Abstract
RGS (regulator of G protein signaling) proteins containing the G protein γ-like (GGL) domain (RGS6, RGS7, RGS9, and RGS11) interact with the fifth member of the G protein β-subunit family, Gβ5. This interaction is necessary for the stability of both the RGS protein and for Gβ5. Consistent with this notion, we have found that elevation of RGS9-1 mRNA levels by transgene expression does not increase RGS9-1 protein level in the retina, suggesting that Gβ5 levels may be limiting. To examine further the interactions of Gβ5 and the GGL domain-containing RGS proteins, we inactivated the Gβ5 gene. We found that the levels of GGL domain-containing RGS proteins in retinas and in striatum are eliminated or reduced drastically, whereas the levels of Gγ2 and RGS4 proteins remain normal in the absence of Gβ5. The homozygous Gβ5 knockout (Gβ5–/–) mice derived from heterozygous knockout mating are runty and exhibit a high preweaning mortality rate. We concluded that complex formation between GGL domain-containing RGS proteins and the Gβ5 protein is necessary to maintain their mutual stability in vivo. Furthermore, in the absence of Gβ5 and all four RGS proteins that form protein complexes with Gβ5, the animals that survive into adulthood are viable and have no gross defects in brain or retinal morphology.
First discovered functionally as negative regulators of G protein signaling in Saccharomyces cerevisiae (Sst2p) (1) and Caenorhabditis elegans (EGL10) (2), RGS (regulator of G protein signaling) proteins accelerate the hydrolysis of GTP by the α-subunits of heterotrimeric G proteins (3). They form a large gene family with a diagnostic ≈120-aa RGS domain in which the GTPase-accelerating activity resides (4). In addition to the RGS domain, most RGS proteins possess additional domains that enable them to interact with other cellular signaling molecules (5). A subgroup of the RGS family, namely RGS9, RGS11, RGS7, and RGS6, possesses a G γ-like (GGL) domain that binds the fifth member of the heterotrimeric G protein β-subunit (Gβ5) both in vitro and in vivo (6, 7, 8, 9, 10). There are five known members of the G protein β-subunit family (11, 12). The first four members, Gβ1–4, are highly similar, sharing 80–90% sequence identity. Gβ5 is the most divergent member of this family, sharing only 50% sequence identity with Gβ1–4. Gβ5 exists in two forms: the common, short-splice form (Gβ5-S) and a unique, long-splice form (Gβ5-L) that exists exclusively in retinal photoreceptors. The long form results from the addition of an N-terminal exon through alternative splicing (11). Gβ5 complexes with either GGL domain-containing RGS proteins or with certain G protein γ-subunits such as Gγ2 (12, 13). Likewise, the GGL domain-containing RGS proteins not only interact with Gβ5, they also can interact with other proteins such as polycystin (for RGS7) (14) and SCG10 (for RGS6) (15). Interestingly, these RGS proteins do not interact with Gβ1–4, indicating that their interaction with Gβ5 is selective and may be important for their in vivo function (6, 16). In C. elegans, the Gβ5 ortholog, GPB-2, is required for the activity of the two GGL domain-containing RGS proteins, EGL-10 and EAT-16 (17, 18, 19). GPB-2 protein level is reduced significantly in eat-16; egl-10 double-mutants. Similarly, the EGL-10 protein is diminished severely in gpb-2 mutants (17). These data indicate that in worms the interactions between GPB-2 and EGL-10 or EAT-16 confer mutual stability to these proteins.
In retinal photoreceptors, the complex of Gβ5 and RGS9-1 modulates the intrinsic GTPase activity of transducin (20, 21). In mice lacking functional RGS9 genes (RGS9–/– mice), the recovery phase of both rod and cone phototransduction is prolonged abnormally because of slowed GTP hydrolysis by rod and cone transducin (22, 23). RGS9 exists in two forms through alternative splicing: a 55-kDa retinal-specific form, RGS9-1, containing a unique C-terminal domain, and an 81-kDa striatal form, RGS9-2, possessing a different and longer C-terminal domain (24, 25). The Gβ5-L protein disappears in RGS9–/– retinas but the Gβ5-L mRNA level is normal, indicating that RGS9-1 is required to maintain a normal Gβ5-L protein level in vivo (22). However, the level of the short Gβ5-S in the striatum appears normal despite the absence of RGS9-2 in the RGS9–/– mice (Fig. 1). The specific loss of Gβ5-L in RGS9–/– mice may be attributed to its exclusive expression in retinal photoreceptors. Interestingly, the RGS9-1/Gβ5-L complex is present more abundantly in cones than in rods (26, 27). Such a difference has been suggested to account, at least in part, for the faster cone-flash responses. In this report, we have overexpressed the RGS9-1 mRNA in mouse retinal photoreceptors and found that elevation of the RGS9-1 mRNA level did not increase the RGS9-1 protein level. In addition, we demonstrated that Gβ5 is required to maintain the in vivo levels of GGL domain-containing RGS proteins but not regular Gγ-subunits or RGS protein without a GGL domain. These data establish in vivo that Gβ5 and GGL domain-containing RGS proteins are obligate partners and support the notion that Gβ5 functions as a component of the GAP (GTPase-accelerating protein) complex, rather than as the β-subunit component of heterotrimeric G proteins.
