Abstract
The contribution of host factors to rabies virus (RV) transcription/replication and axonal/transsynaptic spread is largely unknown. We previously identified several host genes that are up-regulated in the mouse brain during RV infection, including neuroleukin, which is involved in neuronal growth and survival, cell motility, and differentiation, and fibroblast growth factor homologous factor 4 (FHF4), which has been implicated in limb and nervous system development. In this study, we used real-time quantitative RT-PCR to assess the expression of mRNAs specific for neuroleukin, the two isoforms of FHF4 (FHF4-1a and -1b) encoded by the FHF4 gene, and N protein of RV in neurons and astrocytes isolated by laser capture microdissection from mouse brains infected with the laboratory-adapted RV strain CVS-N2c or with a street RV of silver-haired bat origin. Differences in the gene expression patterns suggest that the capacity of RV strains to infect nonneuronal cells and differentially modulate host gene expression may be important in virus replication and spread in the CNS.
Although the predominant effect of rabies virus (RV) infection is the down-regulation of gene expression in the CNS, we have identified a subset of host genes that are induced by the infection (1). Among these are genes relevant to neuronal functions that may be involved in the replication and spread of RV, including neuroleukin (NLK) and fibroblast growth factor homologous factor 4 (FHF4). The member of a family of FHF4-1 genes, the FHF4 gene, which is predominantly expressed in the CNS in mice, encodes two isoforms, FHF4-1a and -1b, through alternative exon usage (2). The FHF4 isoforms both lack secretory signal peptides and accumulate intracellularly but differ functionally and in their cellular distribution (3). NLK has multiple functions, including acting as a growth factor to promote the survival and neurite outgrowth of motor and sensory neurons (4), as an autocrine motility factor to induce cell motility (5), as a maturation factor to mediate the differentiation of myeloid precursor cells to mature monocytes (6), and as a phosphohexose isomerase to catalyze the conversion of glucose-6-phosphate to fructose-6-phosphate (7).
RV strains, such as the laboratory-adapted CVS-N2c RV strain and street viruses of silver-haired bat origin (SHBRV), often have highly disparate growth characteristics in vivo and in vitro (8–10) that may be reflected in their pathogenicity. We speculate that differences in the capacity of RV to replicate and spread is a consequence of the differential induction of factors such as NLK and FHF4-1a and -1b. To test this hypothesis, we have analyzed the expression patterns of these genes in neurons and astrocytes isolated from mice infected with CVS-N2c and SHBRV-17.
Materials and Methods
Virus Infection of Mice and Tissue Preparation. All animal experiments were performed according to procedures approved by the Institutional Review Board's Animal Care and Use Committee. C3H mice 6–8 weeks old were purchased from Taconic Farms and housed in pathogen-free conditions. At 8–10 weeks of age, mice were infected i.m. in the masseter under anesthesia with 106 plaque-forming units of RV strain CVS-N2c or SHBRV-17. At this dose of virus, C3H mice usually survive until days 7–8 postinfection (p.i.) with CVS-N2c and until day 10 p.i. with SHBRV-17. After infection, mice were killed with CO2 at the indicated time points. Brains were removed, immediately frozen in OCT compound (Sakura–Finetek, Torrance, CA), and stored at -80°C. Frozen sections (7 μm thick) of the hippocampus of control (uninfected) and experimental (infected) mice were cut on a ThermoShandon cryostat (Shandon, Pittsburgh) by using disposable blades to avoid cross-contamination among specimens. Sections were mounted on Superfrost Plus glass slides (VWR Scientific) and transferred on dry ice for storage at -80°C.
Nissl Staining of Neurons. Cryostat sections stored at -80°C were immediately immersed in 80% acetone for 2-min fixation. After a brief rinse with water (BioWhittaker), sections were stained with 0.1% aqueous solution of cresyl violet (Sigma) for 1 min, rinsed with water for 30 s, and dehydrated in graded alcohols (90% and 100% ethanol, 30 s each) and xylene (two changes, 4 min each). All reaction steps were performed in RNase-free solutions. Sections were then air-dried under laminar flow for 30 min and immediately used for laser capture microdissection (LCM).
Immunofluorescent Detection of Astrocytes. Frozen sections were immersed in 80% acetone for 2 min, rinsed briefly in 1× PBS (pH 7.4), and incubated for 2 min in PBS containing 2% BSA. To prevent degradation of RNA, all solutions were prepared with diethyl pyrocarbonate-treated water. Sections were stained at room temperature for 10 min with 10 μg/ml anti-glial fibrillary acidic protein (GFAP) conjugated with Alexa Fluor 488 (Molecular Probes). RNasin (Promega) was added to the antibody solution at a concentration of 1 unit/μl. After staining and a brief rinse in 1× PBS, sections were dehydrated as described above, air-dried for 30 min in the dark, and used immediately for LCM.
