Abstract
The majority of DNA in eukaryotic cells exists in the highly condensed structural hierarchy of chromatin, which presents a challenge to DNA repair enzymes in that recognition, incision, and restoration of the original sequence at most sites must take place within these structural constraints. To test base excision repair (BER) activities on chromatin substrates, an in vitro system was developed that uses human uracil DNA glycosylase (UDG), apyrimidinic/apurinic endonuclease (APE), and DNA polymerase β (pol β) on homogeneously damaged, rotationally positioned DNA in nucleosomes. We find that UDG and APE carry out their combined catalytic activities with reduced efficiency on nucleosome substrates (≈10% of that on naked DNA). Furthermore, these enzymes distinguish between two different rotational settings of the lesion on the histone surface, showing a 2- to 3-fold difference in activity between uracil facing ``toward'' and ``away from'' the histones. However, UDG and APE will digest such substrates to completion in a concentration-dependent manner. Conversely, the synthesis activity of pol β is inhibited completely by nucleosome substrates and is independent of enzyme concentration. These results suggest that the first two steps of BER, UDG and APE, may occur ``unassisted'' in chromatin, whereas downstream factors in this pathway (i.e., pol β) may require nucleosome remodeling for efficient DNA BER in at least some regions of chromatin in eukaryotic cells.
Keywords: chromatin, glycosylase, DNA polymerase β, apyrmidinic/apurinic endonuclease
The daily insult to genomic DNA by both exogenous and endogenous genotoxic agents requires that DNA repair pathways constantly survey the genome for potential mutagenic modifications (1–3). Furthermore, the DNA in eukaryotic cells is packaged into heterogeneous, dynamic chromatin fibers. The primary level of packaging in these fibers, the nucleosome core particle (NuCP), consists of an octamer of the four core histones H2A, H2B, H3, and H4, with 147 bp of DNA wound in 1.65 left-handed superhelical turns on the octamer surface (4–6). In bulk (or transcriptionally inactive) chromatin, these nucleosome cores are connected by variable lengths of DNA (from ≈20 to ≈90 bp), called ``linker DNA,'' complexed with linker histones (H1 and/or H5).
Previous studies have demonstrated that chromatin substrates are refractory to DNA repair, transcription, transcription factor binding, and V(D)J recombination (7–10). A major obstacle that chromatin and, more specifically, nucleosomes present to protein binding and enzyme catalysis is that a portion of the DNA is occluded because of steric hindrance by the histone octamer surface (see ref. 5 for review). Also, the malleability of DNA is constrained in chromatin, and any protein/enzyme that requires severe alteration of DNA structure to bind or carry out catalysis may be inhibited dramatically (7). To circumvent these restrictions, modulation of chromatin structure through ATP-dependent nucleosome remodeling and/or posttranslation modification of histones (e.g., acetylation) acts to facilitate access to buried sites in chromatin (see refs. 11 and 12 for reviews). Moreover, additional protein (or protein complexes), not required for activity on naked DNA, may facilitate binding or catalysis in chromatin.
The base excision repair (BER) pathway is responsible for removal of >10,000 DNA lesions daily in each human cell (13, 14). In addition, lesions targeted by the BER pathway are relatively small, causing little (if any) helix distortion, and would seem to pose a problem in the initial step of recognition during BER. Many of the lesions targeted by BER have been shown not to inhibit elongation by RNA polymerases both in vitro and in vivo (15). Lesions of this type can lead to a number of different types of mutations, including transitions, transversions, or deletions (15).
This study examines repair of a guanine–uracil mismatch, which results mainly from two major pathways in the cell: spontaneous deamination of cytosine (16) and chemical deamination of cytosine facilitated by bisulfites and nitric oxide (17, 18). This mismatch has the capacity to form base pairs (19) and should not dramatically affect the structure of DNA in a nucleosome. This aspect of nondistorting DNA lesions makes their recognition and removal more challenging than that of bulky DNA adducts, which can alter nucleosome structure (10, 20). Furthermore, any lesion targeted by BER, such as uracil, can be considered a prototype for BER studies, because, after catalysis by the damage-specific DNA glycosylase, the BER pathway converges to one of two routes for repairing the resulting apyrimidinic/apurinic site (short-patch and long-patch BER; ref. 21).
