surprisingly, the diversity of TEs fossilized in the DMG is higher than in mammalian or vertebrate genomes studied so far. TEs can become stable structural components of the eukaryotic heterochromatin (8, 9, 10). Perhaps the best example in this respect is AT, where TEs are highly compartmentalized: ≈90% of genomic TEs are localized in the paracentromeric heterochromatin, and ≈90% of the paracentromeric heterochromatin is composed of TEs (6, 11). We show that similar compartmentalization characterizes distributions of TEs in the DMG.
Materials and Methods
Computational Analysis. All TEs reported in this article were found by using various methods of computational analysis. We began by compiling DNA sequences of known TEs and other repetitive elements deposited in the DM section of Repbase Update or “RU” (12). Using this database, we annotated TEs in the DM DNA sequences deposited into GenBank and the Berkeley Drosophila Genome Project public database (www.fruitfly.org/sequence/assembly.html). The annotation began with comparing DNA sequences of known TEs against the sequenced portion of DMG by using CENSOR (13). Putative new TEs were identified either as insertions into copies of known TEs or as elements distantly similar to known TEs. These insertions were analyzed in detail for presence of hallmarks such as terminal inverted repeats (TIRs), target site duplications (TSDs), long terminal repeats (LTRs), etc. For each identified insertion, we applied BLASTN (14) or WU BLASTN (http://blast.wustl.edu) to extract DM GenBank sequences whose fragments were similar to the insertion sequence. Subsequently, precise coordinates of these fragments were determined by running the insertion sequence against the extracted GenBank sequences by using CENSOR.
DNA regions of a very low but significant partial identity (60–70%) to the known TE were also analyzed in the same way as the insertions described above. As noted before (6), such regions usually belong to highly diverged families of young TEs.
Using the majority rule applied to multiple aligned copies of TEs, we built their consensus sequences. Copies of TEs created by chromosomal duplications or redundant sequencing were discarded on the basis of similarity between the corresponding flanking regions.
Distantly related proteins were identified by using PSI-BLAST (15). Multiple alignments of protein sequences were created by CLUSTAL-W (16) and edited manually by using GENEDOC (17). Multiple alignments of DNA sequences were obtained by using the VMALN2 and PALN2 programs developed at the Genetic Information Research Institute. We used FGENESH (ref. 18; www.softberry.com) for identifying genes encoded by TEs. Phylogenetic analysis was conducted by using MEGA (19). The ages of TEs were calculated by using the formula t = K/ν, where t is the average time elapsed since the TE copies were transposed in the genome, K is the average divergence of the TE copies from their consensus sequence, and ν is the neutral substitution rate, 0.016 substitution per site per million years (Myr) (20). Ages of retroviruses were estimated on the basis of pairwise divergences of their LTRs by using the formula t = K/2ν, where K is the divergence between LTRs that flanked the same provirus.
Databases. The sequences of TEs reported in this manuscript are deposited in the DM section of RU (ref. 12, www.girinst.org/Repbase_Update.html). TE families originally reported in RU were also included in the data set of TEs maintained by the Drosophila Genome Project Consortium (www.fruitfly.org).
Results and Discussion
Transib. One particular copy of the ProtoP_B transposon (GenBank AE003048, positions 9814–12479) harbors a 1693-bp insertion, positions 9932–11624, which has characteristics of DNA transposons. The insertion is flanked by the 5-bp CAGCG TSD and has a 44-bp TIR. After a search by BLASTN and CENSOR, we identified an additional six copies of the insertion (Fig. 1) scattered throughout the DMG and ≈90% identical to each other. Analysis of the corresponding flanking regions confirmed the presence of 5-bp flanking direct repeats, which can be considered as remnants of 5-bp TSDs induced by transpositions (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). This family of TEs was named Transib1 (after the Trans-Siberian Express running between Moscow and the Pacific Ocean). The 2167-bp Transib1 consensus sequence was reconstructed on the basis of a multiple alignment of Transib1 copies (Fig. 1), and it includes 43-bp TIRs. CENSOR-assisted screening of the DMG has shown that it harbors multiple repetitive elements, which are only ≈60% identical to the Transib1 consensus. On the basis of pairwise nucleotide identities, the identified repetitive elements related to Transib1 were split into three groups called Transib2–4.
Fig. 1.
