Abstract
Protein tyrosine phosphatase RQ (PTPRQ) was initially identified as a protein tyrosine phosphatase (PTPase)-like protein that is upregulated in a model of renal injury. Here we present evidence that, like PTEN, the biologically important enzymatic activity of PTPRQ is as a phosphatidylinositol phosphatase (PIPase). The PIPase specificity of PTPRQ is broader than that of PTEN and depends on different amino acid residues in the catalytic domain. In vitro, the recombinant catalytic domain of PTPRQ has low PTPase activity against tyrosine-phosphorylated peptide and protein substrates but can dephosphorylate a broad range of phosphatidylinositol phosphates, including phosphatidylinositol 3,4,5-trisphosphate and most phosphatidylinositol monophosphates and diphosphates. Phosphate can be hydrolyzed from the D3 and D5 positions in the inositol ring. PTPRQ does not have either of the basic amino acids in the catalytic domain that are important for the PIPase activity of PTEN or the sequence motifs that are characteristic of type II phosphatidylinositol 5-phosphatases. Instead, the PIPase activity depends on the WPE sequence present in the catalytic cleft of PTPRQ, and in the ``inactive'' D2 domains of many dual-domain PTPases, in place of the WPD motif present in standard active PTPases. Overexpression of PTPRQ in cultured cells inhibits proliferation and induces apoptosis. An E2171D mutation that retains or increases PTPase activity but eliminates PIPase activity, eliminates the inhibitory effects on proliferation and apoptosis. These results indicate that PTPRQ represents a subtype of the PTPases whose biological activities result from its PIPase activity rather than its PTPase activity.
The phosphorylation/dephosphorylation of phosphotyrosine in proteins, and of the inositol phosphate moiety of phosphatidylinositol phospholipids, are important mechanisms through which cells regulate proliferation, survival, migration, and secretion. The protein tyrosine phosphatase (PTPase) superfamily of enzymes includes tyrosine-specific, dual specificity, and low molecular weight phosphatases (1, 2). A small number of these PTPase family members, including myotubularin and PTEN, can also hydrolyze the D3 phosphate from phosphatidylinositols, and both of these activities could contribute to the biological activities of these proteins (1, 3–5).
PTPase RQ (PTPRQ) was discovered and cloned in an effort to identify PTPases with up-regulated expression in renal disease (6). The sequence and deduced structure of PTPRQ classifies it as a type III receptor PTPase with 18 fibronectin type III domains and a single catalytic domain. Most receptor PTPases contain two potential catalytic domains, designated D1 and D2, with the D2 domain having little or no measurable enzymatic activity. A phylogenetic evaluation of PTPase sequences revealed that the receptor-like PTPases, previously classified by their extracellular domains into nine distinct subtypes (7), would be classified into virtually identical groups based on the sequence relatedness of their catalytic domains (8). Thus, PTPRQ, DEP1, PTPβ, and GLEPP1 are classified as type R3 PTPases in the phylogenetic classification of catalytic domain sequences and as receptor type III PTPases based on the overall domain structure of the protein.
Here we report that PTPRQ has phosphatidylinositol phosphatase (PIPase) activity against a broad range of phosphatidylinositol phosphates (PIPs). The PIPase activity depends on a WPE motif in the catalytic domain that differs from the WPD motif found in active PTPases, but that is also found in the putatively inactive D2 domains of many double-domain receptor-like PTPases. Amino acid motifs that define the PTEN or myotubularin families of proteins with PIPase activity are not found in PTPRQ. Recombinant PTPRQ cytoplasmic domain (cytoPTPRQ) does have detectable enzymatic activity against tyrosine-phosphorylated peptides and proteins, including autophosphorylated platelet-derived growth factor receptors (PDGFRs), but this PTPase activity is 2 orders of magnitude less than the activity of the recombinant cytoplasmic domain of PTPβ, a similar receptor type III PTPase that we used as a positive control for PTPase activity. When tested in cultured cells, PTPRQ overexpression inhibited proliferation and promoted apoptosis, and this activity depended on its PIPase activity rather than on its PTPase activity. PTPRQ thus represents a unique subtype within the PTPase superfamily. Like the PTEN- and myotubularin-like phosphatases, its predominant biological activity is as a PIPase, but unlike PTEN and myotubularin family members, it dephosphorylates a broader range of PIPs and depends for activity on different amino acid residues in the catalytic domain.