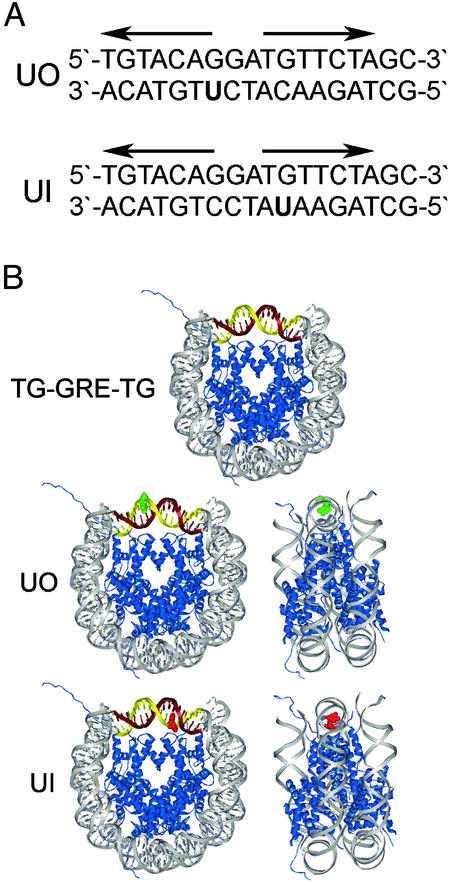
Enzymatic steps in guanine–uracil BER of naked DNA in vitro are well known and have been established in reconstituted systems with purified enzymes (14, 22). Our study divided the BER pathway into its individual components [uracil DNA glycosylase (UDG), apyrmidinic/apurinic endonuclease (APE), and DNA polymerase β (pol β)] and tested which, if any, of the catalytic activities of these enzymes are modulated by nucleosome substrates containing a guanine–uracil mismatch that is rotationally positioned in nucleosomes. Damaged mononucleosomes were constructed with uracil nucleotides at (or near) the nucleosome dyad and were positioned away from (uracil out, UO) and toward (uracil in, UI) the histone octamer surface (Fig. 1). These rotationally positioned substrates were subjected to digestion by UDG/APE and synthesis by pol β and were compared with the naked DNA counterpart.
Fig. 1.
Modified sequence element and assembled nucleosome structure. (A) GRE sequence for UO and UI substrates. Bold arrows are the pseudo-palindrome binding site for the glucocorticoid receptor. (B Top) Nucleosome schematic of the TG motif (gray) flanking the GRE (maroon and gold). (Middle and Bottom) UO and UI (red) substrates (B was adapted from ref. 4). The rotational setting of DNA in these NuCPs has been determined by hydroxyl radical footprinting (10).
Experimental Procedures
Preparation of Oligonucleotide Substrates. Substrates were constructed by annealing and ligating synthetic oligonucleotides containing the TG motif (23), a nucleosome positioning sequence, flanking a glucocorticoid hormone receptor response element (GRE) as described (8–10) with minor modifications. Sequences for the six oligonucleotides, L1, L2, L2UO, L2UI, R1, and R2, are shown in Table 1.
Table 1. Oligonucleotide sequences and abbreviations.
| Abbreviation | Sequence |
|---|---|
| L1* | 5′-TGTTAGAGCCTGTAACTCGGTGTTAGAGCCTGTAAC TCGGTGTTAGAGCCTGTAACTCGGTTGTACAGGATGTTC TAGCCTGTAACAGCC-3′ |
| L2 | 5′-GTTACAGGCTAGAACATCCTGTACAACCGAGTTACA GGCTCTAACACCGAGTTACAGGCTCTAACACCGAGTTAC AGGCTCTAACA-3′ |
| L2UO | 5′-GTTACAGGCTAGAACATCUTGTACAACCGAGTTACA GGCTCTAACACCGAGTTACAGGCTCTAACACCGAGTTAC AGGCTCTAACA-3′ |
| L2UI | 5′-GTTACAGGCTAGAAUATCCTGTACAACCGAGTTACA GGCTCTAACACCGAGTTACAGGCTCTAACACCGAGTTAC AGGCTCTAACA-3′ |
| R1 | 5′-TGTTAGAGCCTGTAACTCGGTGTTAGAGCCTGTAAC TCGGTGTTAGAGCCTGTAACT-3′ |
| R2 | 5′-AGTTACAGGCTCTAACACCGAGTTACAGGCTCTAAC ACCGAGTTACAGGCTCTAACAGGCT-3′ |
Sequence in bold indicates the GRE; bold U indicates the site of a uracil nucleotide; bold, italic GTAC represents an Rsal site; and underlined sequence corresponds to the TG motif.
5′-amino modified nucleotide to avoid radioactive labeling during T4 polynucleotide kinase reaction.
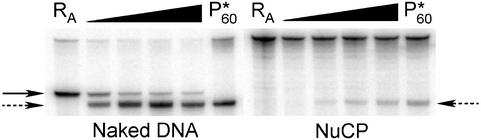
5′ End-Labeling. DNA was end-labeled with 32P, as described (10). To avoid labeling of the undamaged strand, the 5′ terminus of the L1 sequence was synthesized with a 5′-amino modifier (Glen Research, Sterling, VA) attached to block T4 polynucleotide kinase from incorporating a labeled nucleotide. Radiolabeled DNA was extracted with phenol and separated from unincorporated radionucleotides on a G-25 spin column (Amersham Biosciences). Specificity of end-labeling was analyzed by digestion with RsaI (e.g., see Fig. 5 naked DNA, lane RA; nonspecific labeling appears as a doublet).
Fig. 5.
RsaI digestion of UI nucleosome during UDG/APE digestion. Denaturing gel of naked DNA and NuCPs after RsaI and UDG/APE digestion. RA lanes are for naked DNA and NuCP samples digested by RsaI only (solid arrow), and P*60 lanes are for these samples digested for 60 min by UDG and APE only (dashed arrow). Aliquots of samples digested with all three enzymes were taken from 30 sec to 60 min.
