Abstract
The army ant syndrome of behavioral and reproductive traits (obligate collective foraging, nomadism, and highly specialized queens) has allowed these organisms to become the premiere social hunters of the tropics, yet we know little about how or why these strategies evolved. The currently accepted view holds that army ants evolved multiple times on separate continents. I generated data from three nuclear genes, a mitochondrial gene, and morphology to test this hypothesis. Results strongly indicate that the suite of behavioral and reproductive adaptations found in army ants throughout the world is inherited from a unique common ancestor, and did not evolve convergently in the New World and Old World as previously thought. New Bayesian methodology for dating the antiquity of lineages by using a combination of fossil and molecular information places the origin of army ants in the mid-Cretaceous, consistent with a Gondwanan origin. Because no known army ant species lacks any component of the army ant syndrome, this group represents an extraordinary case of long-term evolutionary stasis in these adaptations.
Ever since the original sensational reports by early naturalists of marauding swarms of army ants, these organisms have fascinated animal behaviorists (1), ecologists, social insect biologists (2, 3), conservation biologists (4), and, most recently (5), artificial intelligence researchers. Despite substantial progress in these fields, our understanding of army ant evolution remains poor. Army ant behavioral and reproductive adaptations, among the most spectacular in the animal kingdom, have allowed these organisms to become dominant predators throughout the world's tropics. Yet, we know little about the evolutionary origin and diversification of army ants. This ignorance also hampers interpretation in studies comparing behaviors in army ants living on different continents.
Army ants possess a syndrome of behavioral and reproductive traits, which includes obligate collective foraging, nomadism, and highly modified queens (1, 3, 6). Army ants never hunt or forage solitarily. In contrast to most other ant species, which first send out individual scouts to find food sources and only later recruit others from the colony (7), army ants instead dispatch a mass of cooperative, leaderless foragers to locate and overwhelm prey simultaneously. Army ants also periodically emigrate to new foraging locales and do not construct permanent nests. Their robust queens are permanently wingless and have abdomens capable of pronounced expansion during egg production (i.e., dichthadiigyny). This condition facilitates massive reproductive pulses of up to 3–4 million eggs per month in some species (8) and often results in synchronized brood cycles and colonies with millions of individuals descendant from a single queen. No species of army ant has ever been found to lack any of these three traits.
A fundamental gap in our knowledge of army ant biology is the question of whether this army ant syndrome of behavioral and reproductive traits resulted from a unique set of evolutionary events, or instead, evolved convergently in multiple lineages. Army ants constitute three well defined taxonomic subfamilies (9), two restricted exclusively to the Old World (Aenictinae and Dorylinae) and the other to the New World (Ecitoninae). The prevailing view holds that the army ant syndrome originated several times in independent lineages restricted to the New World and Old World, respectively (3, 10). This polyphyly hypothesis, widely cited in the literature, is founded primarily on the assumption that army ants originated after the breakup of Gondwana, and thus must have evolved independently on separate continents. If true, this would imply multiple origins of army ant behavioral and reproductive adaptations, with their similarities due to convergence.
I assessed the validity of the polyphyly hypothesis by using a combination of genetic, morphological, and fossil data. Members of the subfamily Cerapachyinae have long been implicated as the nearest relatives to army ants (3, 9, 11, 12). I tested whether lineages of army ants evolved from multiple cerapachyine ancestors by generating phylogenetic data from five independent data sources, including three nuclear genes (18S rDNA, 28S rDNA, and wingless), a mitochondrial gene (cytochrome oxidase I), and morphology. To test whether New World and Old World army ants diverged after Gondwanan fragmentation, as the polyphyly hypothesis also predicts, I incorporated fossil information from nine ant taxa together with molecular phylogenetic data in a Bayesian framework, which does not assume a molecular clock.