Materials and Methods
GST-cytoPTPRQ Fusion Proteins and Antibodies. The entire cytoplasmic domain of rat PTPRQ (GenBank accession no. AF063249), beginning with R1932, the first amino acid inside the transmembrane domain, was cloned into pGEX-4T-3 bacterial expression vector (Amersham Pharmacia). As a positive control for an active PTPase, the cytoplasmic domain of the PTPβ from R1621 to the C terminus was cloned into pGEX-4T-3. The QuikChange site-directed mutagenesis kit (Stratagene) was used to generate mutant forms of GST-cytoPTPRQ: GST-cytoPTPRQ(C2203S) and GST-cytoPTPRQ(E2171D). The fusion proteins were expressed in Escherichia coli and purified by glutathione-affinity chromatography essentially as described (9) (for details, see Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org). Polyclonal rabbit antiserum A429 was generated by repeated immunization with 100 μg of purified GST-cytoPTPRQ fusion protein as approved by the University of Washington Committee for Animal Care and Use. The antiserum was affinity purified by binding to GST-cytoPTPRQ fusion protein immobilized on CNBr-Sepharose and eluting the bound antibody with low pH and high salt.
Assays for Phospholipid Dephosphorylation. Lipid phosphatase assays were performed in a 50-μl reaction mixture containing 100 mM Tris·HCl (pH 8.0), 10 mM DTT, 200 μM water-soluble diC8-PIPs (Echelon, Salt Lake City), and 3 μg of fusion protein. diC8-PIPs are PIPs in which the two fatty acyl chains are 8 carbons long, rather than 16 as they would be in cell membranes. Triplicate samples were incubated for 40 min at 37°C and phosphate release was determined by using a malachite green assay (BIOMOL GREEN reagent, Biomol, Plymouth Meeting, PA) in accordance with the manufacturer's instructions. The absorbance at 630 nm was recorded in an ELISA plate reader (EL 312e, Bio-Tek Instruments, Winooski, VT). A standard curve for inorganic phosphate was generated for each assay. Assays with C6 and C16 fluorescent phosphatidylinositol phosphate substrates (Echelon) were performed as described by Taylor and Dixon (10). Briefly, labeled PIPs were incubated with recombinant enzyme and reaction products, along with reference standards, separated by TLC and visualized and quantitated under UV illumination.
Assay for Phosphoinositide 3-Kinase (PI3-Kinase) Activity. Cultured cells were stimulated with 50 ng/ml PDGF-BB for 10 min at 37°C and lysed in 1 ml of Nonidet P-40 lysis buffer containing protease and phosphatase inhibitors. Equal amounts of lysate protein (BCA Protein Assay kit, Pierce) were incubated overnight at 4°C with mAb PR7212 against PDGFR β-subunit (11), then with protein-A Sepharose for 1 h at 4°C. Immunoprecipitates were washed and evaluated for associated PI3-kinase activity by using a competitive ELISA according to the manufacturer's recommendations (Echelon). Briefly, PI3-kinase bound to the beads was incubated for 2 h with 10 μM diC8PI(4,5)P2 substrate at room temperature in 50 μl of buffer containing 5 mM Hepes (pH 7), 25 μM ATP, and 2.5 mM MgCl2. The beads were removed by centrifugation and the supernatant, or known concentrations of phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3], were incubated for 1 h with 50 μl of PI(3,4,5)P3-binding reagent then transferred to a detection plate coated with PI(3,4,5)P3. PI(3,4,5)P3 in the test sample binds to the binding reagent and prevents its binding to PI(3,4,5)P3 on the plate. Plate-bound binding reagent was quantitated by using a secondary detection reagent, peroxidase, and peroxidase substrate, with reaction product measured by absorbance at 450 nm.
Mammalian Expression Constructs and DNA Transfections. The entire cytoplasmic domains (beginning with R1932, the first amino acid inside the transmembrane domain) of PTPRQ, PTPRQ(E2171D), and PTPRQ(C2203S) were amplified by using primers designed to include a BamH1 restriction site and a Kozak consensus translational start and were cloned into the pcDNA3.1/Myc-His(+) B mammalian expression vector (Invitrogen) to generate cytoPTPRQ, cytoPTPRQ(E2171D), and cytoPTPRQ(C2203S). Human glioblastoma U87MG and U373MG cells (American Type Culture Collection) were transfected by using SuperFect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's recommendations. The reporter vector pEGFP-C3 (CLONTECH) was cotransfected at 1:4 [enhanced GFP (EGFP)/cytoPTPRQ] ratio. Transfected cells were sorted for EGFP expression and either evaluated to determine the effects of transient expression or selected with 400 μg/ml of G418 (Calbiochem) for 2 weeks, followed by isolation and expansion of isolated clones and evaluation by Western blotting with PTPRQ-specific A429.