Nucleosome Reconstitutions. NuCPs were prepared from pooled chicken erythrocytes (Lampire Biological Laboratories, Pipersville, PA) by following the methods described by Libertini and Small (24) with minor modifications. The damaged and undamaged DNA substrates were reconstituted into mononucleosomes by histone octamer transfer using excess chicken erythrocyte core particle (CE-CP) and incremental changes in ionic strength (20, 25). The reconstitution products were analyzed on native 6% polyacrylamide (0.3% bisacrylamide) gels in 0.5× TBE buffer (45 mM Tris/45 mM boric acid/1 mM EDTA, pH 8.3), the gels were dried and exposed to PhosphorImager screens (Molecular Dynamics), and the 32P-labeled DNA was visualized on a PhosphorImager (model 445-P90, Molecular Dynamics). Images were analyzed with imagequant software (Molecular Dynamics).
UDG/APE Digestion. The UDG and APE reaction mixture contained 50 mM Hepes (pH 7.5), 2 mM DTT, 0.2 mM EDTA (pH 8.0), 100 μg/ml BSA, 10% glycerol (wt/vol), 5 mM MgCl2,4mM ATP, and 0.3 μM dNTPs (22). CE-CP, at ≈300 nM, were added to naked DNA samples to adjust for the excess CE-CP present in reconstituted nucleosome samples. The reactions were initiated by adding UDG and APE to final concentrations of 1 nM each. Incubation was at 37°C for 30 sec to 1 h. For testing the effect of flanking nucleotides on UDG and APE, digestion was performed on naked DNA (UO and UI) under identical reaction conditions, except that enzyme concentrations were 100 pM and incubations were from 30 sec to 10 min. Aliquots were removed at appropriate times and treated with phenol to stop the reaction. The digested DNA was treated and analyzed as described above, except that separation was on a 10% denaturing gel.
pol β Synthesis. DNA synthesis with pol β was performed in a mixture that contained the same buffer as that used for the UDG and APE reaction. CE-CP was added to naked DNA, as described above, and the reactions were initiated by adding UDG, APE, and pol β to final concentrations of 10 nM each. Incubations were at 37°C for 1–4 h. Aliquots were removed at appropriate times and treated with phenol to stop the reaction. The digested DNA was treated and analyzed as described above, except that separation was on an 8% denaturing gel.
An identical experiment to label repair patches was carried out as above, except that radioactive dCTP ([α-32P]dCTP) was added. Samples were incubated at 37°C, and aliquots were removed at appropriate times and treated with 50 mM EDTA, boiled, and loaded directly onto a 10% denaturing gel. The digested DNA was analyzed as described above.
RsaI Restriction Site Protection. The reaction mixture of RsaI and UDG/APE was identical to that used in the UDG/APE digestion alone. CE-CP was added to naked DNA, as described above. Adding 5 units of RsaI initiated the reactions, and incubation was at 37°C for 1 h. An aliquot was removed at the 1-h time point, and the reaction was stopped by phenol extraction. The UDG and APE were then added to the reaction at 1 nM each, and digestion was continued for an additional hour. Aliquots were taken at 1, 10, 30, and 60 min and extracted with phenol to stop the reaction. The digested DNA was treated and analyzed as described above, except that separation was on a 10% denaturing gel.
Results
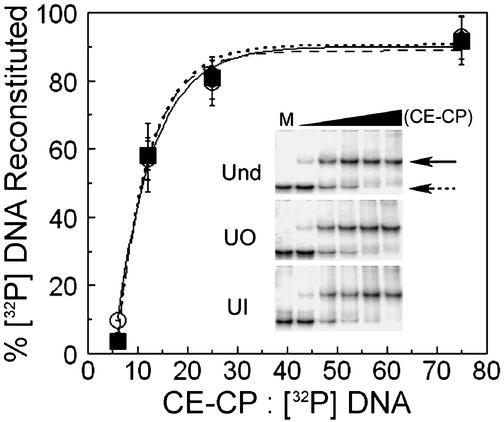
Reconstitution of Nucleosomes with Uracil at the Nucleosome Dyad. To examine whether a guanine–uracil mismatch affects nucleosome formation, reconstitutions were carried out at varying CE-CP concentrations with UO DNA, UI DNA, and undamaged DNA as control. As shown in Fig. 2, there was no marked effect of uracil at either position (UO or UI) on nucleosome formation as assayed by gel shifts. The reconstituted fraction on these gels was quantified and compared for the undamaged and damaged substrates, and the data indicate that inclusion of uracil nucleotides at UO and UI has no effect on nucleosome formation (Fig. 2). Furthermore, the conditions used for nucleosome reconstitutions for various repair experiments did not yield any labeled naked DNA (data not shown).
Fig. 2.
Nucleosome reconstitutions. Percent of NuCPs present for different CE-CP to [32P] DNA ratios. Data represent the mean ± 1 SD (three independent reconstitutions for each substrate) of the ratio of band intensities for naked DNA and NuCPs on native gels (Inset). Undamaged DNA, solid line, ○; UO DNA, dashed line, ▪, and UI DNA, dotted line, ⋄. (Inset) Native gels of nucleosome reconstitutions at varying CE-CP concentration (naked DNA, dashed arrow; NuCP, solid arrow). Lane M is naked DNA only; the black triangle indicates increasing CE-CP concentration.