Fig. 1.
The presence of Gβ5-S in the retina and striatum of RGS9–/– mice. Retinal extracts (20 μg) and striatal extracts (CPu, 100 μg) from wild-type and RGS9–/– mice (indicated, Top) were resolved by 12% SDS/PAGE, and the presence of Gβ5 and RGS9 spliced forms were visualized by Western blots by using generic RGS9 antibody (R4432) and Gβ5 antibody (CT-215). Molecular mass markers (top to bottom: 124, 81, 50, and 42 kDa) are shown on the left. The splice variants of RGS9 and Gβ5 are indicated by arrows (top to bottom, RGS9-2, RGS9-1, Gβ5-L, and Gβ5-S).
Materials and Methods
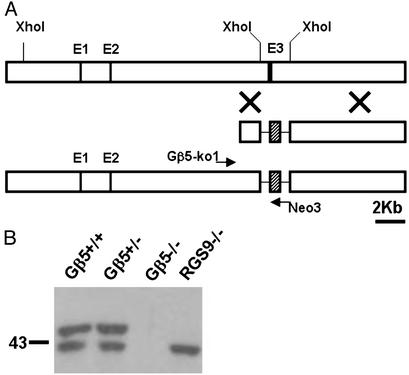
Gene Targeting. All experimental procedures involving the production and use of laboratory mice complied with National Institutes of Health guidelines as approved by the Institutional Animal Care and Use Committee of the California Institute of Technology and the University of Utah. Standard procedures were used to generate Gβ5–/– mice (28). Briefly, the mouse Gβ5 gene was inactivated by replacing a 2.7-kb XhoI genomic fragment encompassing the third exon of the Gβ5 gene with a neomycin (Neo)-resistant marker as shown in Fig. 2 A. The genomic fragments flanking the Neo marker for driving homologous recombination were 1.5 and 7.4 kb in length. The targeting construct, pCKC-Gβ5KO-direct, was introduced into mouse CJ-7 embryonic stem cells by electroporation and selected with G418 for 7 days. Cells in which homologous recombination had occurred were identified by the presence of a 2.1-kb PCR product by using primers Gβ5KO1: 5′-ACA GTC CTA ATG GCC CAG GTG and Neo3: 5′-CGA GGA TCT CGT CGT GAC CCA. The PCR conditions used were 94°C for 3 min followed by 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 3 min, followed by a final extension of 10 min at 72°C. The targeted embryonic stem cells were injected into blastocysts derived from C57BL/6 strain, and the chimeric mice were mated with C57BL/6 or 129SvEv mice to generate heterozygous knockout (Gβ5+/–) mice in both outbred and inbred genetic backgrounds, respectively. The outbred and inbred homozygous knockout (Gβ5–/–) mice were produced by intercross of the Gβ5+/– mice within their respective genetic backgrounds.
Fig. 2.
The generation of Gβ5–/– mice. (A) Gβ5 gene-inactivation scheme. (Top) The first half of the mouse Gβ5 gene. (Middle) Targeting construct replacing a 2.7-kb XhoI fragment with a Neo marker (hatched block). (Bottom) The targeted locus can be identified by the presence of a 2.1-kb PCR fragment when amplified by using Neo3 and Gβ5-ko1 primers. (B) Western blotting of 20 μg of retinal extracts by using antibody CT-215 to show the presence of Gβ5 proteins in various genetic backgrounds. The long-spliced form, Gβ5-L, is absent in RGS9–/– mouse retina, whereas both long and short forms of Gβ5 are absent in Gβ5–/– mice.