A transmembrane-anchored form of PTPRQ was constructed from full-length rat PTPRQ cDNA (6) by using specific primers to include 17 aa from the extracellular domain, the transmembrane and cytoplasmic domains, and cloned into the pcDNA3.1/Myc-His(+)B mammalian expression vector. To facilitate proper expression and processing, an 87-bp fragment containing a Kozak consensus translation start sequence plus the PTPRQ signal sequence was added in-frame at the 5′ prime end.
Cell Proliferation. Clones expressing WT or mutant cytoPTPRQ, and mock-transfected cells, were plated in duplicate at 1 × 106/ml in DMEM supplemented with 2% or 10% FBS and incubated from 24 to 72 h. To determine the proliferation index, the cells were cultured in the presence of 0.1 mM BrdUrd (Sigma) for the last 16 h. Cells were harvested and stained with Hoechst 33258 and 7-aminoactinomycin D as described (12). Fifty thousand cells were analyzed per sample with a Coulter Epics Elite (Beckman Coulter) equipped with 10 mW UV excitation and 15 mW 488-nm excitation.
Mitochondrial Membrane Potential. Attached cells were trypsinized, combined with the floating cells, and incubated in the dark for 30 min at 37°C with a combination of two dyes: CMXRos and MitoTracker green (MTG) (Molecular Probes) each at 100 nM final concentration. Hoechst 33342 dye (Sigma) was added to a final concentration of 10 μM. The cells were then analyzed by flow cytometry and gated to exclude cells/debris with <2 M DNA content. The normalized mitochondrial membrane of each cell was calculated by dividing the fluorescence intensity of the membrane potential-sensitive dye (CMXRos) by the fluorescence intensity from the mitochondrial membrane potential-insensitive dye (MTG) as described by Poot and Pierce (13).
Results
In Vitro Evaluation of Enzymatic Activity. The putative catalytic domain of PTPRQ shows homology to many of the conserved features of PTPases, but has Glu (E2171) in place of Asp in the highly conserved WPD motif. This substitution is known to greatly reduce PTPase activity when introduced in other PTPases, suggesting that phosphotyrosine may not be the preferred substrate for PTPRQ. To evaluate the enzymatic activity of PTPRQ in vitro, we expressed, as GST fusion proteins, the cytoplasmic domains of WT PTPRQ (GST-cytoPTPRQ), and a mutant form, GST-cytoPTPRQ(E2171D), in which Glu-2171 was replaced by the canonical Asp. As a negative control, we expressed a mutant form, GST-cytoPTPRQ(C2203S), in which the catalytic cysteine, Cys-2203, was replaced with Ser. As a positive control for an active PTPase, we expressed GST-cytoPTPβ. PTPRQ and PTPβ both belong to the R3 subfamily of receptor-like tyrosine phosphatases that contain a single catalytic domain and an extracellular domain composed entirely of fibronectin type III domain-like repeats (8).
