Abstract
The most familiar form of plant programmed cell death is the hypersensitive response (HR) associated with successful plant immune responses. HR is preceded by an oxidative burst and the generation of both reactive oxygen intermediates (ROI) and NO. The Arabidopsis LSD1 gene encodes a negative regulator of plant programmed cell death that meets several criteria for a regulator of processes relevant to ROI management during pathogen responses. Here we demonstrate that a highly conserved LSD1 paralogue, LOL1, acts as a positive regulator of cell death. Manipulation of LOL1 expression alters both the superoxide-dependent, runaway cell death phenotype of lsd1 plants and the normal HR. We also show that LSD1 and LOL1 have antagonistic effects on copper-zinc superoxide dismutase accumulation, consistent with functions in cell death control via maintenance of ROI homeostasis.
Plant biology is replete with examples of programmed cell death (PCD), yet very little is known about the relevant control mechanisms. The most familiar form of plant PCD is the hypersensitive response (HR) associated with successful plant innate immune responses (1–3). Recognition of a pathogen leads to rapid ion fluxes, production of superoxide, hydrogen peroxide, and other reactive oxygen intermediates (ROI), NO accumulation, mitogen-activated protein kinase signaling, transcriptional reprogramming in and around the infection site, salicylic acid (SA) biosynthesis, and cell collapse (4–6). While the HR is required for disease resistance in some plant–pathogen interactions (7), it may simply reflect the consequence of passing a signal threshold for cell death in others (8). In an effort to dissect the signal transduction pathway leading to HR and resistance, several loss-of-function mutations in Arabidopsis were isolated that express ectopic cell death (9–11) and also induce disease-resistance responses. The proteins encoded by these genes could be true negative regulators of HR and associated disease resistance responses. Alternatively, they could be negative regulators of cellular processes whose loss results in loss of homeostasis and ectopic cell death that activates disease resistance responses (12). The genes defined to date by these mutants are not related to regulators or executioners of cell death in metazoans (13).
Among the genes identified by mutation, Arabidopsis LSD1 encodes a negative regulator of plant PCD that meets several criteria for a regulator of processes relevant to ROI management in response to pathogens (14). A normal HR forms at attempted infection sites in lsd1 null mutants, but cell death subsequently expands beyond the HR boundary to engulf the entire leaf. Additionally, lsd1 plants cannot control cell death initiated by SA and chemicals that mimic its action (9). These chemicals do not cause cell death themselves, but accumulation of SA influences ROI levels locally in WT plants and leads to cell death in the lsd1 mutant (15). The lsd1 “runaway cell death” (rcd) phenotype is activated by a superoxide-dependent signal, as is the oxidative burst associated with WT HR (16). In the WT HR, this superoxide is rapidly converted by the enzyme superoxide dismutase (SOD) to hydrogen peroxide, and the balance of hydrogen peroxide and NO may ultimately control HR (17–19). Thus, we proposed that LSD1 is required for correct interpretation of ROI or ROI-dependent signals emanating from an HR site (16). Consistent with this idea, the up-regulation of cytosolic copper-zinc SOD (CuZnSOD) after SA application to WT plants is lacking in lsd1 (20). Furthermore, the lsd1 cell death phenotype requires function of EDS1 and PAD4, two genes that are also required for specific pathogen resistance (21), and function of NIM1/NPR1, a gene required for systemic induction of defense and normal SA accumulation (22). These phenotypes collectively suggest that LSD1 meets important criteria for a negative regulator of ROI-related cellular responses, including local signaling after pathogen infection (23).
The deduced LSD1 protein is small (189 aa), contains three highly related zinc fingers, and may function as either a transcriptional regulator or a scaffold protein (14). Families of zinc finger proteins often regulate related cellular processes. For example, the mammalian GATA family of transcription factors is important in erythroid and embryonic (24, 25) development, and the mammalian IAP protein family negatively controls PCD (26–28). Thus, we predicted that LSD1-related proteins could be regulators of responses to oxidative stress and, in particular, to the ROI formed during HR. Because the amino acid domains between the zinc-coordinating residues are often critical for function, we reasoned that other Arabidopsis proteins encoding LSD1-like zinc fingers might function like LSD1. Therefore, we searched the complete Arabidopsis genome for LSD1-related proteins by using only the internally conserved zinc finger motif of LSD1 (defined as a C2C2 class zinc finger, consensus CxxCRxxLMYxxGASxVxCxxC; see Fig. 6B, which is published as supporting information on the PNAS web site, www.pnas.org). Of the 104 C2C2 zinc finger proteins identified in the finished Arabidopsis genome (29) only two additional genes also contain multiple internally conserved LSD1-like zinc fingers. We called these LOL1 (LSD-One-Like 1; At1g32540) and LOL2 (At4g21610). There are two additional proteins predicted to encode only one LSD1-like zinc finger (At1g02170 and At4g25110). Here we focus on analysis of LOL1 and provide evidence that it acts antagonistically to LSD1 to regulate oxidative stress-induced cell death.