Reconstruction of the Transib1 (A), Transib2 (B), Transib3 (C), and Transib4 (D) consensus sequences. The consensus sequences are schematically depicted as rectangles capped by black triangles indicating TIRs. Transposasecoding regions are shaded in gray. Their coordinates in the consensus sequences are shown above the rectangles. Transib copies used for reconstruction of the consensus sequences are shown as thick lines beneath the rectangles. Gaps in the lines mark deletions. GenBank accession numbers and the corresponding sequence coordinates of TEs are indicated.
The Transib families are highly divergent from each other; their consensus sequences are only 60–62% identical to each other. At the same time, the identities between the corresponding consensus sequences and copies from the same family are over 90%. The Transib2-4 consensus sequences are 2,844, 2,883, and 2,656 bp long, including 42-, 45-, and 40-bp TIRs, respectively. Analogously to Transib1, Transib2-4 elements have generated 5-bp TSDs upon their insertions into the genome. The identified TSDs and TIRs indicate that Transib1–4 elements are DNA transposons. Moreover, the Transib1–4 consensus sequences encode hypothetical proteins, 461-aa Transib1p, 690-aa Transib2p, 685-aa Transib3p, and 533-aa Transib4p, respectively, which are 32–46% identical to each other (Table 4, which is published as supporting information on the PNAS web site). Such a high identity between the proteins, encoded by the Transib1–4 DNA sequences that are only ≈60% identical to each other, indicates that Transib1–4p proteins were necessary for the Transibs transposition. Because there are no other proteins encoded by these elements, we consider Transib1–4p as predicted transposases. Based on a TBLASTN search, there are about 100 elements in the DMG that code for proteins similar to Transib1p-4p. Autonomous DNA transposons, i.e., elements that encode transposases, usually constitute a minor fraction of DNA transposons fixed in eukaryotic genomes. The major fraction is composed of nonautonomous elements that do not encode the transposase but share common termini with autonomous elements from which they have descended. The same pattern is seen for Transibs (Fig. 1).
We noted a distant ≈60% identity between Transib1 and Hopper, a 1,435-bp nonautonomous DNA transposon identified as a de novo insertion associated with the SxlM4 allele (21). Hopper has 33-bp TIRs similar to these in Transib, and its copies are also flanked by 5-bp TSDs (21) (Fig. 6). Most likely, Hopper is the youngest member of the Transib clade. Hopper elements identified in the sequenced genome do not encode proteins. Presumably, the autonomous Hopper encoding a Transib-like transposase is not fixed in the DMG.
Currently, Anopheles gambiae (AG) is the only other species whose sequenced portion of the genome (22) encodes proteins similar to the Transib transposases. Using WU BLAST, we extracted all DNA fragments encoding proteins similar to the Transib transposases from the public set of assembled AG scaffolds. These fragments were split into several homogenous clusters. On the basis of the multiple alignment of sequences from the most abundant cluster, we reconstructed their 4,303-bp consensus sequence. This consensus sequence is a “recovered” mosquito Transib-like DNA transposon, called Transib1_AG, which encodes a 708-aa transposase (35% identity to Transib2p) and has 9-bp TIRs similar to those in the fly Transib elements (Fig. 7, which is published as supporting information on the PNAS web site). Moreover, Transib1_AG copies also are flanked by the 5-bp TSDs.
Given the multiple copies, TIRs, TSDs, and insertions of some Transib copies into other TEs, there is no doubt that Transib-like elements are molecular fossils of recently active DNA transposons. As there is no significant similarity of the Transib transposases to any known proteins (PSI-BLAST, BLASTP, E < 1), it is certain that Transib1p–Transib4p and Transib1_AGp belong to a previously unrecognized class of DNA transposases, catalysts of Transib “cut and paste” transpositions. The alignment of Transib1–4p, Transib1_AGp, and Transib2_AGp revealed seven conserved motifs, presumably most important for the transposition (Fig. 2). A putative catalytic D(34)D(35)E signal found in many cut-and-paste transposases is also present in the fly Transib transposases (Fig. 2). However, this signal is not present in the mosquito transposases.
Fig. 2.
Multiple alignment of the Transib transposases. Diamonds mark amino acid residues that belong to a putative noncanonical DDE catalytic site. Asterisks mark bp 10, 30, etc. Conserved motifs and residues are underlined and shaded, respectively.
Given the unique conserved transposase, the 5-bp TSD, the specific TIRs, and their presence in the fly and mosquito genomes, we conclude that Transib-like TEs constitute an eighth superfamily of cut-and-paste DNA transposons (Table 1).