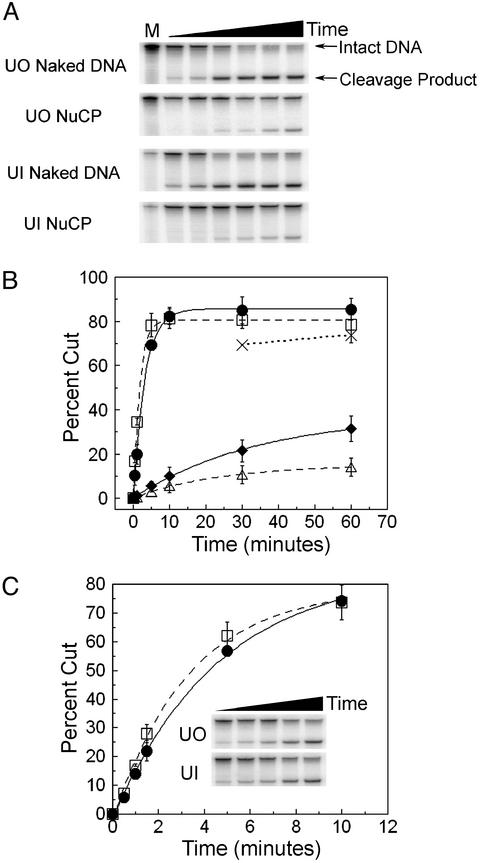
Cleavage of Damaged NuCPs by UDG/APE. Incubation of DNA containing a guanine–uracil mismatch with UDG and APE produces a single-strand break that can be visualized on a denaturing gel (Fig. 3A). At low enzyme concentrations, this band is produced during increasing digestion times and can be quantified in both naked DNA and NuCPs (Fig. 3B). Assaying UDG and APE together on nucleosome substrates indicates that these enzymes proceed at a much slower combined rate than they do on their naked DNA counterparts (Fig. 3 A and B). Indeed, the calculated rates for these digestion curves (Fig. 3B, dashed and solid lines) indicate that nucleosomes are digested at a rate ≈10-fold slower by the combined action of UDG and APE than naked DNA. At higher enzyme concentrations, digestion proceeds to near completion on the chromatin substrates (Fig. 3B, dotted line). The apparently incomplete digestion by UDG and APE of naked DNA in Fig. 3B is due to the small amount of ``resistant'' intact fragment remaining on these gels (Fig. 3A, solid arrow), which is the result of incomplete denaturation of the TG motif sequence (unpublished results).
Fig. 3.
UDG and APE digestion of UO and UI nucleosomes. (A) Denaturing gels of UO and UI naked DNA and NuCPs after incubation with UDG and APE, each at 1 nM. Time is from 30 sec to 1 h and M is mock-treated. (B) Plot of the mean ± 1 SD (three independent experiments) of the percent DNA cut by 1 nM UDG and APE for UO DNA (solid line, •), UI DNA (dashed line, □), UO NuCP (solid line, ⋄), and UI NuCP (dashed line, ▵). Dotted line (×) represents nucleosome samples digested with 10 nM UDG and APE. (C) Plot of the mean ± 1 SD (three independent experiments) of the percent of DNA cut by 100 pM UDG and APE for UO DNA (solid line, □) and UI DNA (dashed line, □). (Inset) Representative gel for digestions of UO and UI naked DNA for times from 30 sec to 10 min.
The rotational setting of nucleotides in relation to the histone octamer surface has been shown to dramatically affect access to the GRE (8) and transcription factor binding sites (26, 27). As shown in Fig. 1B, the damage sites UO and UI differ in rotational setting relative to the histone octamer surface. Digestion by UDG and APE, at low enzyme concentrations, reveals that there is a 2- to 3-fold difference in digestion rate between these two different rotational settings (Fig. 3B). Thus, although UDG and APE can digest either site (UO or UI), their (combined) reaction rate is reduced for uracil facing the histone surface.
The effect that rotational setting of the damage site has on UDG and APE digestion rate could be explained by the different sequence contexts of the UO and UI substrates (Table 1, L2UO and L2UI; ref. 28). Indeed, differences in flanking sequences have been shown to affect UDG digestion kinetics (29). Therefore, we examined the digestion of UO and UI naked DNA at low enzyme concentrations to accurately measure digestion rates. As shown in Fig. 3C, at enzyme concentrations of 100 pM for both UDG and APE, the flanking sequences of the uracil nucleotide had no effect on naked DNA digestion rates. Therefore, the difference in digestion of the two rotational settings (UO and UI) must be due to a variation in accessibility of the uracil residues.