Materials and Methods
Plant Growth and Pathogen Infections. Plants were grown as described (30). All mutants or transgenic lines were generated in the WT genetic background Ws-0. Pseudomonas syringae pv tomato DC3000(avrRpm1) was grown overnight in King's B medium (31) and resuspended in 10 mM MgCl2 to a density of 5 × 107 colony-forming units/ml for HR tests. The bacterial suspension was then infiltrated into the abaxial surface of plant leaves by using a syringe without a needle, until the leaves appeared water-soaked (31). Peronospora parasitica isolates Emco5 and Emwa1 were propagated on the susceptible Arabidopsis ecotype Ws-0 (32). Conidiospores were suspended in water at a concentration of 3 × 104 spores per ml (Emco5) or 2 × 104 spores per ml (Emwa1) and spray-inoculated onto 4-week-old plants (32, 33). Inoculated plants were kept covered with a lid to increase humidity and grown at 19°C with a 9-h light period. Botrytis cinerea isolate B05–10 was propagated on potato dextrose agar (Difco) for 10–14 days at 20°C. Spores were resuspended in 1% glucose at a concentration of 1 × 106 spores per ml. For inoculation of plants, 2-μl droplets of the spore suspension were placed onto the adaxial leaf surface. Inoculated plants were kept under a lid to obtain high humidity and incubated at 20°C with an 8-h light period.
Identification of lol1-1 Mutants. We used two methods to isolate lol1 mutants. First, pooled DNA and mutant seeds from the Feldmann T-DNA insertion collection (34) were provided by the Arabidopsis Biological Resource Center at Ohio State University, Columbus. Using a LOL1 gene-specific (5′-TTCATGGCAATGGTGTGACCCC-3′) and a T-DNA insertion-specific primer (5′-GCTCAGGATCCGATTGTCGTTTCCCGCCTT-3′) we conducted a PCR-based screen and identified a T-DNA insertion 630 bp 3′ of the translational stop codon, designated lol1–1. Second, seven ethyl methanesulfonate point mutation alleles, unfortunately all in LOL1 introns, were isolated for us by the National Science Foundation-sponsored TILLING project (http://tilling.fhcrc.org:9366; ref. 35).
Construction of an lsd1/lol1-1 Double Mutant, Transgenic LOL1 Overexpression Lines, and lol1 Antisense Lines. The lol1-1 mutant was crossed to the lsd1 mutant and a homozygous lsd1/lol1-1 double mutant was identified by genotyping segregating F2 plants. To confirm lsd1 homozygosity we used a triple primer set: 5′-ACCTAACAAAAAGAAAAGTGTGTGAGG-3′, 5′-ATAATAACCCCTACTAGCTCTAACAAG-3′, and 5′-CTGCTACTTTCATCCAAAC-3′ (21). For identification of lol1-1 homozygotes we used primers 5′-TGAGT TATGAGCAATATAGAGGAA-3′ and 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′. To generate overexpression and antisense transgenic lines, we cloned the entire LOL1 coding region downstream of the cauliflower mosaic virus 35S promoter in either the sense or antisense orientation into the binary vector pBAR1-35S (36). These plasmid constructs were first transformed into Agrobacterium tumefaciens GV3101 and then subsequently into LSD1/lsd1 heterozygotes by Agrobacterium-mediated transformation (37). At least six independent lines per construct were identified in isogenic lsd1 and WT Ws-0 backgrounds. All experiments reported were carried out with at least four independent lines per construct per genetic background. All examined lines showed the phenotype we describe for any construct, but results are shown only for the two lines displaying the strongest phenotype per construct in each genetic background. To construct conditionally expressed transgenes, we cloned the entire coding region of LOL1, which had been C-terminally tagged with a hemagglutinin (HA) epitope, into the binary vector pTA7002 (38). This construct was first transformed into A. tumefaciens GV3101 and then subsequently into LSD1/lsd1 heterozygotes by Agrobacterium-mediated transformation (37). Several independent, dexamethasone (DEX)-inducible transgenic lines were identified in either the lsd1 or the Ws-0 backgrounds. Induction with 20 μM DEX was performed as described (39). All lines conditionally expressing LOL1 displayed cell death after induction with DEX, whereas control lines containing only an empty vector construct did not show any DEX-inducible cell death. Results are shown for one line (lsd1/LOL1-HA1) in the lsd1 background and for one line (LOL1-HA19) in the Ws-0 background.
Determination of LOL1 Expression Levels in Transgenic and Mutant Lines. RNA was isolated from WT, transgenic, and mutant lines by using the TRIZOL Reagent (GIBCO/BRL) according to the manufacturer's instructions. LOL1 expression levels in the lol1-1 mutant and LOL1 overexpression lines were determined by Northern hybridization. Fifty micrograms per lane of total RNA was separated on a denaturing gel and transferred to Hybond-N membranes (Amersham Pharmacia). Hybridization was performed in ULTRAhyb (Ambion, Austin, TX) according to the manufacturer's specifications. A LOL1-specific probe was labeled with α-ATP by using the Prime-It II random primer labeling kit (Stratagene). Signal intensities were determined with a PhosphorImager. RNA levels in lol1 antisense lines were determined by RT-PCR (RETROscript, Ambion). RT-PCRs were performed according to the manufacturer's instructions. Plant 18S Competimer Primers (Ambion) were used to coamplify the 18S internal loading control. To amplify the 5′ UTR of endogenous LOL1 transcripts primers 5′-CGAAACGAGATTCTACA AT TATGC-3′ and 5′-AT TCACTCCA AGA AGAATTGC-3′ were used. To label the PCR products, α-ATP was added to the PCRs (95°C30s,55°C30s,72°C 30 s, 32 cycles). PCR products were separated on a standard agarose gel. Signal intensities were measured with a PhosphorImager and standardized against the 18S standard. It is of course possible that protein levels are not directly correlated with mRNA levels in these mutants and transgenic lines.