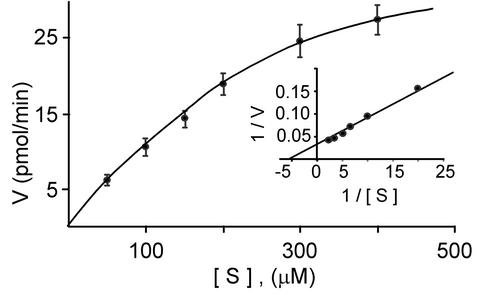
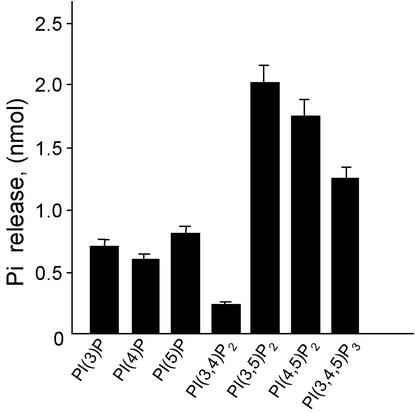
GST-cytoPTPRQ Has PIPase Activity in Vitro. To evaluate the activity of cytoPTPRQ against lipid inositol phosphates, we used diC8-PIPs in which the fatty acid moieties are 8 carbons long rather than 16 as they would be in cell membranes. diC8-PIPs are soluble in aqueous buffer making it possible to vary the substrate concentration and determine kinetic parameters. As shown in Fig. 1, GST-cytoPTPRQ is active against diC8-PI(3,4,5)P3. The values for Km (125 μM) and Vmax (18.3 nmol/min per mg) are comparable to values reported for PTEN (1, 14). Neither GST-cytoPTPβ nor GST-cytoPTPRQ(E2171D) had detectable phosphatase activity against diC8-PI(3,4,5)P3 (data not shown). Fig. 2 shows that WT GST-cytoPTPRQ is able to dephosphorylate other diC8-PIPs and remove the phosphate from the D3, D4, and D5 positions in diC8 monophosphates. HPLC analysis demonstrated that GST-cytoPTPRQ dephosphorylates phosphatidylinositol 3,5-bisphosphate and PI(3,4,5)P3 at positions 3 and 5, and dephosphorylates phosphatidylinositol 4,5-bisphosphate only at position 5 (data not shown). GST-cytoPTPRQ had limited activity against inositol polyphosphates (without fatty acyl chains). WT GST-cytoPTPRQ dephosphorylated inositol tetrakisphosphate, with a specific activity of 4 nmol/min per mg (data not shown). However, GST-cytoPTPRQ was not active against other inositol polyphosphates tested, including inositol 1,4,5-trisphosphate. GST-cytoPTPβ and GST-cytoPTPRQ(E2171D) had no detectable activity against any inositol polyphosphate tested.
Fig. 1.
cytoPTPRQ dephosphorylates PI(3,4,5)P3. The lipid phosphatase activity of GST-cytoPTPRQ fusion protein was assayed by measuring phosphate release from diC8-PI(3,4,5)P3 as described in Materials and Methods. (Inset) A Lineweaver-Burk plot of the data is shown. The data points are the means ± SD of determinations from three experiments.
Fig. 2.
GST-cytoPTPRQ has broad specificity for phosphatidylinositol phosphates. The lipid phosphatase activity of GST-cytoPTPRQ was assayed by measuring phosphate release from diC8-PIPs as described in Materials and Methods. The data are presented as the means ± SD of triplicate determinations.
cytoPTPRQ Has Weak PTPase Activity in Vitro. Because the above results demonstrated that presence of Glu in place of Asp at position 2171 is essential for GST-cytoPTPRQ activity as a PIPase, we evaluated how it affects GST-cytoPTPRQ activity against other possible substrates, including tyrosine-phosphorylated peptides and proteins.
As expected based on mutational analysis of other PTPases, WT GST-cytoPTPRQ had 100-fold less activity than GST-cytoPTPβ against the small artificial substrate para-nitrophenyl phosphate. Activity increased 10-fold when the Glu-2171 was replaced by the canonical Asp and eliminated when the essential Cys-2203 was substituted by Ser (see Table 2, which is published as supporting information on the PNAS web site).
We used the malachite green assay to measure phosphate release from the commonly used tyrosine-phosphorylated peptide substrates RRLIEDAEpYAARG (RR-src) and TSTEPQpYQPGENL (TST). The relative activities of GST-cytoPTPRQ and GST-cytoPTPRQ(E2171D) depended on the substrate and assay pH, but were always <1% of the activity of GST-cytoPTPβ (see Table 3, which is published as supporting information on the PNAS web site). We evaluated 32P release from 32P-phosphorylated reduced and carboxymethylated-maleylated lysozyme protein and 32P-phosphorylated peptide DADEYL (see Supporting Methods). The activity of GST-cytoPTPRQ was 1,000- to 10,000-fold less than the activity of GST-cytoPTPβ (Table 1) but was comparable to published values for PTEN (15). GST-cytoPTPRQ(E2171D) was 10- to 15-fold more active than WT GST-cytoPTPRQ, but was still 200- to 500-fold less active than GST-cytoPTPβ. Neither WT GST-cytoPTPRQ nor cytoPTPRQ(E2171D) had detectable activity against peptides phosphorylated on serine (RRApSVA) or threonine (KRpTIR) (data not shown).