Materials and Methods
Taxon Sampling. Previous morphological work (9) overwhelmingly supports the monophyly of each separate army ant subfamily. For this article, I sampled widely within these groups. I included both Asian and African representatives from the Old World army ant taxa Aenictus and Dorylus, the sole genera of Aenictinae and Dorylinae, respectively. This sampling involves four of the five established species-groups in Aenictus (13) and five of the six currently recognized subgenera of Dorylus, including D. (Dichthadia) laevigatus, which is the sister species to all other members of the genus when all six subgenera are considered (unpublished data). The 14 species included from the New World subfamily Ecitoninae span all five genera. Within Cerapachyinae, I included exemplars from all constituent genera (Acanthostichus, Cerapachys, Cylindromyrmex, Simopone, and Sphinctomyrmex). The genus Cerapachys, by far the largest and most heterogeneous cerapachyine genus, is represented by multiple species from both principle subgroups, the “Cerapachys lineage” and “Phyracaces lineage” (11). Members from four other ant subfamilies (Dolichoderinae, Formicinae, Myrmeciinae, and Ponerinae) served as outgroups.
Data Collection. I generated 3538 base pairs of DNA sequence data from cytochrome oxidase I (COI), 18S rDNA, 28S rDNA, wingless genes, and 116 morphological characters. PCR amplification consisted of 35–40 cycles of 1 min at 94°C, 1 min of an annealing step whose temperature varied according to the primers used, and 1 min 30 sec at 72°C, with an initial denaturation step of 2 min at 94°C and a final extension step of 7 min at 72°C. Temperatures for the annealing step were as follows: COI primers, 45–48°C; 18S and 28S primers, 47–54°C; and wingless primers, 54–58°C. A concentration of 1.5 mM (18S and 28S), 2.0–2.5 mM (COI), or 3.0 mM (wingless) of MgCl2 was used in a final reaction volume of 25 μl. Primer information is available in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. PCR product was purified by using either Microcon 100 microconcentrators (Amicon) or the enzymatic method ExoSap-IT (United States Biochemical). Automated fluorescent dye sequencing reactions were conducted on an Applied Biosystems Prism 377 DNA sequencer (Perkin–Elmer). Both strands were sequenced in all cases. The morphological characters and data matrix are presented in Supporting Text and Table 3, which are published as supporting information on the PNAS web site.
Phylogenetic Analysis. All taxa included in the combined analysis of morphological and molecular data contained complete genetic data except Cerapachys cf. mayri and Dorylus laevigatus (both missing part of 18S); Cerapachys sp. cribrinodis complex, Cerapachys ?kenyensis, and Dorylus sp. (all three missing wingless). A χ2 test of base composition homogeneity across taxa did not reject stationarity of nucleotide base composition among lineages (χ2 = 109.80; d.f. = 123, P = 0.800). The 18S and 28S data sets were aligned with CLUSTAL W (14) using the default parameters, and 265 ambiguously aligned sites were removed before all analyses. Unless stated otherwise, all phylogenetic analyses were conducted by using PAUP* Version 4.0b10 (15). The incongruence length difference (ILD; ref. 16) test was used to quantify character incongruence between data partitions. I ran each test after removing constant and uninformative characters (17, 18) by using 500 repartitions, each consisting of 10 random addition replicates of tree-bisection-reconnection (TBR) branch swapping. The ILD test did not indicate significant heterogeneity between morphological and molecular characters (P = 0.978), nor between partitions within the molecular data (wingless vs. COI, P = 0.916; wingless vs. 18S + 28S, P = 0.116; and 18S + 28S vs. COI, P = 0.418). Recent studies (19–21) have illustrated problems with the ILD in assessing topological incongruence and I did not use this test as a criterion for combining data partitions. However, robust statements of relationships, as judged by bootstrap analyses of individual data sets, are highly concordant among data partitions (results not shown), supporting the rationale behind combining these data. The phylogeny of the combined morphological and molecular data set was inferred under maximum parsimony. Tree searches were conducted by using 100 random addition replicates of TBR branch swapping and no limit to MAXTREES. All characters received equal weight. Branch support was assessed by using 1,000 pseudoreplicates of the nonparametric bootstrap (22).