Transgenic Mice. To express RGS9-1 in mouse photoreceptors, the coding region of the RGS9-1 cDNA was cloned downstream of a 4-kb mouse opsin promoter fragment in the pRho4.2 vector and upstream of the mouse protamine polyadenylation signal by using ClaI/BamHI sites (Fig. 6A). The 4-kb mouse opsin promoter fragment has been used extensively in transgenic studies to drive photoreceptor specific gene expression (29, 30, 31, 32). The 6-kb KpnI/XbaI DNA fragment containing the promoter, the RGS9-1-coding region, and the polyadenylation signal was injected into B6D2 F1 embryos. The founder mice, TG9F2 and TG9F10, were identified by the presence of a 330-bp PCR product by using primers 3ms9: 5′-GGC GTC TGA AAT CGG TAG AGA and RGS9-1: 5′-ATG ACG ATC CGA CAC CAA GGC. The PCR conditions used were as follows: 94°C for 3 min followed by 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, and a final extension period of 10 min at 72°C. To generate TG9F2–/– and TG9F10–/– mice, the TG9F2 and TG9F10 mice were crossed into the RGS9–/– background through two generations of breeding. The wild-type RGS9 allele was identified by the presence of a 480-bp PCR product by using primers DD2: 5′-GTA ACA GCT GCT GTT CCA AAA ATC and RGS9-6: 5′-TGC ATT CTG ACT CCC GTC TCT GGG. The targeted RGS9 allele was identified by the presence of a 300-bp PCR product by using primers RGS9KO1: 5′-CGG CGA GGA TCT CGT CGT GAC and RGS9KO2: 5′-GAA CAA ACG ACC CAA CAC CCG. The PCR conditions used for genotyping RGS9–/– mice were identical to those used for TG9F10.
Fig. 6.
RGS9-1 protein levels in TG9F transgenic mice. The pRho4.2-RGS9-1 transgenic construct shown in A was injected into mouse embryos to produce TG9F2 and TG9F10 mice that express full-length RGS9-1-coding region in the photoreceptors. The retinal levels of RGS9-1 transcripts were measured by dot blot as shown in B. The amounts of total retinal RNAs blotted on the filter (top to bottom) were 1, 0.5, 0.25, and 0.12 μg, respectively. The RGS9-1 message levels (Left) in TG9F2 and TG9F10 retinas were ≈2- and 8-fold more than that of the wild-type control. β-Actin was used as a control for loading (Right). (C) The RGS9-1 protein levels were determined by immunoblots of equal amount of retinal extracts. (D) Western blot analyses showing the amounts of RGS9-1 (Left) and Gβ5 spliced forms (Right) in retinas of indicated mouse lines. To determine whether the transgenic RGS9-1 mRNA could be translated, TG9F2 and TG9F10 mice were bred into the RGS9 knockout background to produce TG9F2–/– and TG9F10–/– mouse lines, respectively. The RGS9-1 protein level in TG9F2–/– retina was ≈20% of the wild-type level, whereas in TG9F10–/– it was 100%. A corresponding restoration of Gβ5-L in both TG9F2–/– and TG9F10–/– mouse retinas was evident. Molecular markers (in kilodaltons) are indicated on the right.
Antibody Production. Polyclonal antibody for RGS11, αs-11, was raised in rabbits against recombinant mouse RGS11 fragment (residues 248–471, including the RGS and the C-terminal domains) and affinity-purified by immobilized antigen. The polyclonal antibody for RGS6, CT-3159, was raised in chicken against RGS6 peptide (CAKKKGKSLAGKRLTG) conjugated to keyhole limpet hemocyanin and affinity-purified by using immobilized peptide on SulfoLink resin (Pierce).
Immunoblots. Approximately 20 μg of total retinal proteins or 100 μg of striatal proteins, determined by Pierce BCA kit by using BSA as a standard, was resolved by SDS/PAGE followed by Western blotting onto nitrocellulose membrane. The dilutions of antibodies used for detection were: CT-215 (anti-Gβ5, used at 1:2,500); CT-317 (RGS9-1 specific, 1:1,000); R4432 (generic RGS9 antibody, 1:5,000); SC-6204 (anti-RGS4, 1:200; Santa Cruz Biotechnology); SC-374 (anti-Gγ2, 1:200; Santa Cruz Biotechnology); 2923AP (anti-RGS7, 1:5,000; Upstate Biotechnology); αs-11 (anti-RGS11, 1:1,000); and CT-3159 (anti-RGS6, 1:1,000). For the detection of RGS6, ≈40 μg of retinal extract was used.