Table 1. Cut-and-paste DNA transposons.
| Superfamily | Homo sapiens | A. thaliana | C. elegans | DM |
|---|---|---|---|---|
| mariner/Tc1 | + | + | + | + |
| hAT | + | + | + | + |
| PiggyBac | + | - | - | + |
| MuDr | ?* | + | +† | ?‡ |
| En/Spm | - | + | - | - |
| Harbinger | +§ | + | + | +§ |
| P | +§ | - | - | + |
| Transib | - | - | - | + |
MuDr-like transposase is not found the human genome. However, Ricksha (unpublished data; RU) has MuDr hallmarks: 9-bp TSD, 70-bp TIR, and 5′-GGG and CCC-3′ termini.
CEMUDR1 (unpublished data; RU).
Putatively, FB belongs to the MuDr superfamily.
Harbinger-and P-like transposases are present as single-copy genes in the DM (unpublished data; RU) and human (27) genomes.
Interestingly, the Transib codon composition is atypical for the DMG. As a result, commonly applied programs, including those used for systematic annotation of the DMG (18), fail to predict Transib1p-4p unless one presumes that the fly genome codes for genes characterized by human-, plant-, or worm-specific features, including their codon composition. Presumably, the atypical codon composition is related to the horizontal transfer of Transibs.
ProtoP. The ≈1100-bp Hoppel (23) or “element 1360” (24) is one of the most abundant TEs in the DMG (25). On the basis of its ≈30-bp TIR, this element was classified as a putative DNA transposon (23, 24). However, its relationship to known superfamilies of DNA transposons was obscure. We found that an ≈2-kb internal portion of one unusually long Hoppel element (GenBank gi 5656725, 634-4052) encoded a protein distantly related to the P transposase (BLASTX, 25–29% similarities to known P transposases, E < 10–20, two stop codons). Surprisingly, Hoppel was the only TE linked to the identified internal portion (Fig. 3). Moreover, the internal sequence does not have any hallmarks of a TE inserted accidentally into a Hoppel element. Therefore, we concluded that the identified long Hoppel element was a fossilized autonomous DNA transposon that gave birth to the nonautonomous Hoppel/1360 elements. Given its antiquity (described below) and the similarity to all known P transposases, we called this transposon ProtoP. The 1,105-bp Hoppel consensus sequence is ≈97% identical to Hoppel elements, indicating that this family is ≈2 Myr old. ProtoP_B is a second family of nonautonomous ProtoP-like transposons described here. ProtoP_B copies are ≈95% identical to the 1,153-bp ProtoP_B consensus sequence (≈3 Myr old). There is only a 5% divergence between the Hoppel and ProtoP_B consensus sequences that contain 191-bp and 136-bp internal regions, respectively, ≈95% identical to different portions of ProtoP (Fig. 3). The 4,480-bp consensus sequence of ProtoP was reconstructed on the basis of a multiple alignment of its 28 copies, which are ≈95% identical to the consensus (Fig. 3). Hoppel and ProtoP_B elements were excluded from the last set of sequences. The ProtoP consensus sequence is 97% and 96% identical to the Hoppel and ProtoP_B consensus sequences, respectively. Therefore, the peak of ProtoP, Hoppel, and ProtoP_B transpositional activity occurred 3–5 Myr ago.
Fig. 3.
Reconstruction of the ProtoP consensus sequence. The consensus sequence is schematically depicted as a rectangle capped by the black triangles indicating TIRs. The region coding for the transposase is shaded in gray. Its coordinates in the consensus sequence are indicated above the rectangle. Copies of ProtoP used for reconstructing the consensus sequences are shown as thick lines beneath the ProtoP, Hoppel, and ProtoP_B consensus sequences. Gaps in the lines mark deletions. GenBank accession numbers and the corresponding sequence coordinates of TEs are indicated.