Detection of Rabies Virus (RV) N Protein. Frozen sections were fixed in 80% acetone for 2 min, rinsed briefly in 1× PBS, and incubated for 2 min in 2% BSA in PBS to block nonspecific binding sites. Sections were then incubated with the FITC-conjugated anti-RV monoclonal antibody (Centocor) for 10 min, rinsed briefly in PBS, dehydrated in alcohols and xylene, and air-dried.
LCM. Microdissection of mouse brain tissue was performed by using a PixCell apparatus (Arcturus Engineering, Mountain View, CA) equipped with the Fluor 300 PixCell II fluorescence package. Neurons and astrocytes from Nissl-stained and immunohistochemically stained sections were captured on thermoplastic polymer-coated caps (CapSure TF-100, Arcturus Engineering) by using a laser beam 7.5 μm in diameter, with 50- to 85-mW power and 300-ms pulse duration. Fifty cresyl violet-stained neurons per cap were collected from the hippocampus. The PixCell II fluorescence package with a blue filter cube (excitation 455–495 nm, emission >510 nm) was used to collect GFAP-stained astrocytes (100 per cap) from the external capsule of the cerebral hemisphere. Caps with cells were placed in Eppendorf tubes containing 100 μl of lysis buffer (Stratagene) and stored in -80°C before RNA isolation.
RNA Isolation from LCM Samples and Frozen Sections. Total RNA from each population of laser-captured cells was isolated with the Absolutely RNA Microprep kit (Stratagene) according to the manufacturer's recommendations. Briefly, the transfer film with adherent cells was incubated in 100 μl of lysis buffer containing 2-mercaptoethanol. Each sample was then mixed with an equal volume of 70% ethanol and applied to a silica-based fiber column. After centrifugation and low-salt buffer washing, samples were digested with DNase I. The column was washed several times with high- and low-salt washing buffers and dried. RNA was eluted with 30 μl of elution buffer (10 mM Tris, pH 7.5) and used directly for reverse transcription. Total RNA from frozen sections was isolated by using the same protocol as above, except that TRIzol reagent (Sigma) was used for RNA extraction from tissue on the slides.
Real-Time Quantitative RT-PCR. First-strand cDNA was synthesized by using the ProSTAR kit (Stratagene). Each RNA sample was mixed with 3 μl of random hexamer primers (100 ng/ml) and incubated at 65°C for 5 min and then at 25°C for 10 min. Reaction mix (7 μl) containing 10× first-strand buffer, ribonuclease inhibitor, 100 mM dNTPs, and reverse transcriptase was added, and after a 1-h incubation at 37°C, 10 μl of the cDNA mixture was used for PCR. Quantitative analysis was performed by using a Bio-Rad iQ Real-Time Detection System and a TaqMan PCR Core Reagent kit (Applied Biosystems) for 40 cycles of a two-step PCR amplification (95°C for 15 s and 60°C for 1 min). Mouse primers and probes for the TaqMan system were designed by using PRIMER3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). To avoid problems associated with DNA contamination, primers were selected that span at least one intron of the genomic sequence. Sequences of the PCR primers (forward and reverse, respectively) and fluorogenic TaqMan probes used for quantification were SHBRV N protein, 5′-TGTGCGCTAACTGGAGTACCA-3′, 5′-GTGCCTACCCTAAATTGCTGA A-3′, 5′-/56-FA M/ CCGAACTTCAGATTCCTAGCTGGAACC/BHQ/-3′; CVS N2c N protein, 5′-CACTTCCGTTCACTAGGCTTGA-3′, 5′-GACCCATGTAGCATCCAACAA-3′, 5′-/56-FAM/TGAACACATGACCGACAGCATTCGA/BHQ/-3′; NLK, 5′-ACCAAGATGATACCCTGTGACT-3′, 5′-AAGAAGTTAGCCAGGAGGATCT-3′, 5′-/56-FAM/ACCCAGCACCCCATACGGAAAGG/BHQ/-3′; glyceraldehyde 3-phosphate dehydrogenase (G3PDH), 5′-TCAAGAAGGTGGTGAAGCAG-3′, 5′-TGGAAGTTGCTGTTGAAGTC-3′,5′-/56-FAM/CCACTGAAGGGCATCTTGGGCTAC/BHQ/-3′; FHF4-1a, 5′-CGGCTTGATCCGTCAGAAACG-3′, 5′-ACCAGGTTGCCATTGAAAAGC3′, 5′-/56-FAM/AGCAGCCCCAGCAAGAACCGC/BHQ/-3′; and FHF4-1b, 5′-TGCCCCTCTTCAGGAGAACT-3′, 5′GAAAAGCAACCCAAGCAGCTT-3′, 5′-/56-FAM/CCACAAGGGCCTCTTCTTTXTCAGGG/BHQ/-3′.