Table 1. PTPase activity of GST-fusion proteins toward tyrosine-phosphorylated reduced and carboxymethylated-maleylated lysozyme and peptide DADEpYL.
| Specific activity, nmol/min per mg
|
|||
|---|---|---|---|
| Substrate | GST-cytoPTPRQ | GST-cytoPTPRQ(E2171D) | GST-cytoPTP β |
| RCML | 0.106 ± 0.05 | 0.916 ± 0.1 | 168 ± 15.4 |
| DADEYL | 0.029 ± 0.0 | 0.433 ± 0.05 | 229 ± 21.3 |
Activity of fusion proteins with indicated substrates are reported as means ± SD (n = 3). Fusion protein concentration was determined by absorbance at 280 nm and verified by the method of Bradford (Bio-Rad). RCML, reduced and carboxymethylated-maleylated lysozyme.
To examine the activity of recombinant GST-cytoPTPRQ toward endogenously tyrosine-phosphorylated proteins, we exposed NIH 3T3 cells to high concentrations of PDGF-BB for 10 min to induce maximal PDGFRβ autophosphorylation. Under these conditions, autophosphorylated PDGFRβ is the major tyrosine-phosphorylated protein in the cell (16) and was easily detected by Western blot analysis using anti-phosphotyrosine antibody PY20. Incubation of cell lysates with increasing amounts of recombinant GST-cytoPTPRQ showed that both WT GST-cytoPTPRQ and GST-cytoPTPRQ(E2171D) can dephosphorylate tyrosine-phosphorylated PDGFRβ in vitro, but GST-cytoPTPRQ is ≈400-fold less active than GST-cytoPTPβ (Fig. 7, which is published as supporting information on the PNAS web site).
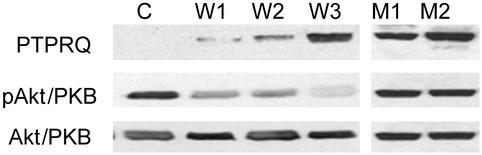
Expression of cytoPTPRQ in Cultured Cells Reduces Phosphorylation of Akt/PKB. The results above suggested that the sequence of WT PTPRQ adapts it to function as a PIPase rather than as a PTPase. To evaluate the ability of cytoPTPRQ to affect PIP levels in cultured cells, we evaluated the phosphorylation status of the serine/threonine kinase Akt/PKB, which is regulated by intracellular levels of PI(3,4,5)P3 (17). The human glioblastoma cell lines U87MG and U373MG do not express the phosphatidylinositol 3-phosphatase PTEN (18, 19), and we did not detect PTPRQ transcripts by RT-PCR (data not shown), or PTPRQ protein, by Western blot analysis with PTPRQ-specific antibody A429 (Fig. 3). Transient expression of WT cytoPTPRQ resulted in a 66% reduction in the level of phosphorylated Akt/PKB with no effect on the level of total Akt/PKB protein (Fig. 8A, which is published as supporting information on the PNAS web site). To determine the effect of stable expression of cytoPTPRQ, we transfected glioblastoma U87MG cells with WT cytoPTPRQ and mutant cytoPTPRQ(E2171D) expressed in the pcDNA3.1Myc/His vector. After a 2-week selection with G-418, we expanded isolated colonies with detectable cytoPTPRQ expression as determined by Western blot analysis. Fig. 3 shows cytoPTPRQ protein levels in three representative clones expressing different levels of WT cytoPTPRQ, and in two clones expressing the cytoPTPRQ(E2171D) mutant, which is active against phosphotyrosine but not against phosphatidylinositols. Expression of increasing levels of WT cytoPTPRQ resulted in a dose-dependent decrease in Akt/PKB phosphorylation, but expression of cytoPTPRQ(E2171D) had no effect. We have not been able, in multiple attempts, to establish cell lines that constitutively overexpress transmembrane-anchored or receptor-like forms of PTPRQ. This finding is consistent with the hypothesis that this localization increases effective activity against membrane PIPs and results in too great a growth disadvantage. In support of this, we found that transient expression of low levels of a transmembrane-anchored form of PTPRQ reduced Akt/PKB phosphorylation as much as did expression of higher levels of cytoplasmic domain form (see Fig. 8A).