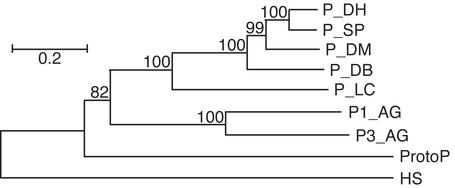
The last estimate is supported by studies of TEs inserted into ProtoP. For example, a ProtoP element identified in the AE002983 GenBank sequence (positions 8879–1919) harbors a copy of the Gypsy6A endogenous retrovirus (unpublished data; RU), including its flanking LTRs (positions 2325–8774). The 14% divergence between these LTRs corresponds to ≈5 Myr elapsed since the insertion of the retrovirus into the ProtoP element. A ProtoP_B copy (AE002743, positions 1–7347) harbors the Invader5 retrovirus (unpublished data; RU). The 15% divergence between its LTRs also indicates that ProtoP transposons colonized the DMG ≈5 Myr ago. Therefore, P-like elements, which were thought to be recent invaders of the DMG (26), colonized this genome several Myr ago. The ProtoP consensus sequence encodes an 864-aa ProtoP1 transposase (positions 1194–3788). Phylogenetic analysis (Fig. 4) shows that the ProtoP transposase separates a cluster formed by insect P elements and a putative human gene derived from a P-like transposase (27). Computer-assisted analysis of ≈30 ProtoP, Hoppel, and ProtoP_B copies and their target sites shows that ProtoP-like elements are flanked by 7-bp TSDs (not shown). However, all studied P elements, including the P1_AG, P2_AG, and P3_AG families present in AG (ref. 22; unpublished data; RU) are characterized by 8-bp TSDs. Usually, the TSD size is one of a few hallmarks of different superfamilies of eukaryotic DNA transposons. For example, 8-bp TSD is a characteristic of hAT-like transposons; 4-bp, piggyBac; 5-bp, Transib; 9-bp, MuDr; 3-bp, En/Spm and Harbinger. mariner/Tc1 is the only superfamily whose members are characterized by either 2-bp or 3-bp TSDs. Presumably, the phylogenetic place of the ProtoP transposase as an “outlier” among other P transposases (Fig. 4) may be related to the TSD size. While this paper was in review, we became aware of a similar report (28) describing a 3,410-bp autonomous Hoppel transposon (nearly identical to ProtoP deposited in RU in 1999). The authors suggested that the intronless P-like transposase is a result of retrotransposition. However, they did not show any evidence supporting this hypothesis. Alternatively, gain or loss of introns is relatively common and may be independent of retrotransposition. For example, the mosquito Helitron transposases are intronless (described below), whereas similar transposases in plants and worms are encoded by multiple exons (7). Similarly, accidental intron gains were reported for plant mariners (29).
Fig. 4.
A phylogenetic tree for P-like transposases. The unrooted tree was constructed by using the neighbor-joining method implemented in MEGA (19). Transposases from the next P-like transposons are shown: P_DH, Drosophila helvetica, GenBank accession no. AAK08181; P_SP, Scaptomyza pallida, joined AAA29959–61; P_DM, DM, A24786; P_DB, Drosophila bifasciata, AAB31526; P_LC, Lucilia cuprina, A46361. P1_AG and P3_AG from A. gambiae and ProtoP from DM are transposases from P elements reported in this manuscript. HS is a putative human gene derived from the P-like transposase (27). The scale of the Poisson correction distances between the protein sequences is indicated. Bootstrap values are shown at the nodes.
Helitrons in Insects. A TBLASTN search for fly proteins similar to those encoded by the plant and worm Helitron rolling-circle DNA transposons (7) did not reveal any evident matches. However, using an approach similar to that described in ref. 7, we identified in AG the insect Helitron transposon, called Helitron1_AG. Its 8,200-bp consensus sequence encodes an intronless 1,700-aa Helitron1_AGp protein composed of the canonical Helitron-like Rep and SF1 helicase domains. Analogously to their plant and worm relatives (7), insect Helitron transposons are characterized by the 5′-TC and CTAG-3′ termini, the 3′-terminal ≈12-bp hairpin, and TSD-free insertions between the target A-3′ and 5′-T nucleotides. The Helitron1_AG consensus sequence is only ≈5% divergent from several Helitron1_AG copies, indicating that these elements were transposed in AG during the last few million years. The AG genome harbors more than 100 copies of different Helitron transposons that belong to more than 10 different families (not shown), characterized by a high (>90%) intra- and low (<70%) interfamily nucleotide identities. Additionally to the Helitron1_AG, we also reconstructed the consensus sequence of a Helitron2_AG family. Surprisingly, we found that a 649-bp portion of the Helitron2_AG (positions 972-1620) was 75% identical to a ≈600-bp fly sequence (AE002840, 9827–9264) (Fig. 7). Because this Helitron-like element is flanked by other DM-specific TEs, sequencing-related artifacts can be discarded. The identified element encodes a portion of the Rep/Helicase protein, and its coding sequence is interrupted by a few stop codons. Therefore, this element is a remnant of a Helitron. Ancestral lineages of DM and AG are thought to have separated ≈250 Myr ago (30). The neutral substitution rate in AG is even higher than in DM (31). Therefore, transposable elements that were present in a lineage ancestral to both AG and DM would not be recognizable anymore. For example, exons from ≈6,000 orthologous genes identified in the mosquito and fly genomes show only a ≈56% nucleotide identity (32). The 75% identity between the mosquito and fly Helitrons indicates that these elements were transferred horizontally rather than transmitted vertically.