Samples were run in duplicate, and a reaction without cDNA was used to establish baseline fluorescence levels. A relative standard curve representing five 10-fold dilutions of total brain cDNA (5, 0.5, 0.05, 0.005, and 0.0005 ng) was used for logarithmic regression analysis of unknown samples. Data are based on a threshold cycle (Ct) in which the signal was higher than that of background. The relative increase in RNA expression was calculated as follows: 2 exp(Ct of lowest expresser of specific mRNA - Ct of experimental specific mRNA)/2 exp(Ct of lowest expresser of G3PDH mRNA - Ct of experimental G3PDH mRNA) (11). A G3PDH Ct value of 30 was obtained from RNA isolated from 10 neurons or 20 astrocytes. No signal was detected when the reverse transcriptase step was omitted. Two independent experiments involving separate cell capture, RNA extraction, and reverse transcription were performed.
Virus Infection of Primary Astrocytes. Primary astrocyte cultures were prepared from cerebral hemispheres of 5-day-old B6129/F1 mice. Briefly, dissected tissue was incubated at 37°C for 30 min in 0.25% trypsin, dispersed with a 19-gauge needle, and centrifuged. The pellet was resuspended in 5 ml of DMEM/F12 medium (Cellgro, Mediatech, Herndon, VA) and cultured in tissue flasks. After the fourth passage, cells were infected with 5 × 103 focus-forming units/ml CVS-N2c or SHBRV-17. Supernatant and cell samples were collected at day 0 (control) and days 1, 2, and 3 p.i. Uninfected cells at all time points and medium with equivalent amounts of virus with no cellular component served as negative and positive controls, respectively. RNA isolation from harvested cells and supernatant and real-time quantitative RT-PCR were performed as described above.
Results
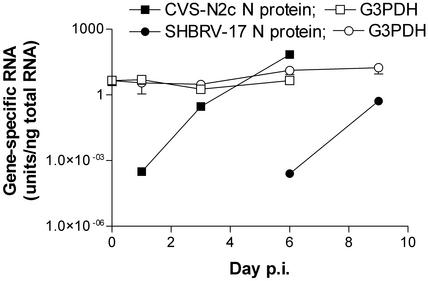
Time Course of the Expression of CVS-N2c and SHBRV-17 N-Protein Genes in Infected Mouse Brain. When coronal sections from the hippocampus of mice were analyzed by quantitative RT-PCR at different times after infection, CVS-N2c N-protein mRNA could be detected at low levels as early as the day after infection, whereas SHBRV-17 N-protein mRNA was not detected until day 6 p.i. (Fig. 1). In both cases, N-protein mRNA expression increased rapidly (>1,000-fold) after initial detection, whereas expression levels of the housekeeping gene G3PDH did not change significantly.
Fig. 1.
Expression levels of rabies CVS-N2c and SHBRV-17 virus N proteins and the housekeeping gene G3PDH in the infected mouse brain. Total RNA was prepared from coronal sections of brain in the same areas of hippocampus of control mice (day 0), of mice infected with CVS-N2c (days 1, 3, and 6), and of mice infected with SHBRV-17 (days 1, 3, 6, and 9) and assessed for rabies N protein-specific and G3PDH RNA by using quantitative RT-PCR as detailed in Materials and Methods. Data are expressed as the mean ± SEM of arbitrary units of specific RNA per ng of total RNA in two replicate sections.
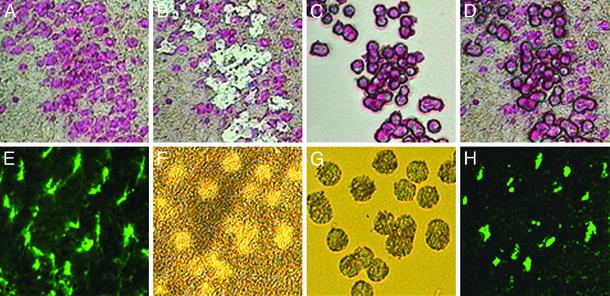
Identification and Laser Capture of Neurons and Astrocytes from Control and RV-Infected Brain. Although rabies N-protein mRNA was detected at days 1 and 3, cells staining strongly positive for rabies N protein could be detected in parallel sections only after 6 and 9 days of infection with CVS-N2c and SHBRV-17, respectively (data not shown). Thus neurons were selected for laser capture at later periods of infection by the presence of rabies N protein, whereas at earlier time points, the cells were identified by Nissl staining. Astrocytes were selected by staining for GFAP. Fig. 2 shows representative views of Nissl- and GFAP-stained sections from control, noninfected tissue, before and after laser capture as well as the isolated cells.