Northern Blots. Total RNAs from retina and striatum were isolated by using the Trizol reagent (Invitrogen) by following the manufacturer's instructions. The RNA was denatured by glyoxal, fractionated on agarose gels, and blotted onto positively charged nylon membranes (Roche, Gipf-Oberfrick, Switzerland). PCR-generated DNA fragments containing the N-terminal coding sequences of RGS9 (1–350), RGS11 (1–350), RGS7 (1–350), and RGS6 (1–400) were used as templates to synthesize radioactive probes by using the Takara Ladderman DNA-labeling kit (Takara Shuzo, Kyoto). Hybridization of the probes to the nylon membranes was carried out in the ExpressHyb solution (CLONTECH) at 68°C for 1 h. The membranes then were washed with 2× SSC/0.1% SDS at room temperature for 30 min, followed by 0.1× SSC/0.1% SDS at 54°C for 30 min. The signals were visualized by autoradiography by using Kodak BioMax MS film.
Results
Lack of Gβ5-L but Not Gβ5-S in RGS9–/– Mice. It has been shown that the Gβ5-L protein, but not its mRNA, is absent in the retina of RGS9–/– mice (22). We found that the level of the short-splice form, Gβ5-S, is not affected by the inactivation of the RGS9 gene in retina or in striatum (Fig. 1). In the retina, Gβ5-S resides in different cell types other than photoreceptors and interacts with other GGL domain-containing RGS proteins such as RGS7 (9). This may stabilize the Gβ5-S protein (10). Similarly, in the striatum, other GGL domain-containing RGS proteins may compensate for the loss of RGS9-2, or perhaps RGS9-2 constitutes only a small fraction of the total GGL domain-containing RGS proteins.
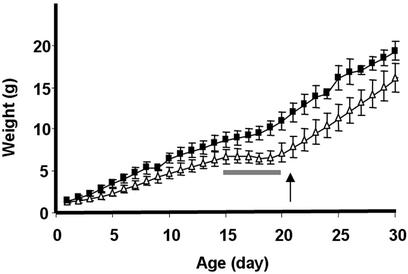
Inactivation of Gβ5 Gene. To examine whether the interaction observed for the RGS9-1/Gβ5-L protein complex in retinal photoreceptors applied to the short-splice variant Gβ5-S and to other GGL domain-containing RGS proteins, we inactivated the Gβ5 gene. As shown in Fig. 2A, a genomic 2.7-kb XhoI fragment containing the third exon of Gβ5 gene was replaced with a 1.1-kb Neo resistance marker in the gene-targeted mice. We made both outbred (mixed background of C57BL/6 and 129SvEv) and inbred (129SvEv) heterozygous knockout (Gβ5+/–) lines. The homozygous Gβ5 knockout (Gβ5–/–) lines were generated by intercross of the Gβ5+/– mice and maintained in both outbred and inbred backgrounds. Inactivating one copy of the Gβ5 gene did not change the Gβ5 protein levels in retina (Fig. 2B) or in brain (data not shown), whereas no Gβ5 protein was detectable in the homozygous knockout mice. The Gβ5–/– mice derived from the mating of heterozygous knockout parents could be identified readily as runty among their normal and Gβ5+/– littermates. More than two-thirds of the Gβ5–/– pups died by the time of weaning at 21 days of age. However, we were able to establish both outbred and inbred homozygous knockout lines by hand-feeding and further breeding of the surviving Gβ5–/– mice. The offspring of the Gβ5–/– mice were smaller in size and displayed slower weight gain in the first month of age (Fig. 3). A period of no weight gain just before weaning (from 15 to 20 days of age) coincided with the time of abnormally high mortality rate observed for the Gβ5–/– mice. At 3 months of age, they lacked obvious morphological abnormalities in retina and brain.
Fig. 3.
The growth of Gβ5–/– mice in the first month of age. The mean body weights of the Gβ5–/– (▵, n = 12) and Gβ5+/+ (▪, n = 4) mice generated in different litters were monitored daily after birth during their first month of growth. All pups were weaned on postnatal day 21 (indicated by an arrow). The smaller body size of the Gβ5–/– pups is evident. In addition, a period of no weight gain, from postnatal day 15 to 20 (indicated by a bar) in the Gβ5–/– pups was noticed. Error bars represent SD.