piggyBac. The 1,881-bp consensus sequence of TE called Looper1_DM, reconstructed from five copies, encodes a 358-aa protein, which is more similar to the piggyBac-like transposase from the human Looper (7) than to the transposase encoded by the moth piggyBac (33, 34). DM was the third species where piggyBac DNA transposons were identified. Recently, piggyBac TEs were also identified in mosquito (22) and fishes (35, 36). These piggyBacs are characterized by ≈15-bp TIRs, including 5′-CCC and GGG-3′, and the TTAA TSDs. Using PSI-BLAST,we found that two hypothetical genes annotated in the DMG (gi 7303440, 21355395) are encoding proteins similar to the piggyBac transposase (PSI-BLAST, E < 10–3). However, because the biological function of these genes is unknown, it is not clear whether they are “host genes” derived from the transposases or remnants of young piggyBacs. Analogously, several hypothetical DM genes are former Harbinger transposases (ref. 6; RU).
DNAREP1_DM. On the basis of a multiple alignment of ≈100 expanded copies of the 300-bp ARS320 repetitive element identified in fly (GenBank J01068), we reconstructed a 600-bp consensus sequence. The ARS320 element matches an internal portion of a 600-bp repetitive element called DNAREP1_DM (positions 100–400). Some DNAREP1_DM copies are inserted into other TEs (not shown). Therefore, DNAREP1_DM is a transposable element. It is the most abundant TE in the DMG. Several thousand copies of DNAREP1_DM are present in the sequenced genome. They are ≈85% identical to the consensus sequence. Therefore, DNAREP1_DM is an old family of TEs, transposed mainly ≈10 Myr ago. Currently, there are no young subfamilies of DNAREP1_DM, i.e., with copies at least 95% identical to each other. It indicates that DNAREP1_DM elements lost their mobility >3 Myr ago. Surprisingly, members of the DNAREP1_DM family do not reveal any hallmarks of DNA transposons and retrotransposons (TIRs, TSDs, 3′-microsatellites, etc). In 1999, we characterized DNAREP1_DM as a putative DNA transposon (unpublished work; RU) because of its conserved termini and multiple internal deletions. Based on the most recent data, another mechanism of DNAREP1_DM transpositions can be suggested. We found that a ≈200-bp 3′ untranslated region (UTR) portion of Penelope, an active Drosophila virilis retrotransposon (37), is ≈72% identical to the DNAREP1_DM consensus sequence (Fig. 8, which is published as supporting information on the PNAS web site). Moreover, the 3′ UTR of Penelope is required for its successful transposition (38). Therefore, we consider DNAREP1_DM as a remnant of Penelope-like retrotransposon that was highly active in the ancestral lineage of DM ≈10 Myr ago. We consider Penelope as a non-LTR retrotransposon. Alternatively, Penelope is also considered as a separate class of retroelements (38). Given that the DNAREP1_DM consensus sequence cannot be expanded, it is unlikely that it is a solo LTR.
Non-LTR Retrotransposons. Members of seven different clades of non-LTR retrotransposons populate the fly genome (Table 2). Remarkably, of all sequenced genomes, no genome contains elements from as many different clades of non-LTR retrotransposons as the DMG does. Among the seven different clades, Jockey is the most diverse and abundant in the DMG. We identified 15 previously undescribed families, bringing the total number to 25 known families (Table 2). Like other TEs, families of non-LTR retrotransposons that belong to the same clade are characterized by <75% inter- and >85% intrafamily DNA identities.
Table 2. Non-LTR retrotransposons in the DMG.