Fig. 2.
LCM of neurons and astrocytes from hippocampus of mouse brain. A Nissl-stained section from the brain of a normal mouse (A), the section after laser microdissection and capture of neurons (B), the isolated neurons on the capture membrane (C), and the neurons overlying the dissected section (D) are shown. A brain section in which fluorescein-conjugated anti-GFAP has been used to identify astrocytes with fluorescent microscopy is shown in E, and the same tissue after laser capture of the stained cells is shown by light microscopy in F; the captured cells under light are shown in G, and fluorescence is shown in H.
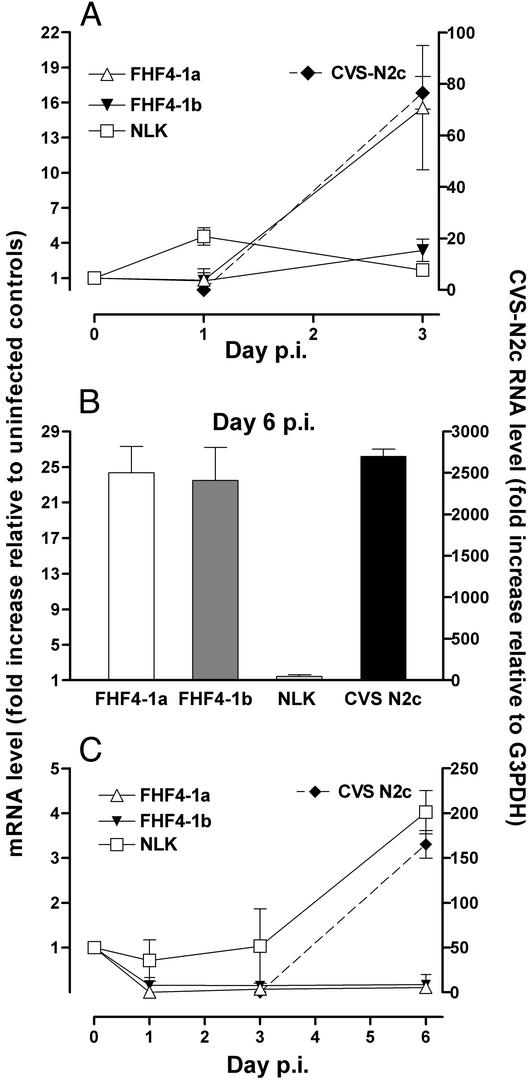
Expression of NLK and FHF4-1a and -1b mRNAs in Neurons and Astrocytes Isolated from the Hippocampus of CVS-N2c-Infected Mice. Real-time quantitative RT-PCR was used to assess the levels of specific mRNAs in neurons and astrocytes isolated from the hippocampus of mice at different times after infection with RV strain CVS-N2c. Although neuronal cell isolation was based on Nissl staining for the first 3 days p.i. with CVS-N2c, at least some of the selected neurons were infected with RV as demonstrated by the presence of rabies N-protein mRNA (Fig. 3A). In neurons, NLK expression was elevated on day 1 but returned to control levels by day 3 p.i. FHF4-1a mRNA was highly up-regulated, and FHF4-1b expression was slightly activated by day 3 of infection, in parallel with rapidly increasing rabies CVS-N2c N-protein mRNA levels. By day 6, near the time when the infection becomes lethal in the mice, both FHF4-1a and FHF4-1b showed high levels of activity in the neurons isolated on the basis of N-protein detection (Fig. 3B). In contrast, FHF4-1a and FHF4-1b expression in astrocytes from the same tissues was down-regulated early in the infection, whereas NLK was up-regulated in the cells late in the infection (Fig. 3C). Rabies N-protein mRNA was also detected in astrocytes prepared at day 6 of infection (Fig. 3C). However, the levels of CVS-N2c N-protein mRNA detected in the astrocytes were ≈100-fold lower than those in neurons from the same sections. This finding suggests that the astrocytes may have become contaminated with N-protein mRNA derived from the surrounding neurons during slide preparation.
Fig. 3.