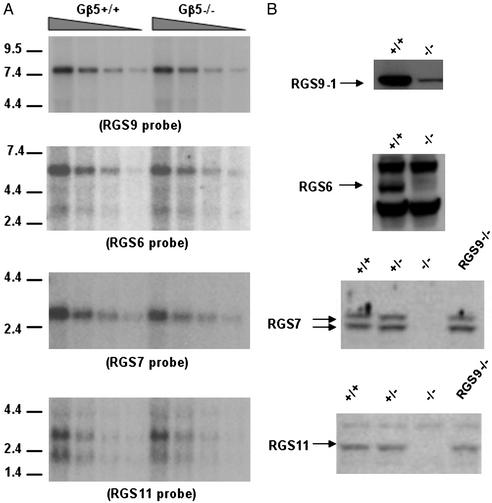
Instability of GGL Domain-Containing RGS Proteins but Not Their Messenger RNA in Gβ5–/– Mice. The inactivation of Gβ5 gene did not affect the steady-state mRNA levels of the GGL domain-containing RGS genes in the retina (Fig. 4A) or in the striatum (Fig. 5A). However, the levels of these RGS proteins fell significantly (e.g., RGS9-1 in the retina; Fig. 4B) or were not detectable (RGS9-2, RGS-6, RGS7, and RGS11; Fig. 4B). In the retina, the RGS9-1 protein level in Gβ5–/– mice was <5% of that in the normal mice. A similar level of residual RGS9-1 also was found in inbred Gβ5–/– mouse retinas (data not shown). In the striatum, both the RGS9-2 and the RGS7 proteins were absent, whereas their messenger RNA levels remained unaffected (Fig. 5). The message levels of RGS6 and RGS11 in the wild-type striatum were reportedly low (33) and fell below our detection limits (data not shown). The absence of RGS7 protein but not its messenger RNA in the cerebellum or in the whole brain also was observed (data not shown). The instabilities were restricted to GGL domain-containing RGS proteins because the level of another RGS protein without the GGL domain, RGS4, was not affected by the loss of Gβ5 (Fig. 5C). The level of Gγ2, a member of the G protein γ-subunit family, which forms a heterodimer with Gβ5 that activates phospholipase in vitro (12, 34), also was not affected by the loss of Gβ5 (Fig. 5C).
Fig. 4.
Instability of GGL domain-containing RGS proteins in retinas of Gβ5–/– mice. (A) Retinal total RNA (3, 1.5, 0.75, and 0.38 μg from left to right) isolated from Gβ5–/– and Gβ5+/+ mice was hybridized with various radioactive probes specific for RGS9, RGS6, RGS7, and RGS11 by Northern blotting (see Materials and Methods). The mRNA levels of the four RGS proteins were comparable between Gβ5–/– and wild-type retinas. Molecular mass markers (in kb) are indicated at right. (B) Western blotting of retinal protein extracts derived from different genetic backgrounds by using antibodies specific for individual RGS proteins demonstrating the loss of RGS6, RGS7, and RGS11 and the reduction of RGS9-1 signals (indicated by arrows) in the Gβ5–/– retina. Approximately 20 μg of retinal extract was analyzed for RGS7, RGS11, and RGS9-1; 40 μg was analyzed for RGS6 expression.
Fig. 5.
The absence of RGS9-2 and RGS7 but not RGS4 and Gγ2 in the striatum of Gβ5–/– mice. (A) Northern blotting of total RNA (3, 1.5, 0.75, and 0.38 μg, left to right) isolated from Gβ5–/– and Gβ5+/+ striatum was hybridized with radioactive RGS9 (Upper) and RGS7 (Lower) specific probes and visualized by autoradiography. The level of the 2.6-kb RGS9-2 transcript and the 2.7-kb RGS7 transcript was similar in Gβ5–/– and in wild-type striatum. Molecular mass markers (in kilobases) are indicated at left. (B and C) Western blotting of 100 μg of striatal extracts by using antibodies for RGS9 (R4432), RGS7 (2923AP), RGS4 (SC-6204), and Gγ2 (SC-374), showing the specific loss of the 82-kDa RGS9-2 and the 55-kDa RGS7 signals in Gβ5–/– mice. The levels of a non-GGL domain RGS protein, RGS4, and a regular γ-subunit, Gγ2, are not affected by the absence of Gβ5. Molecular mass markers (in kilodaltons) are indicated on the left.