Cell Death Measurements. Dead and dying cells were visualized by trypan blue staining as described (33). A protocol adapted from Dellagi et al. (40) was used for conductivity measurements: 48 h after treatment with 150 μM benzo (1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester (BTH), leaf disks (7 mm diameter) were removed from treated leaves with a cork borer, floated in distilled water for 45 min, and subsequently transferred to tubes containing 6 ml of distilled water. Conductivity of the solution was determined with an Orion (Boston) Conductivity Meter at the indicated time points. Means and standard errors were calculated from three replicate measurements per genotype per experiment. For each measurement, we used four leaf disks. The entire experiment was performed four times. Similarly, upon inoculation of leaves with 5 × 107 colony-forming units/ml P. syringae pv tomato DC3000 (avrRpm1) leaf disks were removed and treated as described above. Mean and standard error were calculated from four disks per genotype, with four repetitions within an experiment. The experiment was repeated three times.
Spore Count Assay. Four-week-old plants were spray inoculated with spore suspensions of either P. parasitica isolate Emco5 or P. parasitica isolate Emwa1 (32, 33). At 6 days postinoculation (dpi), all of the leaves of an inoculated plant were harvested and their fresh weight was determined. Spores were resuspended in 300 μl water/100 μg freshweight. Spore concentrations were determined with a hemocytometer (Reichert). Data are presented as spores per ml. Mean and standard error were calculated from four repetitions within one experiment. The experiment was repeated two times.
Protein Extraction and Western Analysis. Protein was isolated in extraction buffer (20 mM Tris·HCl, pH 7.5/150 mM NaCl/1mM EDTA/1% Triton X-100/0.1% SDS) and subsequently separated by standard methods on 12% or 14% SDS/PAGE gels. Western blotting was performed by standard methods with the TRANSBLOT SD system (Bio-Rad). Equal loading and transfer was confirmed by Ponceau S staining. Western blots were developed with either anti-HA antibodies (Roche Diagnostics) or anti-CSD1, anti-CSD2, and anti-MSD1 antibodies (gift of Dan Kliebenstein, University of California, Davis) by standard methods.
Results
LOL1 Is a Member of the LSD1 Gene Family of Zinc Finger Proteins. LOL1 (At1g32540) encodes a protein of 154 aa, containing three LSD1-like zinc fingers (Fig. 6 A and B). Outside of the three zinc fingers, LSD1 and LOL1 share essentially no homology. Surprisingly, LOL1 orthologues are extremely conserved (86–93% identity) among monocotyledonous and dicotyledonous plants that diverged between 170 million and 235 million years ago (41) (Fig. 6C). For comparison, the five genes neighboring LOL1 on Arabidopsis chromosome 1 (At1g32520, At1g32530, At1g32550, At1g32560, At1g32580) are only 50–70% identical to their closest monocot and dicot homologues, whereas housekeeping genes of the Krebs cycle [isocitrate dehydrogenase (At4g35260), succinate dehydrogenase (At3g27380), fumarase (At2g47510), and malate dehydrogenase (At1g04410)] display 65–90% identity. Both LSD1 and LOL1 are absent from bacteria, yeast, and animals. Like LSD1, LOL1 is constitutively expressed in all plant tissues (data not shown). Its expression is unaltered in lsd1 null mutant plants grown under conditions that do not induce rcd and in infected leaves (data not shown).
To elucidate the function of the LOL1 gene, we identified a T-DNA insertion mutant allele, designated lol1-1 (Fig. 6A) and seven ethyl methanesulfonate point mutations by using the TILLING procedure (see Materials and Methods). The latter were all in introns, did not affect mRNA levels, and are not described further. We used Northern analysis to demonstrate that LOL1 mRNA levels in the lol1-1 mutant were reduced to ≈25% of WT levels (Table 1 and Fig. 7, which is published as supporting information on the PNAS web site) despite the T-DNA insertion being 630 nt 3′ of the translational stop site. The lol1-1 mutant was crossed to lsd1 and double mutants were identified (see Materials and Methods; Table 1). We also generated transgenic lines expressing either higher or lower levels of LOL1 mRNA (LOL1-s and lol1-as, respectively) in both the Ws-0 and isogenic lsd1 null backgrounds (see Materials and Methods; Table 1). LOL1 transcripts in lol1-as lines were reduced to 25–60% of WT (Table 1 and Fig. 7). Overexpression in LOL1-s lines resulted in only an ≈2-to 3-fold increase in LOL1 transcript levels, despite the use of a strong viral promoter (Table 1 and Fig. 7). The lol1–1 insertion allele and the various transgenic lines all were developmentally normal. LSD1 transcript levels were unaltered in LOL1-s, lol1-1, and lol1-as lines (data not shown).
Table 1. Mutant and transgenic lines used in this study.
| Line | Mutation or construct | Genetic background | LOL1 expression levels (fold of WT) |
|---|---|---|---|
| lol1-1 | Mutation | Ws-0 | 0.25 |
| lol1-as9 | 35S-antisense | Ws-0 | 0.25 |
| LOL1-s3 | 35S-sense | Ws-0 | 2.00 |
| LOL1-s5 | 35S-sense | Ws-0 | 3.00 |
| LOL1-HA19 | DEX-ind. sense | Ws-0 | >40.00 |
| lsd1/lol1-1 | Mutation | lsd1 | 0.25 |
| lsd1/lol1-as9 | 35S-antisense | lsd1 | 0.25 |
| lsd1/LOL1-s4 | 35S-sense | lsd1 | 2.00 |
| lsd1/LOL1-s5 | 35S-sense | lsd1 | 3.00 |
| lsd1/LOL1-HA1 | DEX-ind. sense | lsd1 | >40.00 |
LOL1 expression levels are derived from the Northern analysis and RT-PCR presented in Fig. 6.