Relative expression levels of FHF4-1a and -1b, NLK, and N-protein RNAs in neurons and astrocytes isolated from CVS-N2c-infected brains. cDNAs synthesized from RNAs prepared from cells captured from uninfected (represented as day 0 p.i.) and infected mouse brains at days 1, 3, and 6 p.i. were analyzed by quantitative PCR as described in Materials and Methods. (A) Expression levels in neurons identified by Nissl staining. (B) Expression levels in neurons at day 6 p.i. identified by antirabies N-protein staining. (C) Gene expression in astrocytes isolated by staining for GFAP. The fold change in mRNA expression relative to uninfected cells for FHF4-1a and -1b and NLK levels (left axes) in neurons and astrocytes at different days p.i. are expressed as the ratio of the G3PDH-normalized value for each infected sample to the mean of the G3PDH-normalized value for uninfected cells. The levels of CVS-N2c N-protein RNA (right axes) are expressed as the ratio of N-protein RNA to G3PDH mRNA in the captured cells.
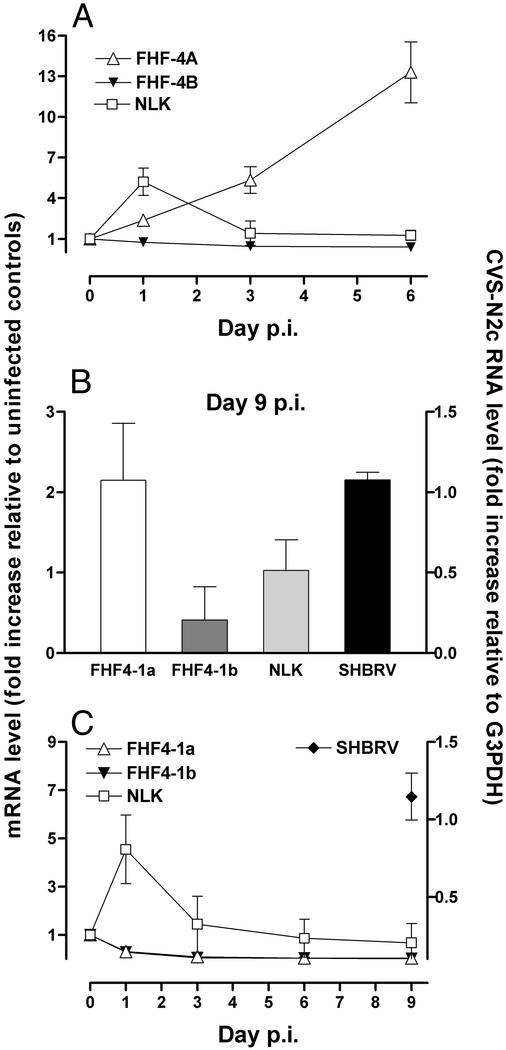
Expression of NLK and FHF4-1a and -1b mRNAs in Neurons and Astrocytes Isolated from the Hippocampus of SHBRV-17-Infected Mice. Real-time quantitative RT-PCR was also used to analyze gene expression in cells isolated by LCM from SHBRV-17 infected mouse brains (Fig. 4). The patterns of gene activity obtained show similarities but also striking differences to those of cells from the CVS-N2c infected brains. As in CVS-N2c infection, neurons of SHBRV-17-infected brain displayed enhanced FHF4-1a gene activity and a transient increase in NLK expression (Fig. 4A). However, unlike CVS-N2c, SHBRV-17 failed to induce FHF4-1b expression in neurons selected on the basis of rabies N-protein expression (Fig. 4B). In addition, NLK expression in astrocytes was up-regulated early in SHBRV-17 infection but decreased as the infection progressed (Fig. 4C). At day 9, rabies N-protein mRNA was detected in astrocytes from the SHBRV-17-infected brain (Fig. 4C).
Fig. 4.
Expression of FHF4-1a and -1b, NLK, and RV N-protein RNAs in neurons and astrocytes isolated from SHBRV-17-infected brains. Total RNA was extracted from neurons and astrocytes isolated by LCM of sections from the brains of mice either uninfected (represented as day 0 p.i.) or infected with SHBRV-17 1, 3, 6, and 9 days prior. Nissl staining was used to identify neurons at days 1, 3, and 6 p.i. (A), whereas staining for rabies N protein was used to select cells at day 9 p.i. (B). Astrocytes were identified by staining with anti-GFAP (C). Relative expression levels of mRNAs specific for FHF4-1a and -1b, NLK (left axes), and rabies N-protein RNA (right axes) were determined as described in the legend of Fig. 3.