Lack of RGS9-1 Overexpression in RGS9-1 Transgenic Mice. In an attempt to alter the response kinetics of rod photoreceptors and to further explore the relationship between Gβ5-L and RGS9-1, we sought to overexpress RGS9-1 in mouse photoreceptors. We made a transgenic construct containing a 4-kb mouse opsin promoter fragment to drive the transcription of RGS9-1 cDNA (Fig. 6A) and used it to generate transgenic mice. Two founders were obtained, TG9F2 and TG9F10, which transmitted the transgene to their offspring. Dot-blot analysis demonstrated that the level of the RGS9-1 mRNA derived from the transgenes was ≈2- to 8-fold higher than that derived from the endogenous RGS9 genes (Fig. 6B). However, we did not observe a corresponding increase in RGS9-1 protein level in the retina (Fig. 6C), and the rod photoresponses appeared normal (data not shown). The lack of RGS9-1 protein overexpression in TG9F2 and TG9F10 transgenic mice may result from the failure of the photoreceptors to translate the transgenic RGS9-1 mRNA. To ensure that the transgenic messages were translated into protein and that the transgenic RGS9-1 protein could interact with endogenous Gβ5-L, we bred the transgenic mice into the RGS9–/– background. As shown in Fig. 6D, the resulting transgenic lines, TG9F2–/– and TG9F10–/–, were found to express RGS9-1 protein in the retinas to ≈20% and 100% of the wild-type RGS9-1 protein level, respectively. The levels of restoration of RGS9-1 protein in TG9F2–/– and TG9F10–/– mice were accompanied by the restoration of corresponding levels of Gβ5-L protein (Fig. 6D), indicating that the transgenic RGS9-1 interacts with endogenous Gβ5-L in vivo to stabilize each other when they can form specific complexes.
Discussion
The Stability of the GGL Domain-Containing RGS Proteins Requires Gβ5. The failure to overproduce RGS9-1 protein in transgenic TG9F10 and TG9F2 mouse retinas despite the elevated mRNA level suggests that the amount of RGS9-1 protein in vivo is not controlled by the levels of its transcripts. The protein level of RGS9-1 apparently is controlled at the translation or posttranslational levels. Several reports show that the regulation can occur at the posttranslational level (10, 35). The disappearance of the GGL domain-containing RGS proteins in the retinas of GB5–/– mice lends further support to that notion. Apparently, Gβ5 and the GGL domain-containing RGS proteins are obligatory partners in vivo. Such a relationship is not seen in other photoreceptor protein complexes such as the catalytic subunits of the cGMP-phosphodiesterase (cG-PDE) found in mouse rods. In rd1 mice carrying a non-sense mutation in the β-subunit of cG-PDE (36, 37), the α-subunit of cG-PDE is still present in rods, although in an inactive form (38, 39). The interaction between GGL domain-containing RGS proteins and Gβ5 as a prerequisite for stability also applies to other regular G protein βγ complexes (40) such as the Gβ1γ7 complex in HEK293 cells (41). The interaction presumably shelters the proteins in these complexes from being targeted for rapid degradation. The mechanisms by which singular components are degraded remain unclear.
The Gβ5–/– Mice. The Gβ5–/– pups have smaller body size (runty) and an unusual mortality rate (66%) before weaning. By hand-feeding the Gβ5–/– pups starting at postnatal day 2 daily with dough food and water, the preweaning mortality can be circumvented, and the animals can survive into adulthood. Given the widespread expression of Gβ5 and the GGL domain-containing RGS proteins in different parts of the nervous system, we were surprised that we were able to establish a Gβ5–/– line. This result indicates that the GGL domain-containing RGS proteins and Gβ5 are not essential for the development or the survival of the animal. However, the smaller body size, slower weight gain, and the abnormally high mortality rate indicate that these protein complexes clearly are important for normal behavioral functions, presumably by regulating G protein signaling in distinct neuronal circuits. By examining the growth curve of the Gβ5–/– mice, a period of no weight gain between postnatal days 15 and 20 was noticed, which coincided with the period of high mortality rate. It is not clear whether this causes the Gβ5–/– mice to die, but the establishment of the Gβ5–/– line enables us to examine this further. The Gβ5–/– mice also can be used to study the physiological roles of the GGL domain-containing RGS proteins under various experimental conditions. An interesting question, for instance, is the function of the residual level (<5%) of RGS9-1 found in the retinal photoreceptors of the Gβ5–/– mice. A thorough electrophysiological analysis currently is underway to elucidate the function, if any, of the residual RGS9-1 in retinal photoreceptors.