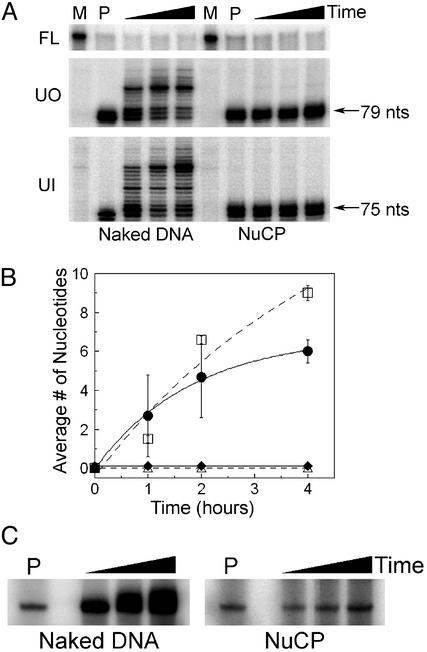
Synthesis by pol β on NuCPs. pol β catalyzes two steps in the short-patch BER pathway: (i) incorporation of the proper nucleotide opposite the undamaged strand and (ii) removal of the sugar phosphate moiety by a deoxyribosephosphodiesterase activity contained within the N-terminal domain of pol β (30, 31). Because this order of reaction has been characterized (22), only the initial step of strand elongation by pol β was assayed. As shown earlier (Fig. 3B, dotted line), digestion by 10 nM UDG and APE at 30 min to 1 h is nearly equivalent for both naked DNA and NuCP DNA samples (Fig. 4A, Top). Furthermore, synthesis of pol β is seen in the naked DNA lanes of UO and UI samples, where the cleaved 147-bp fragments are extended by 1–11 nt (Fig. 4 A Middle Left and Bottom Left and B). However, synthesis is absent in the NuCP lanes of the same substrates (Fig. 4 A Middle Right and Bottom Right and B) and was not detected regardless of time or enzyme concentration. Indeed, direct labeling of repair patches with [α-32P]dCTP verified that pol β readily incorporates nucleotides onto naked DNA, whereas the NuCP samples are incompetent substrates for extension (Fig. 4C).
Fig. 4.
pol β synthesis activity on UO and UI nucleosomes. (A) Denaturing gels of UO and UI naked DNA and NuCPs. Shown is the uncut fraction remaining (Top) and increasing size of fragment (Middle and Bottom) after incorporation of nucleotides by pol β into naked DNA or NuCPs. Lanes M are for mock-treated samples (no enzyme present), and lanes P are for samples digested with UDG and APE only. Incubation times are from 1 to 4 h with all three enzymes. (B) Plot of the mean ± 1 SD (three independent experiments) of the average number of nucleotides inserted by pol β into UO DNA (solid line, •), UI DNA (dashed line, □), UO NuCP (solid line, ⋄), and UI NuCP (dashed line, ▵). (C) Denaturing gels of UI naked DNA and NuCPs showing the 5′ end-labeled cleavage product and subsequent addition of radionucleotides to the 3′ terminus. P represents incubation with UDG and APE alone, and the black triangles represent increasing incubation time (30 min to 2 h) with UDG, APE, pol β, and [α-32P]dCTP.
RsaI Restriction Site Protection During UDG/APE Digestion. Transient exposure, or displacement, of DNA away from the histone surface during digestion with UDG/APE could explain the limited access of uracil to UDG and APE in nucleosomes (Fig. 3). To test this hypothesis, the RsaI site (Table 1, bold italics), which is within one helical turn of the guanine–uracil mismatch (Table 1), was digested with RsaI in the presence of UDG and APE. As shown in Fig. 5, the RsaI site is readily digested in naked DNA samples (naked DNA lane RA, solid arrow) and further digested by UDG and APE (naked DNA, dashed arrow). The NuCP, however, is not digested by RsaI, indicating that this site is protected completely in the nucleosome (Fig. 5 Right, lane RA). Importantly, this site remains protected from RsaI while the damage site is digested by UDG and APE (Fig. 5, dashed arrow).
Discussion
In this study, the catalytic activity of individual human BER enzymes was monitored in a specifically damaged oligonucleotide reconstituted into NuCPs. The first two enzymes in BER, UDG and APE, are able to carry out their respective catalytic activities on nucleosome substrates at reduced rates compared with those on naked DNA (Fig. 3 A and B). Furthermore, these enzymes distinguished between the different rotational settings of uracil, with UO being digested ≈2- to 3-fold more rapidly than UI (Fig. 3B). Conversely, the next step in BER, synthesis by pol β, is inhibited completely, regardless of rotational setting (Fig. 4). Previous work with nucleosome substrates has shown that BER intermediates (various flap substrates) and completion of the BER pathway (nicked substrates) can be carried out by FEN1 and DNA ligase I, respectively (32, 33). Thus, the major point of restriction of BER in chromatin must be the synthesis step of pol β.