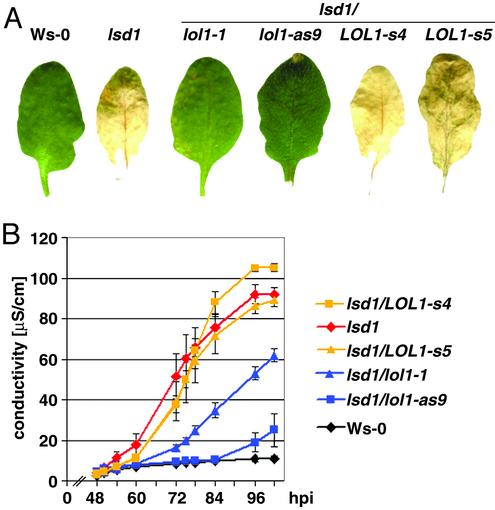
LOL1 Function Is Required for Full lsd1 rcd. We addressed whether manipulation of LOL1 mRNA levels altered either inducible rcd in the lsd1 background or the WT response to pathogens. BTH induces rcd in lsd1 plants (42). We sprayed the lsd1/lol1-1, lsd1/lol1-as, and lsd1/LOL1-s lines with BTH and monitored rcd. By 7 days post-BTH spray, rcd devastated lsd1 and lsd1/LOL1-s leaves; the tissue was collapsed and completely dried. In contrast, lsd1/lol1-1 and lsd1/lol1-as lines were predominantly healthy and green (Fig. 1A). We quantified cell death by monitoring cellular ion leakage, a measure of membrane damage (Fig. 1B) (40, 43, 44). WT tissue did not exhibit any significant cell death and thus no increase in ion leakage, whereas lsd1 mutant tissue expressed maximal ion leakage at 96 h after BTH treatment. In contrast, the reduction of LOL1 transcript levels in tissues of the lsd1/lol1-as lines or lsd1/lol1-1 significantly reduced ion leakage compared with either lsd1 or the lsd1/LOL1-s lines (Fig. 1B).
Fig. 1.
LOL1 function is required for lsd1 rcd. Four-week-old plants were sprayed with 150 μM BTH. (A) Representative pictures of leaves were taken 7 dpi. Genotypes are indicated above the leaves. This experiment was performed four times, using a total of ≈40 plants and ≈200 leaves per genotype. (B) Leaf disks were removed for conductivity measurements 48 h after BTH treatment. Mean and standard error were calculated from four disks per genotype, with three repetitions within an experiment (a total of 12 disks per genotype). Symbols represent the genotypes indicated at right.
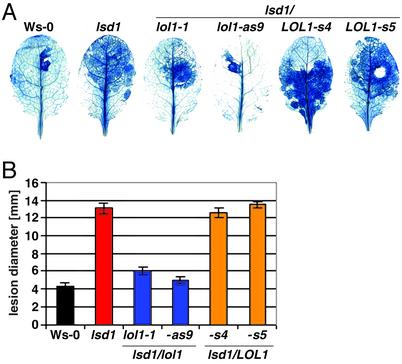
We infected lsd1/lol1 double mutants with the necrotrophic pathogen Botrytis cinerea. The lsd1 rcd phenotype is accelerated after B. cinerea infection, presumably because of increased ROI production associated with this interaction (45). We drop-inoculated 4-week-old plants with B. cinerea and visualized cell death by lactophenol trypan blue staining (Fig. 2A) (46). The B. cinerea isolate used in this experiment is moderately pathogenic on Ws-0, and the dark blue staining is thus limited to the site of infection. In contrast, lsd1 leaves are killed by rcd and fungal proliferation, and the dark blue staining zones spread. The lsd1/lol1-as lines exhibited significantly reduced rcd, whereas the lsd1/LOL1-s lines were as susceptible as lsd1. We measured the lesion diameter of 20 leaves per genotype (Fig. 2B). Reduction of LOL1 function clearly attenuated lsd1 rcd.
Fig. 2.
LOL1 function is required for lsd1 rcd. Four-week-old plants were inoculated with 2-μl droplets of B. cinerea isolate B05-10 (gift of S. Lam, Syngenta Biotechnology, Research Triangle Park, NC). Inoculated leaves were removed and stained with lactophenol trypan blue at 3 dpi. (A) Representative leaves stained with lactophenol trypan blue. Genotypes are indicated above leaves. The white zone in the lsd1/LOL1-s5 leaf indicates full maceration of the tissue. (B) As lesions develop from the site of infection in a more or less circular manner, lesion diameter was determined by measuring the longest transect across the lactophenol trypan blue stains with a caliper. Mean and standard error were calculated from 20 leaves per genotype. The experiment was repeated four times with similar results.
Collectively, the data in Figs. 1 and 2 demonstrate that LOL1 positively regulates the lsd1 rcd phenotype. In these assays, we did not observe any obvious enhanced cell death in the lsd1/LOL1-s overexpression lines compared with lsd1. We speculated that expression of sufficient LOL1 to enhance lsd1 rcd would be lethal (see below). Further, manipulation of LOL1 mRNA levels in a WT background did not induce an rcd phenotype in these assays, suggesting that the regulatory function of LSD1 is epistatic to that of LOL1.