To generate a topology for the divergence dating analysis, I conducted maximum likelihood (ML) analysis by using molecular data from the same four genes for 49 taxa. All included taxa contained complete sequences for all gene segments used (COI, 18S, 28S, and wingless) except for 10 taxa missing COI (see Table 2). I used MODELTEST 3.06 (23) to initially estimate ML values under 56 different substitution models, which were then subjected to hierarchical likelihood ratio (LR) tests to determine the most appropriate model (24). The selected model was a general time-reversible model with γ-distributed rate heterogeneity and a proportion of invariant sites (GTR + G + I), which was used to infer the ML tree after multiple rounds of TBR searches. An LR test (25) rejected rate consistency among lineages, i.e., rejected a treewide molecular clock (–2 ln LR = 309.606; d.f. = 47; P < 0.005). Bayesian posterior probabilities of clades were estimated under a general time-reversible model with site-specific rates among genes and codon positions by using the program MRBAYES 2.0 (26). I performed multiple searches starting from different random trees to ensure convergence of Markov chain Monte Carlo runs; each run consisted of four simultaneous chains run for 1,000,000 cycles, sampled every 100 cycles, with a burnin of 100,000 cycles.
Divergence Date Analysis. Using the inferred ML topology, I estimated branch lengths and divergence dates with a computer program package (ESTBRANCHES and MULTIDIVTIME) that implements the Bayesian divergence dating method developed by Thorne and coworkers (27–29). This method allows for rate variation both among genes and among lineages. Branch lengths were estimated separately for each of three gene partitions (18S + 28S, wingless, and COI) by using an F84 + γ model (complexity of the model is limited by the dating program) with Apis mellifera as the outgroup (GenBank accession nos. U89834, X89529, AF214668, AJ307465, and AY222546). Placement of the root was accomplished using three additional vespid out-groups and a modified 18S/28S/morphological data set. To infer posterior values for divergence times, I established 120 million years ago (Mya) (160–92 Mya) as the mean and range for the prior date of the root node; this time span encompasses all realistic dates for the origin of ants (30–32). I also constrained nine internal nodes in the tree with minimum ages based on the fossil record (see legend to Fig. 2 for details). I generated the prior on the rate at the root node by using the penalized likelihood approach provided by the program R8S (33). In conducting the Bayesian divergence dating analysis, I performed multiple searches starting from different random parameter values, with each run consisting of an initial burnin of 10,000 cycles followed by 100,000 cycles sampled every 100 times.
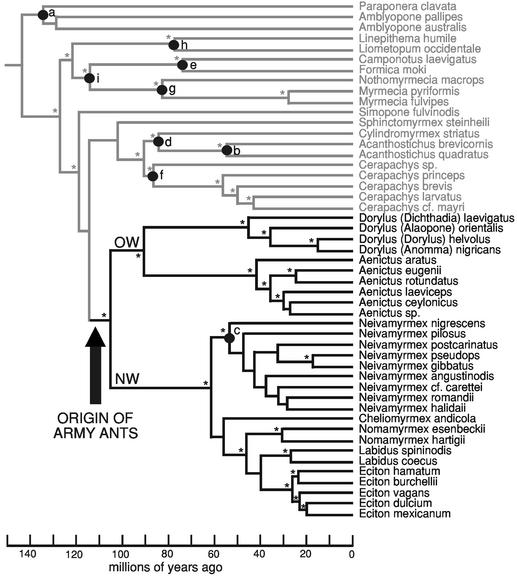
Fig. 2.
Bayesian divergence dating analysis. NW, New World; OW, Old World. Divergence dates were estimated on the ML phylogeny derived from COI, 18S rDNA, 28S rDNA, and wingless genes (–ln L = 26603.88301). Clades marked with asterisks had a posterior probability of >95% after independent Bayesian phylogenetic analysis. Lowercase letters at nodes indicate minimum age constraints obtained from the fossil record: a–c, 20 Mya (34, 35); d, 25 Mya (36, 37); e–f, 42 Mya (38); g, 50 Mya (39); h, 65 Mya (40); i, 92 Mya (32). Army ant taxa are shown in thick type. Branch lengths are drawn scaled to estimated mean values of absolute time. The origin of army ants is estimated at 105 Mya (±11 SD).