The RGS9-1 Transgenic Mice. In C. elegans, overexpression of either EGL-10 or EAT-16 produced noticeable effects in egg-laying behavior (17). These data suggest that in worms, the GPB-2 level is not limiting the amounts of functional EGL-10/GPB-2 and EAT-16/GPB-2 protein complexes. Consistent with the notion is that overexpression of GPB-2 has no obvious effect in related behavior (19), although an effect in locomotion was noticed (18). The lack of RGS9-1 protein overexpression despite elevated mRNA level in retinas of the TG9F10 mice suggests that Gβ5 limits the amount of the RGS9-1/Gβ5-L complex. Thus simultaneous expression of both proteins may be necessary to achieve overexpression of this complex in photoreceptors. We notice that the endogenous retinal RGS9-1 mRNA (Fig. 3) is significantly longer than the striatal RGS9-2 mRNA (Fig. 4), whereas the RGS9-1 protein is smaller than the RGS9-2 protein (Fig. 1). The role of the varied length of the noncoding region of endogenous RGS9 mRNA is not known. However, we could detect only 20% normal level of RGS9-1 protein whereas the level of RGS9-1 transgenic mRNA in the TG9F2–/– retina is 200% of the normal level. Because the transgene contains only the coding region of the RGS9-1 and apparently its mRNA is not translated as efficiently as the endogenous one, this suggests that the noncoding region of the endogenous RGS9-1 mRNA may contain important elements for efficient translation. However, the partial restoration of retinal RGS9-1/Gβ5-L protein level in TG9F2–/– mice provides additional proof that these two proteins form an obligatory complex and are required for their mutual stability.
Acknowledgments
We thank Alan John Watson, Marie Burns, and Wolfgang Baehr for helpful discussion and Yi-Hui Hu and the Caltech Transgenic Core Facility for technical assistance. C.-K.C. is a recipient of the Research to Prevent Blindness Career Development Award. This work was supported by National Institutes of Health Grants EY013811 (C.-K.C.) and AG12288 (M.I.S.).
Abbreviations: RGS, regulator of G protein signaling; GGL, G protein γ-like; Neo, neomycin.
References
- 1.Dohlman, H. G., Song, J., Ma, D., Courchesne, W. E. & Thorner, J. (1996) Mol. Cell. Biol. 16, 5194–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koelle, M. R. & Horvitz, H. R. (1996) Cell 84, 115–125. [DOI] [PubMed] [Google Scholar]
- 3.Berman, D. M., Wilkie, T. M. & Gilman, A. G. (1996) Cell 86, 445–452. [DOI] [PubMed] [Google Scholar]
- 4.Ross, E. M. & Wilkie, T. M. (2000) Annu. Rev. Biochem. 69, 795–827. [DOI] [PubMed] [Google Scholar]
- 5.Zheng, B., De Vries, L. & Gist Farquhar, M. (1999) Trends Biochem. Sci. 24, 411–414. [DOI] [PubMed] [Google Scholar]
- 6.Snow, B. E., Krumins, A. M., Brothers, G. M., Lee, S. F., Wall, M. A., Chung, S., Mangion, J., Arya, S., Gilman, A. G. & Siderovski, D. P. (1998) Proc. Natl. Acad. Sci. USA 95, 13307–13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snow, B. E., Betts, L., Mangion, J., Sondek, J. & Siderovski, D. P. (1999) Proc. Natl. Acad. Sci. USA 96, 6489–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, J. H. & Simonds, W. F. (2000) J. Neurosci. 20, RC59, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabrera, J. L., de Freitas, F., Satpaev, D. K. & Slepak, V. Z. (1998) Biochem. Biophys. Res. Commun. 249, 898–902. [DOI] [PubMed] [Google Scholar]
- 10.Witherow, D. S., Wang, Q., Levay, K., Cabrera, J. L., Chen, J., Willars, G. B. & Slepak, V. Z. (2000) J. Biol. Chem. 275, 24872–24880. [DOI] [PubMed] [Google Scholar]
- 11.Watson, A. J., Aragay, A. M., Slepak, V. Z. & Simon, M. I. (1996) J. Biol. Chem. 271, 28154–28160. [DOI] [PubMed] [Google Scholar]
- 12.Watson, A. J., Katz, A. & Simon, M. I. (1994) J. Biol. Chem. 269, 22150–22156. [PubMed] [Google Scholar]
- 13.Jones, M. B. & Garrison, J. C. (1999) Anal. Biochem. 268, 126–133. [DOI] [PubMed] [Google Scholar]