LOL1 Is a Positive Regulator of Cell Death. We next assessed whether misregulation of LOL1 could influence HR in a WT background. We used P. syringae pv tomato DC3000(avrRpm1) to trigger HR (through the action of the RPM1 disease resistance gene, ref. 47) in Ws-0, the LOL1-s and lol1-as lines, and lol1-1. We quantified ion leakage over the time course of HR (Fig. 3). The onset of HR using this assay was at 2 hours postinoculation (hpi), and maximum ion leakage was reached at 6 hpi. Thus, this in vitro assay correlates with the observed onset of RPM1-dependent HR in vivo, where AvrRpm1 is delivered into the host cell at 1–2 hpi (48, 49), tissue collapse is visible at ≈3 hpi, and full tissue collapse is evident by 6 hpi (50). The time course of cell collapse was accelerated in LOL1-s lines and slightly reduced in lol1-as lines (Fig. 3). These data suggest that LOL1 also acts as a positive regulator of HR-associated signaling.
Fig. 3.
Modest LOL1 overexpression enhances HR. Four-week-old plants were infiltrated with 5 × 107 colony-forming units/ml of P. syringae pv tomato DC3000(avrRpm1) (53). Immediately afterward, leaf disks were removed and processed as in Fig. 1B. Mean and standard error were calculated from four disks per genotype, with four repetitions within an experiment. The experiment was performed three times with similar results. Control leaves infiltrated with 10 mM MgCl2 did not show HR or increased conductivity (data not shown). Genotypes and transgenic line designations are indicated at right.
We presume that the lol1-as lines displayed no significant diminution in ion leakage because the input signal levels from the high-dose bacterial inoculum irreversibly committed the cells to HR. Thus, we additionally analyzed infected leaves for growth of P. syringae pv tomato DC3000 (avrRpm1) by using a 500-fold lower inoculum. In support of our signal threshold hypothesis, lol1-as lines exhibited slightly reduced resistance, allowing ≈0.5 logs more bacterial growth than WT (data not shown).
If LOL1 functions as a positive regulator of cell death (Figs. 1, 2, 3), then the LOL1-s and lol1-as lines might exhibit enhanced and suppressed basal disease resistance to virulent pathogens, respectively. We inoculated Ws-0, LOL1-s, and lol1-as lines with two virulent isolates (Emwa1 and Emco5) of the oomycete pathogen P. parasitica and monitored production of asexual spores as a measure of susceptibility. In LOL1-s lines, susceptibility was reduced by 20–80%, whereas lol1-as lines showed an increase in susceptibility by 20–100% (Table 2, which is published as supporting information on the PNAS web site).
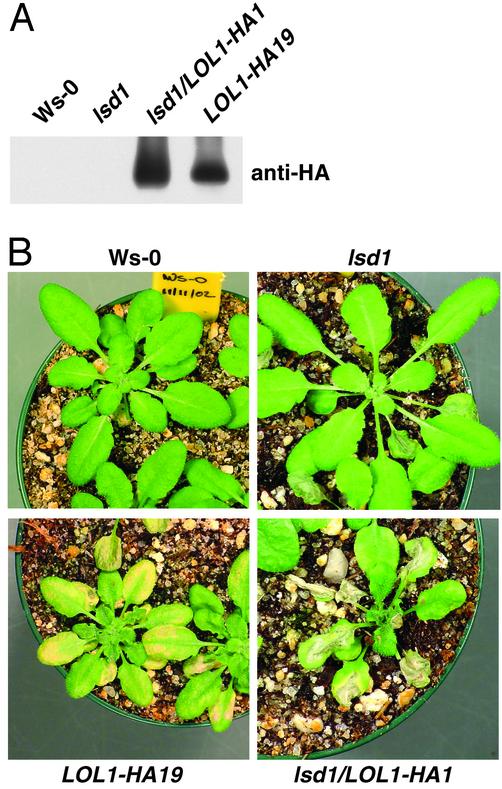
Conditional Overexpression of LOL1 Is Sufficient for Induction of Cell Death in WT Plants and Enhances rcd in lsd1. Constitutive overexpression of LOL1 resulted in transgenic lines that up-regulate LOL1 transcription only 2- to 3-fold (Table 1 and Fig. 7). We thus observed only subtle cell death phenotypes in these lines (Fig. 3). We made transgenic lines that conditionally express a LOL1-HA epitope-tagged fusion under the control of a DEX-inducible promoter (see Materials and Methods). We generated multiple, independent LOL1-HA lines in both Ws-0 and lsd1 genetic backgrounds (see Materials and Methods and Table 1). RNA blot analysis revealed that lines lsd1/LOL1-HA1 and LOL1-HA19 up-regulate LOL1 transcription levels >40-fold at 12 h after induction with DEX (Table 1 and Fig. 7). These lines also accumulate large amounts of HA epitope-tagged LOL1 at 24 hpi (Fig. 4A). We observed cell death at 24 hpi by using Trypan blue staining. Extensive cell death and chlorosis in both lsd1/LOL1-HA1 and LOL1-HA19 plants was apparent at 4 dpi (Fig. 4B). At 7 dpi, DEX-treated lsd1/LOL1-HA1 and LOL1-HA19 plants are dead (data not shown). Throughout this time course, cell death in the lsd1 background is enhanced compared with WT. Thus, conditional overexpression of LOL1 is sufficient to induce cell death in both WT and lsd1 plants. This finding is consistent with the data in Figs. 1, 2, 3 using the low-level overexpression transgenic lines.