Results and Discussion
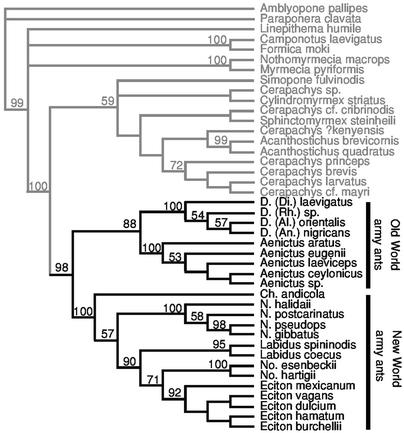
Phylogenetic analysis of the combined molecular and morphological data (Fig. 1) reveals robust support for army ant monophyly (98% bootstrap). This result is corroborated by ML and Bayesian (99% posterior value) analyses of molecular data alone (Fig. 2). A single origin of army ants implies that they inherited their behavioral and reproductive adaptations from a unique common ancestor, and that these traits evolved only once in this group.
Fig. 1.
Phylogenetic relationships among army ants and cerapachyine relatives. Al., Alaopone; An., Anomma; Ch., Cheliomyrmex; D., Dorylus; Di., Dichthadia; N., Neivamyrmex; No., Nomamyrmex; and Rh., Rhogmus. The phylogeny is the strict consensus of the two most parsimonious trees (length = 5,592, consistency index = 0.313, retention index = 0.536) resulting from equal weights analysis of 3,654 characters from COI, 18S rDNA, 28S rDNA, wingless, and morphological data. Numbers above branches indicate clades with >50% bootstrap support. Army ant taxa are shown in thick type.
The majority of contemporary army ant species is found in the African and American tropics, suggesting a Gondwanan origin. New World and Old World army ant taxa form robust sister groups (Fig. 1), reinforcing the notion that the breakup of Gondwana into its South American and African constituents caused this dichotomy. I conducted Bayesian dating analysis to test whether the date for army ant origination is consistent with this tectonic event. This analysis (Fig. 2) estimates the age for the most recent common ancestor of the Old World and New World army ant lineages at 105 Mya (±11 SD). This date is remarkably congruent with the geological timing of the complete separation between Africa and South America, which occurred ≈100 Mya (41–43).
Both phylogenetic and divergence dating analyses provide results that strongly conflict with the polyphyly hypothesis, and instead support a single origin of army ants. Although Brown (11) proposed that two or possibly three army ant lineages evolved from separate cerapachyine ancestors, he was unable to identify any putative sister groups within Cerapachyinae to support this view. Similarly, Gotwald (3, 10) hypothesized that army ants evolved on three separate occasions, with Ecitoninae arising in situ in South America, Dorylus in Africa, and Aenictus in Asia. This argument was not based on a phylogenetic hypothesis, but rather, depended on a postulated date for the origin of ants in the early Tertiary or late Cretaceous, thus concluding that army ant lineages must have arose after the breakup of Gondwana. Recent fossil discoveries, however, implicate an early Cretaceous origin of ants (30–32, 44), now making Gondwanan origins for major ant taxa a legitimate possibility.
Army ant distribution, relationships, and divergence dates all suggest a pattern of Gondwanan diversification. The population biology of army ants predisposes them to such vicariant diversification, which involves isolation by physical barriers of formerly continuously distributed taxa. Army ant species ranges can be quite broad over continuous land masses. For example, some individual species within four of the five genera of Ecitoninae are distributed continuously from the southern United States to Argentina (45). Jump dispersal across substantial barriers, however, appears quite difficult for army ant colonies. All army ant queens remain flightless throughout their lives and found new colonies by splitting from a mature colony and traveling on the ground with a large retinue of workers to a nearby location. This mode of colony foundation makes long-distance dispersal across significant ocean barriers highly unlikely, a conclusion bolstered by the absence of army ants from islands such as Madagascar (46), Polynesia (47), and Cuba (48). Limited dispersal capability also contributes to their acute sensitivity to habitat fragmentation, as pointed out by many conservation biologists (4, 49–51).