These results address two aspects of accessibility to rotationally positioned damage in a nucleosome. First, the access of UDG and APE to UI requires substantial ``torsional flexibility'' of nucleosome DNA (Fig. 3). This observation agrees with previous work demonstrating that DNA bound to a nucleosome has torsional flexibility similar to that of DNA free in solution (34, 35). Second, the crystal structures for UDG, APE, and pol β (36–38) lend insight into the degree of bending of DNA that can be tolerated in nucleosomes. UDG and APE are relatively small enzymes that bend the helix axis of DNA ≈20° and 35°, respectively, on binding (36, 37). This distortion is considerably less than that of a nicked DNA molecule bound to pol β, which causes a bend of ≈90° (38, 39). Moreover, the catalytic activity of the V(D)J recombinase enzymes recombination-activating gene (RAG) 1 and RAG2 was tested on a nucleosome substrate, in combination with the nonhistone proteins HMG1 and HMG2 (7). Together, these proteins induce a severe helical bend in the DNA to carry out catalysis during V(D)J recombination (40). When the recombination signal sequences, recognized by RAG1 and RAG2, were packaged into a nucleosome, the catalytic activity of RAG1 and RAG2 was inhibited completely (7). Taken together, these findings suggest that torsional flexibility of DNA is significant on the histone surface and that it allows access of DNA glycosylases and APE to DNA lesions. Furthermore, nucleosomes must be able to accommodate moderate bending (20° to 35°) of DNA. Bending beyond this point, however, must result in a severe energy penalty incompatible with preserving a canonical nucleosome structure.
To relieve the restriction imposed by nucleosomes on pol β, additional activities may be required to modify nucleosomes, such as those of ATP-dependent nucleosome remodeling factors and/or x-ray cross complementation group protein 1 (XRCC1), Werner syndrome protein (WRN), and poly(ADP-ribose) polymerase (PARP)-1 and -2 (12, 41–43). The proteins XRCC1, PARP-1 and -2, and WRN have been shown to modulate BER by interacting with nicked DNA, pol β, FEN1, and DNA ligase III (43–46). Interestingly, PARP and XRCC1 also associate with chromatin and proteins involved in chromatin architecture, suggesting a connection between these proteins and facilitation of BER in chromatin (47–49). However, our initial findings indicate that inclusion of XRCC1 or WRN during the reaction does not facilitate repair of these nucleosome substrates (unpublished data). In addition, we have found that removal of the histone tails with limited trypsin digestion has no effect on UDG and APE activity, RsaI accessibility, or pol β synthesis in these nucleosomes (unpublished data). Such ``tailless'' histones have been used to mimic the high acetylation state of histone octamers (see ref. 50 for review). Finally, previous work has demonstrated that protected lesions in nucleosomal DNA can be exposed to nucleotide excision repair enzymes or UV photolyase by the addition of ATP-dependent remodeling factors in vitro (51–53). Although no direct interaction has been shown between pol β and a nucleosome remodeling factor, proteins such as XRCC1, WRN, and/or PARP may mediate such an interaction.
A recent study by Nilsen et al. (54) also showed that enzymatic processes in the BER pathway proceed with reduced activity on nucleosome substrates. This study used the 5S rDNA nucleosome positioning sequence with uracil residues at sites more than two or five helical turns away from the dyad center. As observed by these authors (54), a feature of 5S rDNA nucleosomes, demonstrated previously by others (55–59), is the presence of multiple translational settings of the DNA on the histone octamer surface. Thus, the transient exposure of uracil by variability in translational setting of the 5S rDNA, as well as increased ``breathing'' of DNA away from the dyad center of nucleosomes (8, 52), makes the results of Nilsen et al. (54) more difficult to interpret. Indeed, it was demonstrated that different translational settings of DNA on a histone octamer alter the affinity of transcription factors (8, 9), even when the rotational setting remains constant (26). Furthermore, release of stretches of DNA away from the histone surface leads to the same transient exposure to restriction enzymes (60). In the present study, the TG motif was used for positioning uracil at the nucleosome dyad, because these sequences ``lock down'' the translational setting of DNA in nucleosomes more so than that of 5S rDNA (61). This is demonstrated by the complete protection of the RsaI site when DNA is bracketed with TG motifs and packaged into a nucleosome (Fig. 5), whereas the restriction enzyme sites used to probe the stability of the 5S rDNA nucleosome were only partially protected (54).
Even though distinct differences exist between the nucleosomes used in the present study and those used by Nilsen et al. (54), the results obtained in these studies are complementary. Taken together, they suggest that nucleosome stability plays a critical role in recognition of DNA damage and completion of BER. For instance, transient exposure of DNA away from the histones has been described as a dynamic equilibrium, with the DNA being more constrained near the nucleosome dyad (60). The 5S rDNA is less constrained on the histone surface than the TG motif in nucleosomes (61). In addition, the torsional flexibility of DNA is directly addressed in the present study by the use of rotationally positioned damage sites (UO and UI) and by monitoring the digestion by UDG and APE. Finally, the flexibility of DNA along the helix axis is addressed in both studies when the synthesis by pol β (after cleavage by UDG and APE) is monitored. The complete lack of pol β synthesis observed in this study and the partial inhibition of pol β synthesis seen by Nilsen et al. (54) may reflect differences in stability between the two nucleosome substrates. Furthermore, recent studies have identified mouse genomic DNA repeats with equivalent or superior ability to form nucleosomes as the TG motif sequences (61). Therefore, nucleosomes used in this study represent a more ``closed'' chromatin structure in intact cells than do 5S rDNA nucleosomes, and such stable structures may require additional activities for completion of DNA repair.