Fig. 3.
cytoPTPRQ expression reduces Akt/PKB phosphorylation. Stable clones established from U87MG glioblastoma cells transfected with empty vector (C), WT cytoPTPRQ (clones W1, W2, W3), or mutant cytoPTPRQ(E2171D) (clones M1, M2) were evaluated for cytoPTPRQ expression level by Western blotting with the anti-PTPRQ antiserum A429 (Top), for phosphorylated Akt/PKB (pAkt/PKB) level with antibodies to pSer473 (Middle), and for total Akt/PKB level with a pan-Akt/PKB antibody (Bottom).
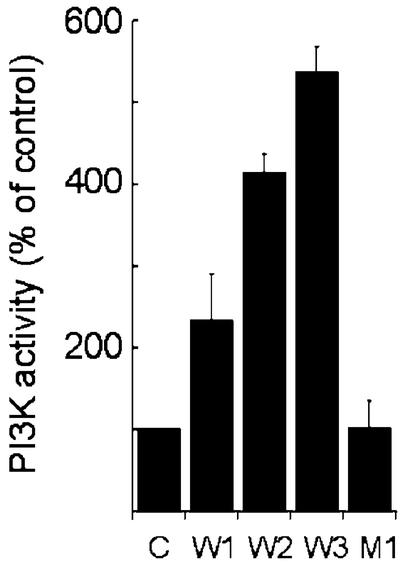
As noted above, PTPRQ is able to dephosphorylate tyrosine-phosphorylated PDGFRs in vitro, although with very low activity compared with standard PTPases. Receptor dephosphorylation would reduce binding and consequent activation of PI3-kinase and would provide a pathway through which PTPRQ could reduce Akt/PKB phosphorylation other than via action as a PIPase to reduce PI(3,4,5)P3 levels. To evaluate this possibility, we stimulated control and cytoPTPRQ-expressing cells with PDGF and determined the activity of PI3-kinase associated with the activated PDGFR. Fig. 4 shows that expression of WT, but not mutant, cytoPTPRQ results in increased activity of PDGFR-associated PI3-kinase, and that clones that express higher levels of cytoPTPRQ and that have lower levels of phospho-Akt/PKB (Fig. 3) have higher levels of PDGFR-associated PI3-kinase activity. This result is opposite to what would be expected if PTPRQ were reducing Akt/PKB phosphorylation by inhibiting PI3-kinase activity or its recruitment to activated receptors, e.g., via dephosphorylation of autophosphorylated PDGFRs. It is consistent with the report that intracellular PI(3,4,5)P3 acts as a competitive inhibitor of p85/PI3-kinase binding to pY on activated receptors (20) because the proposed PIPase activity of PTPRQ would keep PI(3,4,5)P3 levels low and reduce their interference with PI3-kinase binding to PDGFRs.
Fig. 4.
PDGFR-associated PI3-kinase activity is increased in cells expressing WT cytoPTPRQ. Stable clones of U87MG cells transfected with control plasmid (C), constitutively expressing different levels of WT cytoPTPRQ (W1, W2, W3) or mutant cytoPTPRQ(E7121D) (M1), were stimulated with 50 ng/ml PDGF-BB for 10 min. Cells were lysed, and PDGFRs and associated proteins were immunoprecipitated from equal amounts of total protein by using a mAb (11) against the PDGFR β-subunit. The activity of PI3-kinase associated with the PDGFR was assayed as described in Materials and Methods. Results are presented as mean ± SD (n = 3) of PI3-kinase activity as a percentage of activity in the control cells.