Myrmecologists have noted that a few species outside the three army ant subfamilies also engage in group predation, forage in an army ant-like fashion, frequently change nest sites, or have dichthadiiform queens (Table 1). These observations have motivated statements that such species should also be considered army ants, which might qualify any conclusions regarding a unique origin of the army ant syndrome. Group foraging, however, can be used to describe other foraging strategies such as slave raiding or mass recruitment to food sources previously located by individual scouts. This latter behavior is common among ants with large colony sizes (7). By contrast, the quality that sets army ants apart from virtually all other ant species is obligate collective foraging. Army ants never scout or forage on an individual basis; rather, the location, conquest, and retrieval of prey is always conducted by leaderless groups (1, 3, 73). The fundamental inability of army ant foragers to operate individually is reflected in the phenomenon of circular milling (74), whereby a disturbance of their pheromonal communication results in a mass of foragers literally running around in a circle, each ant slavishly following its predecessor, until they all expire. Although nomadism and dichthadiiform queens also occur in a few other ant taxa, the only ants known conclusively to possess these two traits, together with obligate collective foraging, are army ants (Table 1).
Table 1. Ant taxa reported to display some army ant-like traits.
| Taxon | Obligate collective foraging | Nomadism | Dichthadiigyny | References |
|---|---|---|---|---|
| Army ants (Aenictinae, Dorylinae, Ecitoninae) | Yes | Yes | Yes | 1, 2, 3 |
| Cerapachys spp. | No | Possibly | No | 52, 53, 54 |
| Leptanilla japonica | No | Yes | Yes | 55 |
| Leptogenys distinguenda, Leptogenys nitida, and several other Leptogenys spp. | Yes | Yes | No | 6, 56, 57, 58 |
| Other Leptogenys spp. | No | Yes | No | 56, 59, 60 |
| Onychomyrmex spp. | ? | Yes ? | Yes | 61, 62, 63 |
| Pachycondyla analis, Pachycondyla commutata, Pachycondyla laevigata | No | Yes | No | 64, 65, 66 |
| Pheidologeton diversus, Pheidologeton silenus | Yes | No | No | 67, 68, 69 |
| Simopelta spp. | Yes ? | ? | Yes | 70, 71 |
| Sphinctomyrmex spp. | No | Yes ? | Yes (partial) | 11, 72 |
Only army ants are known to possess all three components. A question mark indicates uncertainty due to lack of information.
Hence, among ant species studied to date, only the three army ant subfamilies (Aenictinae, Dorylinae, and Ecitoninae) can be said to possess the army ant syndrome. The possibility, of course, cannot be ruled out that poorly studied groups may share this syndrome. A few cryptic and geographically restricted species in the ponerine genera Onychomyrmex and Simopelta remain contenders, and definitive studies on these ants are badly needed. Even allowing the possibility of future discovery of the army ant syndrome in these or other ants, only in army ants would this adaptive strategy have evolved on such a grand ecological and biogeographical scale. Furthermore, not only is the full integration of these traits apparently unique to army ants, but none of their several hundred known species lacks any of these three components.
The previous hypothesis of a polyphyletic origin of army ants was based on what was then known of the ant fossil record and subjective impressions about the rate of ant diversification, without supporting phylogenetic data. The current study provides robust phylogenetic evidence and divergence date estimates that instead support a monophyletic origin for army ants. Obligate collective foraging, nomadism, and dichthadiigyne queens evolved together only once in the history of life, ≈100 Mya in the ancestral army ant. These roving army ant colonies became the premiere collective hunters of the tropics, capturing prey typically unavailable to other insects: social wasps, large arthropods, and even small vertebrates, but at the cost of requiring expansive, contiguous foraging ranges. After these adaptations became fully integrated into the lifestyle of army ants, no extant lineage subsequently lost any of these traits, suggesting that extreme specialization has prevented the evolution of alternative strategies. This is perhaps the most striking case of long-term evolutionary stasis in the behavior of a social insect, with the exception of the entrenchment of eusociality itself. Future work must determine whether ant lineages that show some, but not all, components of this adaptive syndrome display similar evolutionary canalization.