Fig. 4.
Conditional overexpression of LOL1 is sufficient to induce cell death. Four-week-old plants were sprayed with 20 μM DEX. (A) Twenty-five micrograms of protein isolated 24 h after DEX-treatment was separated on a 12% SDS/PAGE gel. The Western blot was probed for the HA epitope tag. Genotypes and transgenic status are indicated above each lane. (B) Pictures of representative plants (genotypes listed above or below photo) were taken 4 days after DEX treatment. This experiment was performed two times. Additional independent transgenic lines (two lsd1/LOL1-HA lines and three LOL1-HA lines) displayed the same phenotype (data not shown).
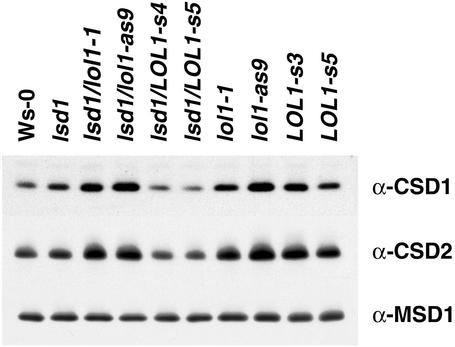
SOD Accumulation Is Antagonistically Regulated in lsd1/lol1-as and lsd1/LOL1-s Lines. We previously demonstrated that (i) although LSD1 can positively regulate CSD1 and CSD2 accumulation, it is not required for basal CSD expression, and (ii) cell death results in LSD1-independent accumulation of CSD1 and CSD2 (20). If LOL1 antagonizes LSD1, one might expect this to be reflected at the level of CSD1 and CSD2 accumulation. We observed CSD1 and CSD2 expression in Ws-0 and lsd1 (Fig. 5). In lsd1/lol1-1 and the lsd1/lol1-as line, we noted elevated levels of CSD1 and CSD2, presumably because LOL1 levels are lowered. Consistent with these results, we also observed higher than WT accumulation of CSD1 and CSD2 when LOL1 levels were lowered in an LSD1 background. Conversely, low-level LOL1 overexpression in lsd1 resulted in diminution of CSD1 and CSD2 levels, again presumably because of the negative regulatory capacity of LOL1. Thus, in a sensitized lsd1 background, the antagonistic effect of LOL1 on CSD1 and CSD2 accumulation is easily observed. In contrast, the LOL1-s lines also expressed slightly elevated levels of CSD1 and CSD2 (Fig. 5). At first glance this appears inconsistent with the other data in Fig. 5. We propose, however, that cells in the LOL1-s lines are constitutively poised to undergo cell death (see Fig. 3). This activates the previously described CSD1 and CSD2 up-regulation that is independent of LSD1 (20). In sum, the data in Fig. 5 are consistent with our model that LSD1 and LOL1 act antagonistically to control ROI-related stress and subsequent cell death.
Fig. 5.
Antagonism between LSD1 and LOL1 function leads to misregulation of SOD accumulation. Twenty-five micrograms of protein isolated from untreated plants was separated on a 14% (CSD1) or 12% (CSD2, MSD1) SDS/PAGE gel. Western blots were probed with antisera against CSD1, CSD2, or MSD1. [MnSOD expression levels have been shown to be unaffected by cell death. It thus is suitable as a loading control (20).] Genotypes and transgenic line designations are indicated above the Western blots.
Discussion
Our results clearly establish LOL1 as a positive regulator of plant PCD in three contexts. First, LOL1 is required for full rcd in an lsd1 background. Second, very modest overexpression of LOL1 levels in WT plants enhances pathogen-driven HR. Third, conditional overexpression of LOL1 to high levels is sufficient to induce cell death in both WT and lsd1 plants. These are probably not the only contexts where LOL1 regulates PCD. For example, our inability to isolate full loss-of-function alleles, even as heterozygotes, strongly suggests that LOL1 functions during gametophyte or seedling development. Our inability to recover strong constitutive overexpression phenotypes originally suggested that this, too, would be lethal, an idea confirmed by the use of conditional LOL1 overexpression lines.
Both the deduced LOL1 and LSD1 proteins feature three plant-specific versions of the (Cys-X-X-Cys)2 type zinc finger motif. LOL1 is astonishingly conserved throughout monocot and dicot species (86–93% identity, Fig. 6C). In contrast, LSD1 displays only 49–72% identity to its orthologues in various monocot and dicot species (M. Ellerström and J.L.D., unpublished work) as does the final member of this gene family, LOL2 (P.E. and J.L.D., unpublished work). Because these three proteins share a common, functionally relevant domain, we envision that they collaborate to integrate many signals that impinge on ROI homeostasis in plants, including pathogen infection.
Two models of how LOL1 and LSD1 regulate PCD are conceivable. First, LOL1 and LSD1 may function as antagonistic transcriptional regulators or scaffolds, competing for the same promoter elements and/or accessory transcription factors on cell death execution genes. Alternatively, PCD control in plants might be analogous to the control of cell death in metazoan systems, where caspase function is modulated by inhibitor of apoptosis proteins (IAPs). These zinc finger proteins function as an apopstat and maintain a threshold for cell death execution (51). IAPs themselves can be inhibited by unrelated IAP-binding proteins such as DIABLO in mammals and REAPER/HID/GRIM in Drosophila (28, 52). In plants, a threshold for the commitment to PCD after HR is apparently maintained by LSD1 (9, 16). In this scenario, the balance of LSD1 and LOL1 could regulate cell death commitment: an excess of LSD1 would antagonize the cell death machinery whereas LOL1 excess would activate it. A direct interaction of LSD1 and LOL1, as described in this model, is supported by yeast two-hybrid data (P.E. and J.L.D., unpublished work). Our results demonstrate that related proteins act antagonistically to control ROI-mediated cell death in plants; they set the stage for detailed examination of the mechanism by which balances and imbalances among the members of the small LSD1 gene family control this important facet of plant physiology.