| Clade | Families identified in DM |
|---|---|
| Jockey | Fw, Jockey, Helena, BS, G, Doc, Strider, X, Het-A, TART, RU: Jockey2, Doc2_DM-Doc5_DM, G2_DM-G7_DM, Bs3_DM, Bs4_DM, Fw2_DM, Fw3_DM |
| CR1 | RU: DMCR1A |
| I | I, RU: IVK_DM,* DMRT1A,** DMRT1B,** DMRT1C |
| R1 | R1, RU: R1-2_DM |
| LOA | Bilbo, RU: Baggins1 |
| R2 | R2 |
| Penelope | RU: DNAREP1_DM |
LTR Retrotransposons. We identified and characterized 28 families of LTR retrotransposons in DM (Table 3). All fly LTR retrotransposons belong to Gypsy, Copia, or Bel superfamilies (39), of which Gypsy is the most diverse and abundant. Surprisingly, the DMG does not contain nonautonomous LTR retrotransposons, present in mammals (40) and plants (6). On the basis of sequence identities and structural commonalities, Gypsy can by divided further into four major groups (Table 3), called Gypsy, MDG1, MDG3, and Osvaldo. The best-studied Gypsy group is composed of canonical Gypsy-like families, including Gypsy, 297, 17.6, and Zam. The Osvaldo group includes retrotransposons similar to Osvaldo and Ulysses, discovered in Drosophila buzzatii (41) and D. virilis (42). Gypsy12 is an Osvaldo family we found in DM. The ≈2,300-bp Gypsy12 LTR is the longest LTR identified in DM. Some members of the Gypsy and Osvaldo groups encode env, the surface envelope proteins, characteristic for animal retroviruses. Independently, the Gypsy superfamily was split into four groups (43), on the basis of the phylogenetic study of reverse transcriptase, RNase H, and integrase domains, and by us (unpublished work; RU 1999) on the basis of the structural commonalities (Table 3). In RU, the Accord, Tabor, and Invader groups were introduced instead of Gypsy, MDG1, and MDG3, respectively (43). Members of the Bel superfamily (44) encode one long polyprotein composed of gag, protease, reverse transcriptase, RNase H, and endonuclease domains. This superfamily was also called BCDRP (after Bel–Catch–Diver–Roo–Pao) and included Diver in fly and Catch in fugu (unpublished work; RU 1999).
Table 3. LTR retrotransposons in the DMG.
| Superfamily/group | Family | TSD, bp | PBS* | LTR termini | env |
|---|---|---|---|---|---|
| Gypsy/Gypsy | Gypsy, 297, 17.6, Tirant, Zam, Idefix, Nomad, Ninja, Burdock, HMS Beagle, Transpac, Springer; RU: Gypsy2-Gypsy4, Gypsy5,IGypsy6, Gypsy7, Gypsy9, Gypsy10, Quasimodo,IIAccord, Accord2, GtwinIII | 4 | -1 | AG.. .YT | + |
| Gypsy/MDG1 | MDG1, 412, Blood, Stalker; RU: Tabor,IVStalker, Stalker2, Gypsy11 | 4 | +1 | TG.. .CA | - |
| Gypsy/MDG3 | MDG3, Micropia, Blastopia; RU: Invader1-Invader6 | 4 | 5-8 | TG.. .CA | - |
| Gypsy/Osvaldo | Circe; RU: Gypsy8, Gypsy12 | 4 | 35 | TG.. .CA | - |
| Bel | Bel, Roo, Batumi, Max; RU: Diver,VDiver2, RooA | 5 | 1-3 | TG.. .CA | - |
| Copia | Copia, 1731; RU: Copia2_DM | 5 | 5-7 | TG.. .CA | - |
Families following “RU:” are reported in this article and RU. Subsequent nomenclature changes are marked as superscripts I-V and the secondary names are I, nik; II, antonia; III, hamilton; IV, wolfman; and V, mazi (50).
Start position of primer binding site (PBS); an internal portion starts at 1.
Abundance of TEs. TEs account for 6% and 60% of the sequenced DM euchromatin and heterochromatin regions, respectively. Given that heterochromatin constitutes ≈30% of the DMG (5), TEs make up ≈22% of the whole genome. Our estimates are three times higher than those reported recently (25). The difference can be attributed to the different sets of TEs used as query sequences. Whereas the previous report (25) included TEs from 39 families, we used elements representing ≈150 families. Also, CENSOR and WU BLAST, used in our studies, are more sensitive than BLAST used previously (25). As a result, we identified numerous families of TEs that are only 60% identical to known elements and are represented by only a few truncated copies. There is a strong increase in the TE density on the acrocentric chromosomes 2 (Fig. 5) and 3 (not shown) in ≈500-kb regions separating euchromatin and paracentromeric heterochromatin, which is mostly not sequenced. This chromosomal distribution of TEs is very similar to that observed in AT (6). Given the fact that ≈80% of the paracentromeric/centromeric heterochromatin in DMG has not yet been sequenced, the percentage of TEs may be even higher.
Fig. 5.
Density of TEs across chromosome 2. It was calculated as a percentage of TE-derived sequences per 100 kb in nonoverlapping windows. The ≈8-megabase (Mb) centromeric region is shaded in gray.