Supplementary Material
Acknowledgments
I thank Phil Ward for his guidance throughout all stages of this project; Michael Sanderson, who provided molecular lab facilities used to gather some of these data; all those who provided specimens for molecular work, especially Stephanie Berghoff, Brian Fisher, Steve Heydon, John Lattke, Andy Suarez, Phil Ward, Alex Wild, and Seiki Yamane; Bryan Danforth, Laura Harrington, Michael Sanderson, H. Bradley Shaffer, Phil Ward, and two anonymous reviewers, who commented on earlier versions of this manuscript. The following curators provided access to museum collections and/or loans of material: Barry Bolton (British Museum of Natural History, London), Roberto Brandão (Museu de Zoologia da Universidade de São Paulo, São Paulo, Brazil), Stefan Cover (Museum of Comparative Zoology, Harvard University, Cambridge, MA), William Gotwald (personal collection), Roy Snelling (Los Angeles County Museum, Los Angeles), and Steve Shattuck (Australian National Insect Collection, Canberra City, Australian Capital Territory, Australia). This work was supported by the University of California (Davis, CA) and National Science Foundation Grants INT-9904233 (to P. S. Ward and S.G.B.) and DEB-9903650 (to P. S. Ward).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mya, million years ago; COI, cytochrome oxidase; ML, maximum likelihood.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF398151, AF398162, AY218304–AY218321, AY218336–AY218353, and AY233467–AY233725).
References
- 1.Schneirla, T. C. (1971) Army Ants: A Study in Social Organization (Freeman, San Francisco).
- 2.Hölldobler, B. & Wilson, E. O. (1990) The Ants (Harvard Univ. Press, Cambridge, MA).
- 3.Gotwald, W. H., Jr. (1995) Army Ants: The Biology of Social Predation (Cornell Univ. Press, Ithaca, NY).
- 4.Boswell, G. P., Franks, N. R. & Britton, N. F. (2000) in Behaviour and Conservation, eds. Gosling, L. M. & Sutherland, W. J. (Cambridge Univ. Press, Cambridge, U.K.), pp. 141–158.
- 5.Camazine, S., Deneubourg, J.-L., Franks, N. R., Sneyd, J., Theraulaz, G. & Bonabeau, E. (2001) Self-Organization in Biological Systems (Princeton Univ. Press, Princeton).
- 6.Wilson, E. O. (1958) Evolution (Lawrence, Kans.) 12, 24–36. [Google Scholar]
- 7.Beckers, R., Goss, S., Deneubourg, J.-L. & Pasteels, J. M. (1989) Psyche 96, 239–256. [Google Scholar]
- 8.Raignier, A. & van Boven, J. K. A. (1955) Ann. Mus. R. Congo Belge Nouv. Ser. Quarto Sci. Zool. 2, 1–359. [Google Scholar]
- 9.Bolton, B. (1990) J. Nat. Hist. 24, 1339–1364. [Google Scholar]
- 10.Gotwald, W. H., Jr. (1979) Ann. Entomol. Soc. Am. 72, 462–467. [Google Scholar]
- 11.Brown, W. L., Jr. (1975) Search Agric. 5, 1–116. [Google Scholar]
- 12.Bolton, B. (1990) J. Nat. Hist. 24, 53–68. [Google Scholar]
- 13.Wilson, E. O. (1964) Pac. Insects 6, 427–483. [Google Scholar]
- 14.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swofford, D. L. (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b10.