Supplementary Material
Acknowledgments
We thank Dr. Dan Kliebenstein for anti-CSD1, anti-CSD2, and anti-MSD1 antibodies; Shruti Chudasama for technical assistance; Drs. John McDowell, Robert Dietrich, Jeff Chang, and Mats Ellerström for useful comments on the manuscript; and Dr. Todd Vision for suggestions on LOL1 sequence comparisons. This work was supported by postdoctoral fellowships from the Deutsche Akakademische Austauschdienst and the Schweizer Nationalfonds (to P.E.), National Institutes of Health Grant R01-GM057171-01 (to J.L.D.), and Research Experience for undergraduates supplements from the National Science Foundation (to A.A.M. and V.R.F.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BTH, benzo (1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester; DEX, dexamethasone; dpi, days postinoculation; HA, hemagglutinin; hpi, hours postinoculation; HR, hypersensitive response; PCD, programmed cell death; rcd, runaway cell death; ROI, reactive oxygen intermediates; SA, salicylic acid; SOD, superoxide dismutase.
References
- 1.Dangl, J. L., Dietrich, R. A. & Thomas, H. (2000) in Biochemistry and Molecular Biology of Plants, eds. Buchanan, B., Gruissem, W. & Jones, R. (ASPP Press, Rockville, MD), pp. 1044-1100.
- 2.Morel, J.-B. & Dangl, J. L. (1997) Cell Death Diff. 19, 17-24. [DOI] [PubMed] [Google Scholar]
- 3.Shirasu, K. & Schulze-Lefert, P. (2000) Plant Mol. Biol. 44, 371-385. [DOI] [PubMed] [Google Scholar]
- 4.Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826-833. [DOI] [PubMed] [Google Scholar]
- 5.Feys, B. J. & Parker, J. E. (2000) Trends Genet. 16, 449-455. [DOI] [PubMed] [Google Scholar]
- 6.McDowell, J. M. & Dangl, J. L. (2000) Trends Biochem. Sci. 25, 79-82. [DOI] [PubMed] [Google Scholar]
- 7.Peterhänsel, C., Freialdenhoven, A., Kurth, J., Kolsch, R. & Schulze-Lefert, P. (1997) Plant Cell 9, 1397-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendahmane, A., Kanyuka, K. & Baulcombe, D. C. (1999) Plant Cell 11, 781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietrich, R. A., Delaney, T. P., Uknes, S. J., Ward, E. R., Ryals, J. A. & Dangl, J. L. (1994) Cell 77, 565-577. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg, J. T. & Ausubel, F. M. (1993) Plant J. 4, 327-342. [DOI] [PubMed] [Google Scholar]
- 11.Walbot, V., Hoisington, D. A. & Neuffer, M. G. (1983) in Genetic Engineering of Plants, eds. Kosuge, T. & Meredith, C. (Plenum, New York), Vol. 3, pp. 431-442. [Google Scholar]
- 12.Dangl, J. L., Dietrich, R. A. & Richberg, M. H. (1996) Plant Cell 8, 1793-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, A. M. & Dangl, J. L. (1996) Trends Plant Sci. 1, 114-119. [Google Scholar]
- 14.Dietrich, R. A., Richberg, M. H., Schmidt, R., Dean, C. & Dangl, J. L. (1997) Cell 88, 685-694. [DOI] [PubMed] [Google Scholar]
- 15.Shirasu, K., Nakajima, H., Rajasekhar, V. K., Dixon, R. A. & Lamb, C. (1997) Plant Cell 9, 261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jabs, T., Dietrich, R. A. & Dangl, J. L. (1996) Science 273, 1853-1856. [DOI] [PubMed] [Google Scholar]
- 17.Delledonne, M., Xia, Y., Dixon, R. A. & Lamb, C. (1998) Nature 394, 585-588. [DOI] [PubMed] [Google Scholar]
- 18.Delledonne, M., Zeier, J., Marocco, A. & Lamb, C. (2001) Proc. Natl. Acad. Sci. USA 98, 13454-13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendehenne, D., Pugin, A., Klessig, D. F. & Durner, J. (2001) Trends Plant Sci. 6, 177-183. [DOI] [PubMed] [Google Scholar]
- 20.Kliebenstein, D. J., Dietrich, R. A., Martin, A. C., Last, R. L. & Dangl, J. L. (1999) Mol. Plant Microbe Interact. 12, 1022-1026. [DOI] [PubMed] [Google Scholar]
- 21.Rustérucci, C., Aviv, D. H., Holt, B. F., III, Dangl, J. L. & Parker, J. E. (2001) Plant Cell 13, 2211-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aviv, D. H., Rusterucci, C., Holt, B. F., III, Dietrich, R. A., Parker, J. E. & Dangl, J. L. (2002) Plant J. 29, 381-391. [DOI] [PubMed] [Google Scholar]
- 23.Loake, G. (2001) Curr. Biol. 11, R1028-R1031. [DOI] [PubMed] [Google Scholar]