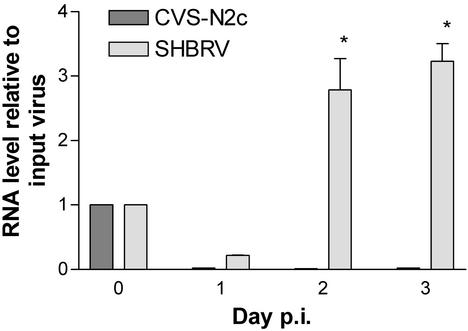
Analysis of Expression of RV N-Protein mRNAs in Primary Astrocyte Cultures. Unlike CVS-N2c-infected tissues in which rabies N-protein mRNA was detected in astrocytes at substantially lower levels than in the surrounding neurons, the levels of N-protein mRNA detected in astrocytes from SHBRV-17-infected brains were virtually the same as those in neurons. This is particularly noteworthy because the neurons were selected by immunohistochemical detection of rabies N-protein expression, whereas the astrocytes were identified phenotypically. Thus, it seemed unlikely that the astrocyte preparation could have been contaminated with neuron-derived rabies N-protein mRNA to an extent equivalent to the level of expression of this mRNA in the surrounding SHBRV-17-infected neurons. Quantitative RT-PCR analysis of rabies N-protein mRNA was used to assess whether CVS-N2c or SHBRV-17 might infect primary astrocytes in culture. N-protein specific mRNA levels higher than those of the input virus preparation were only detected in astrocytes incubated for 2–3 days after the addition of SHBRV-17 (Fig. 5). N-protein mRNA present in the input CVS-N2c material rapidly disappeared in culture (Fig. 5) suggesting that SHBRV-17, but not CVS-N2c, may be able to infect astrocytes in vivo.
Fig. 5.
N-protein RNA is expressed by primary astrocytes infected with SHBRV-17 but not CVS-N2c in culture. Primary mouse astrocytes were infected with either CVS-N2c or SHBRV-17, and N-protein mRNA levels were assessed by real-time RT-PCR and normalized to G3PDH mRNA levels in the sample as described in Materials and Methods. The results are expressed as the fold increase in N-protein RNA over time of culture with the amount of N protein-specific RNA in the input virus, represented as day 0, assigned a value of 1. SHBRV-17 N-protein RNA levels at days 2 and 3 of culture were significantly greater than the input level by ANOVA with Bonferroni's multiple comparison test (*, P < 0. 05).
Discussion
RVs exist in nature as a wide variety of genotypically distinct strains associated with different host species (12). In animal models and in vitro, diverse RV isolates display phenotypic differences that may or may not be related to their pathogenesis for humans (9). For example, the characteristics of the replication and spread of SHBRV, the predominant cause of human rabies in North America over the last 5–10 years, in the infected mouse brain differ from those of a laboratory-adapted strain originating from a dog (10). The RV strains used in this investigation differ greatly in their capacity to replicate and spread in mice. For example, RV N-protein mRNA was detected in brain sections at day 1 p.i. with CVS-N2c but only at day 6 p.i. with SHBRV-17.
Our previous microarray analysis of the effects of infection with CVS-N2c on gene expression in the mouse CNS revealed that the expression of the majority of host genes is down-regulated, but that of a limited subset of genes, including those encoding the growth factors FHF4 and NLK, is enhanced (1). To investigate whether the replication and spread of particular RVs in CNS tissue might be related to their capacity to induce growth factors, we assessed the effects of CVS-N2c versus SHBRV-17 infection on NLK and FHF4-1a and -1b expression in neurons and astrocytes isolated from the hippocampus, an area through which both viruses spread (10, 13). Both viruses induced FHF4-1a expression in neurons that, at least in the case of SHBRV-17, evidently did not depend on the cells being infected, because rabies N protein mRNA was not detected in the cells until several days after FHF4-1a became elevated. Both viruses also induced a transient increase in NLK expression by neurons, which evidently occurred before the cells became infected. However, only infection with CVS-N2c induced high levels of FHF-4b expression in neurons.
The patterns of growth factor expression in astrocytes from the hippocampi of mice infected with the two viruses showed similarities as well as distinct differences. Expression of both FHF4-1a and -1b in astrocytes was rapidly down-regulated by day 1 p.i. with either virus. However, NLK expression in astrocytes from CVS-N2c-infected brains remained unchanged for the first 3 days of infection and then became highly elevated at day 6. In contrast, NLK expression by astrocytes from SHBRV-17 infected brain was elevated transiently at day 1 of infection, returning to basal levels by days 3–6. We speculate that the differential response of astrocytes to infection with the two viruses, with respect to NLK expression, might reflect low-level replication of SHBRV-17, but not CVS-N2c, in astrocytes. Nevertheless, many of the changes in growth factor expression by both neurons and astrocytes are likely due to the response of noninfected cells to signals originating from infected neurons, because no evidence of rabies infection was detected at many of the time points at which altered growth factor gene activity was seen.