- 16.Farris, J. S., Källersjö, M., Kluge, A. G. & Bult, C. (1995) Cladistics 10, 315–319. [Google Scholar]
- 17.Cunningham, C. W. (1997) Mol. Biol. Evol. 14, 733–740. [DOI] [PubMed] [Google Scholar]
- 18.Lee, M. S. Y. (2001) Mol. Biol. Evol. 18, 676–680. [DOI] [PubMed] [Google Scholar]
- 19.Dolphin, K., Belshaw, R., Orme, C. D. L. & Quicke, D. L. J. (2000) Mol. Phylogenet. Evol. 17, 401–406. [DOI] [PubMed] [Google Scholar]
- 20.Barker, F. K. & Lutzoni, F. M. (2002) Syst. Biol. 51, 625–637. [DOI] [PubMed] [Google Scholar]
- 21.Darlu, P. & Lecointre, G. (2002) Mol. Biol. Evol. 19, 432–437. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein, J. (1985) Evolution (Lawrence, Kans.) 39, 783–791. [DOI] [PubMed] [Google Scholar]
- 23.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 24.Posada, D. & Crandall, K. A. (2001) Syst. Biol. 50, 580–601. [PubMed] [Google Scholar]
- 25.Felsenstein, J. (1988) Annu. Rev. Genet. 22, 521–565. [DOI] [PubMed] [Google Scholar]
- 26.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 27.Thorne, J. L., Kishino, H. & Painter, I. S. (1998) Mol. Biol. Evol. 15, 1647–1657. [DOI] [PubMed] [Google Scholar]
- 28.Kishino, H., Thorne, J. L. & Bruno, W. J. (2001) Mol. Biol. Evol. 18, 352–361. [DOI] [PubMed] [Google Scholar]
- 29.Thorne, J. L. & Kishino, H. (2002) Syst. Biol. 51, 689–702. [DOI] [PubMed] [Google Scholar]
- 30.Grimaldi, D., Agosti, D. & Carpenter, J. M. (1997) Am. Mus. Novit. 3208, 1–43. [Google Scholar]
- 31.Rust, J. & Andersen, N. M. (1999) Zool. J. Linn. Soc. 125, 331–348. [Google Scholar]
- 32.Grimaldi, D. & Agosti, D. (2000) Proc. Natl. Acad. Sci. USA 97, 13678–13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderson, M. J. (2002) Mol. Biol. Evol. 19, 101–109. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, E. O. (1985) Science 229, 265–267. [DOI] [PubMed] [Google Scholar]
- 35.de Andrade, M. L. (1998) Mitt. Schweiz. Entomol. Ges. 71, 269–274. [Google Scholar]
- 36.de Andrade, M. L. (1998) Rev. Suisse Zool. 105, 581–664. [Google Scholar]
- 37.de Andrade, M. L. (2001) Beitr. Entomol. 51, 51–63. [Google Scholar]
- 38.Dlussky, G. M. (1997) Paleontol. J. 31, 616–627. [Google Scholar]
- 39.Baroni Urbani, C. (2000) Eclogae Geol. Helv. 93, 471–480. [Google Scholar]
- 40.Dlussky, G. M. (1999) Paleontol. Zh. 4, 73–76. [Google Scholar]
- 41.Parrish, J. C. (1993) in The Africa-South America Connection, eds. George, W. & Lavocat, R. (Clarendon, Oxford), pp. 8–27.
- 42.Pitman, W. C. I., Cande, S., Labrecque, J. & Pindell, J. (1993) in Biological Relationships Between Africa and South America, ed. Goldblatt, P. (Yale Univ. Press, New Haven, CT), pp. 15–34.
- 43.McLoughlin, S. (2001) Aust. J. Bot. 49, 271–300. [Google Scholar]
- 44.Dlussky, G. M. (1999) Paleontol. Zh. 3, 62–66. [Google Scholar]
- 45.Watkins, J. F., II. (1976) The Identification and Distribution of New World Army Ants (Dorylinae: Formicidae) (Baylor Univ. Press, Waco, TX).