- 14.Kim, E., Arnould, T., Sellin, L., Benzing, T., Comella, N., Kocher, O., Tsiokas, L., Sukhatme, V. P. & Walz, G. (1999) Proc. Natl. Acad. Sci. USA 96, 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, Z., Chatterjee, T. K. & Fisher, R. A. (2002) J. Biol. Chem. 277, 37832–37839. [DOI] [PubMed] [Google Scholar]
- 16.Levay, K., Cabrera, J. L., Satpaev, D. K. & Slepak, V. Z. (1999) Proc. Natl. Acad. Sci. USA 96, 2503–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chase, D. L., Patikoglou, G. A. & Koelle, M. R. (2001) Curr. Biol. 11, 222–231. [DOI] [PubMed] [Google Scholar]
- 18.Robatzek, M., Niacaris, T., Steger, K., Avery, L. & Thomas, J. H. (2001) Curr. Biol. 11, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Linden, A. M., Simmer, F., Cuppen, E. & Plasterk, R. H. (2001) Genetics 158, 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, W., Cowan, C. W. & Wensel, T. G. (1998) Neuron 20, 95–102. [DOI] [PubMed] [Google Scholar]
- 21.Makino, E. R., Handy, J. W., Li, T. & Arshavsky, V. Y. (1999) Proc. Natl. Acad. Sci. USA 96, 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, C. K., Burns, M. E., He, W., Wensel, T. G., Baylor, D. A. & Simon, M. I. (2000) Nature 403, 557–560. [DOI] [PubMed] [Google Scholar]
- 23.Lyubarsky, A. L., Chen, C.-K., Naarendorp, F., Zhang, X., Wensel, T., Simon, M. I. & Pugh, E. N., Jr. (2001) Mol. Vis. 7, 71–78. [PubMed] [Google Scholar]
- 24.Zhang, K., Howes, K. A., He, W., Bronson, J. D., Pettenati, M. J., Chen, C., Palczewski, K., Wensel, T. G. & Baehr, W. (1999) Gene 240, 23–34. [DOI] [PubMed] [Google Scholar]
- 25.Rahman, Z., Gold, S. J., Potenza, M. N., Cowan, C. W., Ni, Y. G., He, W., Wensel, T. G. & Nestler, E. J. (1999) J. Neurosci. 19, 2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, X., Wensel, T. G. & Kraft, T. W. (2003) J. Neurosci. 23, 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowan, C. W., Fariss, R. N., Sokal, I., Palczewski, K. & Wensel, T. G. (1998) Proc. Natl. Acad. Sci. USA 95, 5351–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez-Solis, R., Davis, A. C. & Bradley, A. (1993) Methods Enzymol. 225, 855–878. [DOI] [PubMed] [Google Scholar]
- 29.Lem, J., Applebury, M. L., Falk, J. D., Flannery, J. G. & Simon, M. I. (1991) Neuron 6, 201–210. [DOI] [PubMed] [Google Scholar]
- 30.Chen, J., Flannery, J. G., LaVail, M. M., Steinberg, R. H., Xu, J. & Simon, M. I. (1996) Proc. Natl. Acad. Sci. USA 93, 7042–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodford, B. J., Chen, J. & Simon, M. I. (1994) Exp. Eye Res. 58, 631–635. [DOI] [PubMed] [Google Scholar]
- 32.Mendez, A., Burns, M. E., Roca, A., Lem, J., Wu, L. W., Simon, M. I., Baylor, D. A. & Chen, J. (2000) Neuron 28, 153–164. [DOI] [PubMed] [Google Scholar]
- 33.Gold, S. J., Ni, Y. G., Dohlman, H. G. & Nestler, E. J. (1997) J. Neurosci. 17, 8024–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Posner, B. A., Mukhopadhyay, S., Tesmer, J. J., Gilman, A. G. & Ross, E. M. (1999) Biochemistry 38, 7773–7779. [DOI] [PubMed] [Google Scholar]
- 35.He, W., Lu, L., Zhang, X., El-Hodiri, H. M., Chen, C. K., Slep, K. C., Simon, M. I., Jamrich, M. & Wensel, T. G. (2000) J. Biol. Chem. 275, 37093–37100. [DOI] [PubMed] [Google Scholar]
- 36.Pittler, S. J. & Baehr, W. (1991) Proc. Natl. Acad. Sci. USA 88, 8322–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowes, C., Li, T., Danciger, M., Baxter, L. C., Applebury, M. L. & Farber, D. B. (1990) Nature 347, 677–680. [DOI] [PubMed] [Google Scholar]
- 38.Lee, R. H., Navon, S. E., Brown, B. M., Fung, B. K. & Lolley, R. N. (1988) Invest. Ophthalmol. Visual Sci. 29, 1021–1027. [PubMed] [Google Scholar]
- 39.Lolley, R. N. & Lee, R. H. (1989) Prog. Clin. Biol. Res. 314, 155–168. [PubMed] [Google Scholar]
- 40.Simonds, W. F., Butrynski, J. E., Gautam, N., Unson, C. G. & Spiegel, A. M. (1991) J. Biol. Chem. 266, 5363–5366. [PubMed] [Google Scholar]
- 41.Wang, Q., Mullah, B. K. & Robishaw, J. D. (1999) J. Biol. Chem. 274, 17365–17371. [DOI] [PubMed] [Google Scholar]