Acknowledgments
We thank Drs. Brian J. Vande Berg and Rajendra Prasad for helpful discussions and for graciously providing human UDG, APE, and pol β; Dr. Gerhardt Munske for oligonucleotide synthesis; Dr. Lisa Gloss for help with the calculation of rate constants for UDG and APE; and current and past members of the Smerdon laboratory, especially Drs. Joe Kosmoski and Antonio Conconi, for helpful discussions and guidance during this study. This study was supported by National Institutes of Health Grant ES04106 from the National Institute of Environmental Health Sciences.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NuCP, nucleosome core particle; BER, base excision repair; UDG, uracil DNA glycosylase; APE, apyrimidinic/apurinic endonuclease; pol β, DNA polymerase β; UO, uracil out; UI, uracil in; GRE, glucocorticoid hormone receptor response element; CE-CP, chicken erythrocyte core particle; RAG, recombination-activating gene.
References
- 1.Hoeijmakers, J. H. J. (2001) Nature 411, 366–374. [DOI] [PubMed] [Google Scholar]
- 2.Wood, R. D., Mitchell, M., Sgouros, J. & Lindahl, T. (2001) Science 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- 3.Hanawalt, P. C. (1998) Mutat. Res. 400, 117–125. [DOI] [PubMed] [Google Scholar]
- 4.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 5.Wolffe, A. P. (1998) Chromatin: Structure and Function (Academic, San Diego).
- 6.van Holde, K. E. (1988) Chromatin (Springer, New York).
- 7.Golding, A., Chandler, S., Ballestar, E., Wolffe, A. P. & Schissel, M. S. (1999) EMBO J. 18, 3712–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, Q. & Wrange, O. (1995) Mol. Cell. Biol. 8, 4375–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, Q. & Wrange, O. (1993) Genes Dev. 7, 2471–2482. [DOI] [PubMed] [Google Scholar]
- 10.Kosmoski, J. V. & Smerdon, M. J. (1999) Biochemistry 38, 9485–9494. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 12.Becker, P. B. & Horz, W. (2002) Annu. Rev. Biochem. 71, 247–273. [DOI] [PubMed] [Google Scholar]
- 13.Ames, B. N., Shigenaga, M. K. & Hagen, T. M. (1993) Proc. Natl. Acad. Sci. USA 90, 7915–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindahl, T. (1993) Nature 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 15.Doetsch, P. W. (2002) Mutat. Res. 510, 131–140. [DOI] [PubMed] [Google Scholar]
- 16.Frederico, L. A., Kunkel, T. A. & Shaw, B. R. (1993) Biochemistry 32, 6523–6530. [DOI] [PubMed] [Google Scholar]
- 17.Wink, D. A., Kasprzak, K. S., Maragos, C. M., Elespuru, R. K., Misra, M., Dunams, T. M., Cebula, T. A., Koch, W. H., Andrews, A. W., Allen, J. S. & Keefer, L. K. (1991) Science 254, 1001–1003. [DOI] [PubMed] [Google Scholar]
- 18.Chen, H. & Shaw, B. R. (1993) Biochemistry 32, 3535–3539. [DOI] [PubMed] [Google Scholar]
- 19.Strobel, S. A. & Cech, T. R. (1995) Science 267, 675–679. [DOI] [PubMed] [Google Scholar]
- 20.Mann, D. B., Springer, D. L. & Smerdon, M. J. (1997) Proc. Natl. Acad. Sci. USA 94, 2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 22.Srivastava, D. K., Vande Berg, B. J., Prasad, R., Molina, J. T., Beard, W. A., Tomkinson, A. E. & Wilson, S. H. (1998) J. Biol. Chem. 273, 21203–21209. [DOI] [PubMed] [Google Scholar]
- 23.Shrader, T. E. & Crothers, D. M. (1990) J. Mol. Biol. 216, 69–84. [DOI] [PubMed] [Google Scholar]
- 24.Libertini, L. J. & Small, E. W. (1980) Nucleic Acids Res. 8, 3517–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson, R. T. & Kunzler, P. (1979) Nucleic Acids Res. 6, 1387–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterton, H. G. & Hapgood, J. (1996) Nucleic Acids Res. 24, 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godde, J. S., Nakatani, Y. & Wolffe, A. (1995) Nucleic Acids Res. 23, 4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, S. & Smerdon, M. J. (1999) J. Biol. Chem. 274, 12201–12204. [DOI] [PubMed] [Google Scholar]
- 29.Bellamy, S. R. W. & Baldwin, G. S. (2001) Nucleic Acids Res. 29, 3857–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto, Y. & Kim, K. (1995) Science 269, 699–702. [DOI] [PubMed] [Google Scholar]
- 31.Piersen, C. E., Prasad, R., Wilson, S. H. & Lloyd, R. S. (1996) J. Biol. Chem. 271, 17811–17815. [DOI] [PubMed] [Google Scholar]