- 24.Charron, F., Paradis, P., Bronchain, O., Nemer, G. & Nemer, M. (1999) Mol. Cell. Biol. 19, 4355-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pevny, L., Simon, M. C., Robertson, E., Klein, W. H., Tsai, S. F., D'Agati, V., Orkin, S. H. & Costantini, F. (1991) Nature 349, 257-260. [DOI] [PubMed] [Google Scholar]
- 26.Deveraux, Q. L. & Reed, J. C. (1999) Genes Dev. 13, 239-252. [DOI] [PubMed] [Google Scholar]
- 27.Miller, L. K. (1999) Trends Cell Biol. 9, 323-328. [DOI] [PubMed] [Google Scholar]
- 28.Verhagen, A. M., Coulson, E. J. & Vaux, D. L. (2001) Genome Biol. 2, 3009.1-3009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C.-J., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O. J., Samaha, R. R., et al. (2000) Science 290, 2105-2110. [DOI] [PubMed] [Google Scholar]
- 30.Dangl, J. L., Lehnackers, H., Kiedrowski, S., Debener, T., Rupprecht, C., Arnold, M. & Somssich, I. E. (1991) in Advances in Molecular Genetics of Plant–Microbe Interactions, eds. Hennecke, H. & Verma, D. P. S. (Kluwer Academic, Dordrecht, The Netherlands), Vol. 1, pp. 78-83. [Google Scholar]
- 31.Debener, T., Lehnackers, H., Arnold, M. & Dangl, J. L. (1991) Plant J. 1, 289-302. [DOI] [PubMed] [Google Scholar]
- 32.Dangl, J. L., Holub, E., Debener, T., Lehnackers, H., Ritter, C. & Crute, I. R. (1992) in Methods in Arabidopsis Research, eds. Koncz, C., Chua, N.-H. & Schell, J. (World Scientific, Singapore), pp. 393-418.
- 33.Koch, E. & Slusarenko, A. J. (1990) Plant Cell 2, 437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinney, E. C., Ali, N., Traut, A., Feldmann, K. A., Belostotsky, D. A., McDowell, J. M. & Meagher, R. B. (1995) Plant J. 8, 465-477. [DOI] [PubMed] [Google Scholar]
- 35.Colbert, T., Till, B. J., Tompa, R., Reynolds, S., Steine, M. N., Yeung, A. T., McCallum, C. M., Comai, L. & Henikoff, S. (2001) Plant Physiol. 126, 480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDowell, J. M., Dhandaydham, M., Long, T. A., Aarts, M. G. M., Goff, S., Holub, E. B. & Dangl, J. L. (1998) Plant Cell 10, 1861-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bechtold, N., Ellis, J. & Pelletier, G. (1993) C. R. Acad. Sci. 316, 1194-1199. [Google Scholar]
- 38.Aoyama, T. & Chua, N.-H. (1997) Plant J. 11, 605-612. [DOI] [PubMed] [Google Scholar]
- 39.Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R. T., Katagiri, F. & Dangl, J. L. (2000) Cell 101, 353-363. [DOI] [PubMed] [Google Scholar]
- 40.Dellagi, A., Brisset, M. N., Paulin, J. P. & Expert, D. (1998) Mol. Plant-Microbe Interact. 11, 734-742. [DOI] [PubMed] [Google Scholar]
- 41.Yang, Y. W., Lai, K. N., Tai, P. Y. & Li, W. H. (1999) J. Mol. Evol. 48, 597-604. [DOI] [PubMed] [Google Scholar]
- 42.Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K.-H., Oostendorp, M., Staub, T., Ward, E., Kessman, H. & Ryals, J. (1996) Plant Cell 8, 629-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker, C. J., Mock, N., Glazener, J. & Orlandi, E. W. (1993) Physiol. Mol. Plant Pathol. 43, 81-94. [Google Scholar]
- 44.Torres, M. A., Dangl, J. L. & Jones, J. D. G. (2002) Proc. Natl. Acad. Sci. USA 99, 523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Govrin, E. M. & Levine, A. (2000) Curr. Biol. 10, 751-757. [DOI] [PubMed] [Google Scholar]
- 46.Keogh, R. C., Deverall, B. J. & McLeod, S. (1980) Trans. Br. Mycol. Soc. 74, 329-333. [Google Scholar]
- 47.Grant, M. R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R. W. & Dangl, J. L. (1995) Science 269, 843-846. [DOI] [PubMed] [Google Scholar]
- 48.Ritter, C. & Dangl, J. L. (1995) Mol. Plant-Microbe Interact. 8, 444-453. [DOI] [PubMed] [Google Scholar]
- 49.Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A. & Mansfield, J. (2000) Plant J. 23, 441-450. [DOI] [PubMed] [Google Scholar]
- 50.Dangl, J. L., Ritter, C., Gibbon, M. J., Mur, L. A., Wood, J. R., Goss, S., Mansfield, J., Taylor, J. D. & Vivian, A. (1992) Plant Cell 4, 1359-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Budihardjo, I., Oliver, H., Lutter, M., Luo, X. & Wang, X. (1999) Annu. Rev. Cell Dev. Biol. 15, 269-290. [DOI] [PubMed] [Google Scholar]
- 52.Hay, B. A. (2000) Cell Death Differ. 7, 1045-1056. [DOI] [PubMed] [Google Scholar]
- 53.Ritter, C. & Dangl, J. L. (1996) Plant Cell 8, 251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.