- 46.Fisher, B. L. (1997) J. Nat. Hist. 31, 269–302. [Google Scholar]
- 47.Morrison, L. W. (1996) Ecography 19, 73–84. [Google Scholar]
- 48.Fontenla Rizo, J. L. (1997) Cocuyo (Havana) 6, 18–21. [Google Scholar]
- 49.Partridge, L. W., Britton, N. F. & Franks, N. R. (1996) Proc. R. Soc. London Ser. B 263, 735–741. [Google Scholar]
- 50.Terborgh, J., Lopez, L., Tello, J., Yu, D. & Rita Bruni, A. (1997) in Tropical Forest Remnants: Ecology, Management, and Conservation of Fragmented Communities, eds. Laurance, W. F. & Bierregaard, R. O., Jr. (Univ. of Chicago Press, Chicago), pp. 256–274.
- 51.Suarez, A. V., Bogler, D. T. & Case, T. J. (1998) Ecology 79, 2041–2056. [Google Scholar]
- 52.Wilson, E. O. (1958) Insectes Soc. 5, 129–140. [Google Scholar]
- 53.Hölldobler, B. (1982) Psyche 89, 3–23. [Google Scholar]
- 54.Ravary, F. & Jaisson, P. (2002) Insectes Soc. 49, 114–119. [Google Scholar]
- 55.Masuko, K. (1990) Insectes Soc. 37, 31–57. [Google Scholar]
- 56.Maschwitz, U., Steghaus-Kovac, S., Gaube, R. & Hänel, H. (1989) Behav. Ecol. Sociobiol. 24, 305–316. [Google Scholar]
- 57.Duncan, F. D. & Crewe, R. M. (1994) Oecologia 97, 118–123. [DOI] [PubMed] [Google Scholar]
- 58.Witte, V. & Maschwitz, U. (2000) Insectes Soc. 47, 76–83. [Google Scholar]
- 59.Maschwitz, U. & Schönegge, P. (1983) Oecologia 57, 175–182. [DOI] [PubMed] [Google Scholar]
- 60.Dejean, A. & Evraerts, C. (1997) J. Insect Behav. 10, 177–191. [Google Scholar]
- 61.Wheeler, W. M. (1916) Bull. Mus. Comp. Zool. 60, 45–54. [Google Scholar]
- 62.Brown, W. L., Jr. (1960) Bull. Mus. Comp. Zool. 122, 145–230. [Google Scholar]
- 63.Hölldobler, B., Palmer, J. M., Masuko, K. & Brown, W. L., Jr. (1989) Zoomorphology 108, 255–261. [Google Scholar]
- 64.Longhurst, C. & Howse, P. E. (1979) Insectes Soc. 26, 204–215. [Google Scholar]
- 65.Longhurst, C., Baker, R. & Howse, P. E. (1979) J. Chem. Ecol. 5, 703–725. [Google Scholar]
- 66.Hölldobler, B. & Traniello, J. F. A. (1980) J. Chem. Ecol. 6, 883–893. [DOI] [PubMed] [Google Scholar]
- 67.Moffett, M. W. (1988) in Advances in Myrmecology, ed. Trager, J. C. (E. J. Brill, Leiden, The Netherlands), pp. 355–370.
- 68.Moffett, M. W. (1988) Ann. Entomol. Soc. Am. 81, 356–361. [Google Scholar]
- 69.Moffett, M. W. (1988) J. Insect Behav. 1, 309–331. [Google Scholar]
- 70.Borgmeier, T. (1950) Rev. Entomol. 21, 369–380. [Google Scholar]
- 71.Gotwald, W. H., Jr., & Brown, W. L., Jr. (1967) Psyche 73, 261–277. [Google Scholar]
- 72.Buschinger, A., Peeters, C. & Crozier, R. H. (1989) Psyche 96, 287–300. [Google Scholar]
- 73.Rettenmeyer, C. W. (1963) Univ. Kans. Sci. Bull. 44, 281–465. [Google Scholar]
- 74.Schneirla, T. C. (1944) Am. Mus. Novit. 1253, 1–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.