- 32.Huggins, C. F., Chafin, D. R., Aoyagi, S., Henricksen, L. A., Bambara, R. A. & Hayes, J. J. (2002) Mol. Cell 10, 1201–1211. [DOI] [PubMed] [Google Scholar]
- 33.Chafin, D. R., Vitolo, J. M., Henricksen, L. A., Bambara, R. A. & Hayes, J. J. (2000) EMBO J. 19, 5492–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashikawa, I., Kinosita, K., Jr., Ikegami, A., Nishimura, Y., Tsuboi, M., Watanabe, K., Iso, K. & Nakano, T. (1983) Biochemistry 22, 6018–6026. [DOI] [PubMed] [Google Scholar]
- 35.Wang, J., Hogan, M. & Austin, R. H. (1982) Proc. Natl. Acad. Sci. USA 79, 5896–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mol, C. D., Izumi, T., Mitra, S. & Tainer, J. A. (2000) Nature 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 37.Slupphaug, G., Mol, C. D., Kavli, B., Arvai, A. S., Krokan, H. E. & Tainer, J. A. (1996) Nature 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 38.Sawaya, M. R., Prasad, R., Wilson, S. H., Kraut, J. & Pelletier, H. (1997) Biochemistry 36, 11205–11215. [DOI] [PubMed] [Google Scholar]
- 39.Krahn, J. M., Beard, W. A., Miller, H., Grollman, A. P. & Wilson, S. H. (2003) Structure (Cambridge, Mass.) 11, 121–127. [DOI] [PubMed] [Google Scholar]
- 40.Aidinis, V., Bonaldi, T., Beltrame, M., Santagata, S., Bianchi, M. E. & Spanopoulou, E. (1999) Mol. Cell. Biol. 19, 6532–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson, C. L. (2002) EMBO Rep. 3, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ura, K. & Hayes, J. J. (2002) Eur. J. Biochem. 269, 2288–2293. [DOI] [PubMed] [Google Scholar]
- 43.Harrigan, J. A., Opresko, P. L., Von Kobbe, C., Kedar, P. S., Prasad, R., Wilson, S. H. & Bohr, V. A. (March 27, 2003) J. Biol. Chem., 10.1074/jbc.M213103200. [DOI] [PubMed]
- 44.Kubota, Y., Nash, R. A., Klungland, A., Schar, P., Barnes, D. E. & Lindahl, T. (1996) EMBO J. 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 45.Marintchev, A., Robertson, A., Dimitriadis, E. K., Prasad, R., Wilson, S. H. & Mullen, G. P. (2000) Nucleic Acids Res. 28, 2049–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidal, A. E., Boiteux, S., Hickson, I. D. & Radicella, J. P. (2001) EMBO J. 20, 6530–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griesenbeck, J., Oei, S. L., Mayer-Kuckuk, P., Ziegler, M., Buchlow, G. & Schweiger, M. (1997) Biochemistry 36, 7297–7304. [DOI] [PubMed] [Google Scholar]
- 48.Bernstein, C., Bernstein, H., Payne, C. M. & Garewal, H. (2002) Mutat. Res. 511, 145–178. [DOI] [PubMed] [Google Scholar]
- 49.Masson, M., Niedergang, C., Schreiber, V., Muller, S., Menissier-de Murcia, J. & de Murcia, G. (1998) Mol. Cell. Biol. 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner, B. M. & O'Neill, L. P. (1995) Semin. Cell Biol. 6, 229–236. [DOI] [PubMed] [Google Scholar]
- 51.Hara, R. & Sancar, A. (2002) Mol. Cell. Biol. 22, 6779–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ura, K., Araki, M., Saeki, H., Masutani, C., Ito, T., Iwai, S., Mizukoshi, T., Kaneda, Y. & Hanaoka, F. (2001) EMBO J. 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaillard, H., Fitzgerald, D. J., Smith, C. L., Peterson, C. L., Richmond, T. J. & Thoma, F. (March 11, 2003) J. Biol. Chem., 10.1074/jbc.M300770200. [DOI] [PubMed]
- 54.Nilsen, H., Lindahl, T. & Verreault, A. (2002) EMBO J. 21, 5943–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, X., Mann, D. B., Suquet, C., Springer, D. L. & Smerdon, M. J. (2000) Biochemistry 39, 557–566. [DOI] [PubMed] [Google Scholar]
- 56.Pennings, S., Meersseman, G. & Bradbury, E. M. (1991) J. Mol. Biol. 220, 101–110. [DOI] [PubMed] [Google Scholar]
- 57.Dong, F., Hansen, J. C. & van Holde, K. E. (1990) Proc. Natl. Acad. Sci. USA 87, 5724–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flaus, A., Luger, K., Tan, S. & Richmond, T. J. (1996) Proc. Natl. Acad. Sci. USA 93, 1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panetta, G., Buttinelli, M., Flaus, A., Richmond, T. J. & Rhodes, D. (1998) J. Mol. Biol. 282, 683–697. [DOI] [PubMed] [Google Scholar]
- 60.Anderson, J. D., Thastrom, A. & Widom, J. (2002) Mol. Cell. Biol. 22, 7147–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Widlund, H. R., Cao, H., Simonsson, S., Magnusson, E., Simonsson, T., Nielsen, P. E., Kahn, J. D., Crothers, D. M. & Kubista, M. (1997) J. Mol. Biol. 267, 807–817. [DOI] [PubMed] [Google Scholar]