Current information about the functions of NLK and FHF4-1a and -1b, together with the results of our analysis of their expression patterns in the mouse brain, allows some speculation on the potential impact of the expression of these factors on RV replication and spread. NLK, which is involved in neuronal growth and survival, motility, and differentiation (4, 14, 15), was transiently up-regulated at day 1 of infection in neurons from brains of mice infected with either CVS-N2c or SHBRV-17 and in astrocytes from the SHBRV-17-infected brain. Moreover, late in infection, higher levels of NLK expression were present only in astrocytes from mouse brain infected with CVS-N2c, which replicates and spreads more rapidly than SHBRV-17. These observations raise the possibility that NLK, which is likely produced by astrocytes later in infection in response to some impact of the virus on neuronal function, facilitates RV replication and spread through an effect on neurons. For instance, NLK may promote the spread of the virus by inducing neurons to form new axonal connections through activation of terminal axonal sprouting (16). Alternatively, NLK might increase neuronal metabolism and prolong the survival of infected neurons, allowing them to produce more virus. In either case, the curtailed NLK production by astrocytes in SHBRV-17-infected brains, possibly caused by infection of these cells, might ultimately contribute to a reduction in the spread and replication of the virus.
FHF4-1a and -1b differ in their expression patterns and subcellular localization (17). FHF4-1a localizes primarily to the nucleus, whereas FHF4-1b appears exclusively in the cytosol of NIH 3T3 cells (2). In adult mice, FHF4-1b seems to be the predominant form of FHF-4 expressed in the brain (3). In our studies, FHF4-1b was expressed at ≈100-fold higher levels in neurons and 7-fold higher levels in astrocytes than FHF-4a in normal mouse brain. Because FHF4-1b is localized in axonal projections, it may be important for axonal or synaptosomal function or for neurotransmisson (18). Thus, the increased levels of FHF4-1b, seen only in CVS-N2c-infected neurons, may contribute to the greater replication and spread of this strain of RV as compared with SHBRV-17. FHF4-1a, through interaction with the mitogen-activated protein kinase scaffold protein IB2, is believed to modulate intracellular signaling pathways (19). The fact that FHF4-1a expression was up-regulated in neurons from brains infected with either RV, but to a considerably higher level in those from CVS-N2c-infected brains, raises the possibility that FHF4-1a, like FHF4-1b, contributes to the replication and spread of the virus. However, elevated expression of these growth factors in association with CVS-N2c infection might reflect the greater impact of CVS-N2c than SHBRV-17 on neurons because of its more rapid replication and spread. Further experiments are required to distinguish between these possibilities.
Although N-protein mRNA was detected in astrocytes captured from slides prepared from both CVS-N2c- and SHBRV-17-infected brains, we concluded that its presence in the CVSN2c samples was more likely to be the result of contamination because of the substantially higher mRNA levels seen in adjacent neurons. However, we considered that astrocytes from SHBRV-17-infected brains may become infected with the virus as roughly equivalent N-protein mRNA levels were detected in neurons and astrocytes. Indeed, analysis revealed that SHBRV-17 but not CVS-N2c can infect primary astrocytes in vitro. This finding is consistent with previous findings, suggesting that viruses associated with the silver-haired bat have a capacity to replicate in nonneuronal cells in vitro that is not seen with street viruses isolated from dogs (9). Another study has also provided evidence that a skunk-derived street RV infects Bergman glial cells in the cerebellum (20). In the case of SHBRV-17, the capacity to replicate in nonneuronal cells, which may potentiate the spread of a small amount of virus between a host and human, might be relevant to the apparently unique characteristics of SHBRV strains in causing rabies in humans without known exposure (21).
Acknowledgments
We thank Dr. Bernhard Dietzschold for providing virus stocks and Rhonda B. Kean for assistance with the manuscript. This study was supported by Public Health Service Grants AI-09706 and AI-41544, and by a grant to the Biotechnology Foundation Laboratories from the Commonwealth of Pennsylvania.
Abbreviations: RV, rabies virus; NLK, neuroleukin; FHF4, fibroblast growth factor homologous factor 4; SHBRV, silver-haired bat rabies virus; p.i., postinfection; LCM, laser capture microdissection; GFAP, glial fibrillary acidic protein; G3PDH, glyceraldehyde 3-phosphate dehydrogenase.