Age of TEs. All TEs identified in DMG are younger than 20 Myr, analogous to those in AT (6). Importantly, both genomes lack families with copies <80% identical to the family consensus, and they also do not contain LTR retrotransposons whose flanking LTRs are <80% identical to each other. It is unlikely that high rates of point mutation and short deletion in the fly genome (20, 45) are factors that alone can explain the age distribution of TEs. For example, despite the 2- to 3-fold difference in the mutation rates, there is no noticeable difference between the TE age distributions in AT and DM. Also, if the mutation rate is the major factor, one has to see families with copies <80% identical to their consensus sequence. Most likely, this phenomenon can be explained by relatively infrequent >100-kb deletions in paracentromeric heterochromatin composed mainly of TEs, which have been accumulated in heterochromatin as a result of a strong selection against their presence in the gene-rich euchromatin. It is unlikely that paracentromeric heterochromatin attracts de novo insertions of TEs as was suggested (46). For example, studies of de novo insertions of hAT and En/Spm DNA transposons in AT show (47, 48) that they are targeted preferentially outside paracentromeric heterochromatin, despite the apparent long-time accumulation of the same class TEs in heterochromatin (6, 11).
Supplementary Material
Acknowledgments
We thank Margaret Kidwell and reviewers for helpful comments, Dominique Anxolabehere for sharing unpublished results, and Jolanta Walichiewicz and Michael Jurka for assistance in preparing the manuscript. This work was supported by the Grant 2 P41 LM06252-04A1 from the National Institutes of Health.
Abbreviations: TE, transposable element; TIR, terminal inverted repeat; TSD, target site duplication; RU, Repbase Update; DM, Drosophila melanogaster; DMG, DM genome; AG, Anopheles gambiae; AT, Arabidopsis thaliana; Myr, million years.
References
- 1.Berg, D. E. & Howe, M. H., eds. (1987) Mobile DNA (Am. Soc. Microbiol., Washington, DC).
- 2.Kidwell, M. G. & Lisch, D. R. (2001) Evol. Int. J. Org. Evol. 55, 1–24. [DOI] [PubMed] [Google Scholar]
- 3.Fedoroff, N. V. (1999) Ann. N.Y. Acad. Sci. 870, 251–264. [DOI] [PubMed] [Google Scholar]
- 4.Craig, N. L. (1995) Science 270, 253–254. [DOI] [PubMed] [Google Scholar]
- 5.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 6.Kapitonov, V. V. & Jurka, J. (1999) Genetica 107, 27–37. [PubMed] [Google Scholar]
- 7.Kapitonov, V. V. & Jurka, J. (2001) Proc. Natl. Acad. Sci. USA 98, 8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pimpinelli, S., Berloco, M., Fanti, L., Dimitri, P., Bonaccorsi, S., Marchetti, E., Caizzi, R., Caggese, C. & Gatti, M. (1995) Proc. Natl. Acad. Sci. USA 92, 3804–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapitonov, V. V., Holmquist, G. P. & Jurka, J. (1998) Mol. Biol. Evol. 15, 611–612. [DOI] [PubMed] [Google Scholar]
- 10.Ananiev, E. V., Phillips, R. L. & Rines, H. W. (1998) Proc. Natl. Acad. Sci. USA 95, 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Arabidopsis Genome Initiative (2000) Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 12.Jurka, J. (2000) Trends Genet. 16, 418–420. [DOI] [PubMed] [Google Scholar]
- 13.Jurka, J., Klonowski, P., Dagman, V. & Pelton, P. (1996) Comput. Chem. 20, 119–121. [DOI] [PubMed] [Google Scholar]
- 14.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 15.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholas, K. B., Nicholas, H. B. & Deerfield, D. W. (1997) EMBNET News 4, 1–4. [Google Scholar]
- 18.Salamov, A. A. & Solovyev, V. V. (2000) Genome Res. 10, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 20.Li, W.-H. (1997) Molecular Evolution (Sinauer, Sunderland, MA).
- 21.Bernstein, M., Lersch, R. A., Subrahmanyan, L. & Cline, T. W. (1995) Genetics 139, 631–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M., Wides, R., et al. (2002) Science 298, 129–149.12364791 [Google Scholar]
- 23.Kurenova, E. V., Leibovich, B. A., Bass, I. A., Bebikhov, D. V., Pavlova, M. N. & Danilevskaia, O. N. (1990) Genetika 26, 1701–1712. [PubMed] [Google Scholar]
- 24.Kholodilov, N. G., Bolshakov, V. N., Blinov, V. M., Solovyov, V. V. & Zhimulev, I. F. (1988) Chromosoma 97, 247–253. [DOI] [PubMed] [Google Scholar]
- 25.Bartolome, C., Maside, X. & Charlesworth, B. (2002) Mol. Biol. Evol. 19, 926–937. [DOI] [PubMed] [Google Scholar]
- 26.Clark, J. B. & Kidwell, M. G. (1997) Proc. Natl. Acad. Sci. USA 94, 11428–11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagemann, S. & Pinsker, W. (2001) Mol. Biol. Evol. 18, 1979–1982. [DOI] [PubMed] [Google Scholar]
- 28.Reiss, D. E., Quesneville, H., Noaud, D., Andrieu, O. & Anxolabehere, D. (2003) Mol. Biol. Evol. 20, 869–879. [DOI] [PubMed] [Google Scholar]
- 29.Feschotte, C. & Wessler, S. R. (2002) Proc. Natl. Acad. Sci. USA 99, 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaunt, M. W. & Miles, M. A. (2002) Mol. Biol. Evol. 19, 748–761. [DOI] [PubMed] [Google Scholar]
- 31.Sharakhov, I. V., Serazin, A. C., Grushko, O. G., Dana, A., Lobo, N., Hillenmeyer, M. E., Westerman, R., Romero-Severson, J., Costantini, C., Sagnon, N., et al. (2002) Science 298, 182–185. [DOI] [PubMed] [Google Scholar]
- 32.Zdobnov, E. M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R. R., Christophides, G. K., Thomasova, D., Holt, R. A., Subramanian, G. M., et al. (2002) Science 298, 149–159. [DOI] [PubMed] [Google Scholar]
- 33.Fraser, M. J., Ciszczon, T., Elick, T. & Bauser, C. (1996) Insect Mol. Biol. 5, 141–151. [DOI] [PubMed] [Google Scholar]
- 34.Cary, L. C., Goebel, M., Corsaro, B. G., Wang, H. G., Rosen, E. & Fraser, M. J. (1989) Virology 172, 156–169. [DOI] [PubMed] [Google Scholar]
- 35.Smit, A. F. A. (2002) Repbase Rep. 2 (1), 35. [Google Scholar]
- 36.Kapitonov, V. V. & Jurka, J. (2002) Repbase Rep. 2 (6), 21. [Google Scholar]
- 37.Evgen'ev, M. B., Zelentsova, E. S., Shostak, N. G., Kozitsina, M., Barskyi, V., Lankenau, D. H. & Corces, V. G. (1997) Proc. Natl. Acad. Sci. USA 94, 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyatkov, K. I., Shostak, N. G., Zelentsova, E. S., Lyozin, G. T., Melekhin, M. I., Finnegan, D. J., Kidwell, M. G. & Evgen'ev, M. B. (2002) Proc. Natl. Acad. Sci. USA 99, 16150–16155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eickbush, T. & Malik, H. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 1111–1144.
- 40.Smit, A. F. (1999) Curr. Opin. Genet. Dev. 9, 657–663. [DOI] [PubMed] [Google Scholar]
- 41.Pantazidis, A., Labrador, M. & Fontdevila, A. (1999) Mol. Biol. Evol. 16, 909–921. [DOI] [PubMed] [Google Scholar]
- 42.Scheinker, V. S., Lozovskaya, E. R., Bishop, J. G., Corces, V. G. & Evgen'ev, M. B. (1990) Proc. Natl. Acad. Sci. USA 87, 9615–9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malik, H. S. & Eickbush, T. H. (1999) J. Virol. 73, 5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malik, H. S., Henikoff, S. & Eickbush, T. H. (2000) Genome Res. 10, 1307–1318. [DOI] [PubMed] [Google Scholar]
- 45.Petrov, D. A. (2001) Trends Genet. 17, 23–28. [DOI] [PubMed] [Google Scholar]
- 46.Dimitri, P. & Junakovic, N. (1999) Trends Genet. 15, 123–124. [DOI] [PubMed] [Google Scholar]
- 47.Tissier, A. F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M. A., Murphy, G. & Jones, J. D. (1999) Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parinov, S., Sevugan, M., De, Y., Yang, W. C., Kumaran, M. & Sundaresan, V. (1999) Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berezikov, E., Bucheton, A. & Busseau, I. (2000) Genome Biol. 1, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowen, N. J. & McDonald, J. F. (2001) Genome Res. 11, 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.