References
- 1.Prosniak, M., Hooper, D. C., Dietzschold, B. & Koprowski, H. (2001) Proc. Natl. Acad. Sci. USA 98, 2758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto, S., Mikami, T., Ohbayashi, N., Ohta, M. & Itoh, N. (1998) Biochim. Biophys. Acta 1398, 38-41. [DOI] [PubMed] [Google Scholar]
- 3.Wang, Q., McEwen, D. G. & Ornitz, D. M. (2000) Mech. Dev. 90, 283-287. [DOI] [PubMed] [Google Scholar]
- 4.Gurney, M. E., Heinrich, S. P., Lee, M. R. & Yin, H. S. (1986) Science 234, 566-574. [DOI] [PubMed] [Google Scholar]
- 5.Timar, J., Toth, S., Tovari, J., Paku, S. & Raz, A. (1999) Clin. Exp. Metastasis 17, 809-816. [DOI] [PubMed] [Google Scholar]
- 6.Chiao, J. W., Xu, W., Seiter, K., Feldman, E. & Ahmed, T. (1999) Leuk. Res. 23, 13-18. [DOI] [PubMed] [Google Scholar]
- 7.Niinaka, Y., Paku, S., Haga, A., Watanabe, H. & Raz, A. (1998) Cancer Res. 58, 2667-2674. [PubMed] [Google Scholar]
- 8.Morimoto, K., Hooper, D. C., Carbaugh, H., Fu, Z. F., Koprowski, H. & Dietzschold, B. (1998) Proc. Natl. Acad. Sci. USA 95, 3152-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietzschold, B., Morimoto, K., Hooper, D. C., Smith, J. S., Rupprecht, C. E. & Koprowski, H. (2000) J. Hum. Virol. 3, 50-57. [PubMed] [Google Scholar]
- 10.Reid, J. E. & Jackson, A. C. (2001) J. Neurovirol. 6, 511-517. [DOI] [PubMed] [Google Scholar]
- 11.Hooper, D. C., Kean, R. B., Scott, G. S., Spitsin, S. V., Mikheeva, T., Morimoto, K., Bette, M., Rohrenbeck, A. M., Dietzschold, B. & Weihe, E. (2001) J. Immunol. 167, 3470-3477. [DOI] [PubMed] [Google Scholar]
- 12.Dietzschold, B., Rupprecht, C. E., Fu, Z. F. & Koprowski, H. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M., Howley, P. M., Chanock, R. M., Monath, T. P., Melnick, J. L., Roizman, B. & Straus, S. E. (Lippincott–Raven, Philadelphia), pp. 1137-1159.
- 13.Yan, X., Prosniak, M., Curtis, M. T., Weiss, M. L., Faber, M., Dietzschold, B. & Fu, Z. F. (2001) J. Neurovirol. 7, 518-527. [DOI] [PubMed] [Google Scholar]
- 14.Niizeki, H., Kobayashi, M., Horiuchi, I., Akakura, N., Chen, J., Wang, J., Hamada, J. I., Seth, P., Katoh, H., Watanabe, H., et al. (2002) Br. J. Cancer 86, 1914-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo, Y., Long, J. M., Lu, C., Chan, S. L., Spangler, E. L., Mascarucci, P., Raz, A., Longo, D. L., Mattson, M. P., Ingram, D. K., et al. (2002) J. Neurochem. 80, 354-361. [DOI] [PubMed] [Google Scholar]
- 16.Gurney, M. E. (1987) Immunol. Rev. 100, 203-223. [DOI] [PubMed] [Google Scholar]
- 17.Munoz-Sanjuan, I., Fallon, J. F. & Nathans, J. (2000) Mech. Dev. 95, 101-112. [DOI] [PubMed] [Google Scholar]
- 18.Wang, Q., Bardgett, M. E., Wong, M., Wozniak, D. F., Lou, J., McNeil, B. D., Chen, C., Nardi, A., Reid, D. C., Yamada, K., et al. (2002) Neuron 35, 25-38. [DOI] [PubMed] [Google Scholar]
- 19.Schoorlemmer, J. & Goldfarb, M. (2001) Curr. Biol. 11, 793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, A. C., Phelan, C. C. & Rossiter, J. P. (2000) Can. J. Vet. Res. 64, 226-228. [PMC free article] [PubMed] [Google Scholar]
- 21.Morimoto, K., Patel, M., Corisdeo, S., Hooper, D. C., Fu, Z. F., Rupprecht, C. E., Koprowski, H. & Dietzschold, B. (1996) Proc. Natl. Acad. Sci. USA 93, 5653-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]