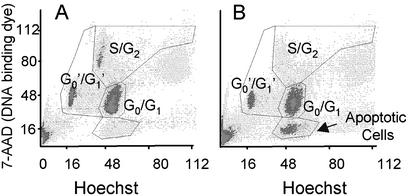
Expression of cytoPTPRQ Inhibits the Proliferation of Glioblastoma Cells and Leads to G0/G1 Arrest. Because cell proliferation is positively regulated by PIPs, the PIPase activity of WT PTPRQ would be predicted to inhibit proliferation. We compared the growth rates (increase in cell number) of mock-transfected glioblastoma cells with the three clones constitutively expressing cytoPTPRQ. All three clones expressing WT cytoPTPRQ had significantly reduced proliferative rates compared with control cells (data not shown). Constitutive expression of mutant cytoPTPRQ(E2171D), which retains PTPase activity but has lost PIPase activity, did not alter the growth rate of the cells. We evaluated the proliferation of transiently transfected cells by using BrdUrd incorporation followed by staining with Hoechst 33258 and 7-aminoactinomycin D (12). As shown in Fig. 5, expression of cytoPTPRQ decreased the percentage of proliferating cells (in S+G2 and G0′/G1′) by ≈50%, and resulted in the appearance of a new population of apoptotic cells with <2 N DNA content. Treatment of control cultures with the PI3-kinase inhibitor LY294002 resulted in a comparable decrease in the percentage of proliferating cells and did not further decrease proliferation of cells expressing cytoPTPRQ (data not shown). This finding is consistent with an action of PTPRQ via hydrolysis of PIP product(s) of PI3-kinase.
Fig. 5.
Transient expression of cytoPTPRQ promotes apoptosis and cell cycle arrest. U87MG cells were cotransfected with EGFP (transfection control) and either empty vector (A) or cytoPTPRQ (B). Cells were maintained in 2% serum for 48 h and labeled with BrdUrd for the last 16 h. Cells were trypsinized and incubated with 7-aminoactinomycin D (7-AAD) and Hoechst 33358, and EGFP-positive cells were evaluated by flow cytometry as described in Materials and Methods. In the scatter plots, 7-AAD fluorescence provides information about the position of the cells within each cell cycle, whereas quenching of Hoechst fluorescence provides information about the number of cell cycles traversed during the labeling period.
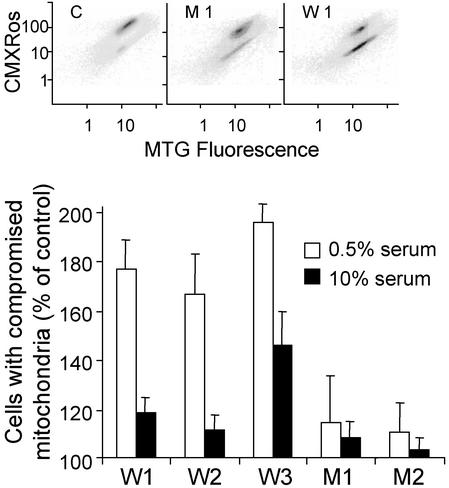
Expression of PTPRQ Induces Apoptotic Changes. To determine whether PTPRQ expression promotes early apoptotic changes, we evaluated the ability of cytoPTPRQ to reduce mitochondrial membrane potential, a change associated with proapoptotic events. The U87MG clones constitutively expressing WT cytoPTPRQ showed a 50–100% increase in the number of cells with compromised mitochondrial membrane potential (Fig. 6). The effect on membrane potential was greater when the cells were cultured in 0.5% serum than in 10% serum (Fig. 6), just as the inhibition of proliferation was greater in low serum (data not shown). Expression of mutant cytoPTPRQ(E2171D), which retains PTPase activity but has lost PIPase activity, did not alter mitochondrial membrane potential in the two clones examined (Fig. 6), supporting the hypothesis that the biological effects of PTPRQ derive from its activity as a PIPase rather than as a PTPase. When assayed by annexin V binding in transient transfection assays, the proapoptotic activity of a transmembrane-anchored form of PTPRQ (210% of control) was substantially greater than that of the cytoplasmic domain (137% of control), consistent with activity against membrane PIPs (see Fig. 8B).
Fig. 6.
Constitutive cytoPTPRQ expression results in reduced mitochondrial membrane potential. Clones of U87MG cells transfected with control plasmid (C) or constitutively expressing different levels of WT cytoPTPRQ (W1, W2, W3) or mutant cytoPTPRQ(E7121D) (M1, M2) were maintained in 0.5% or 10% serum and labeled with a combination of CMXRos and MitoTracker green (MTG) as described in Materials and Methods. The scatter plots show the distribution of cells with high CMXRos values (normal membrane potential) and low CMXRos values (characteristic of early apoptotic cells) for three representative clones. The bar graphs show the calculated percentages (mean ± SD, n = 4) of cells with compromised mitochondria for clones maintained in 0.5% serum (open bars) or 10% serum (solid bars).
Discussion
Although PTPRQ is classified as a receptor type III PTPase based on the overall domain structure of the protein (7), and recombinant cytoPTPRQ does have low but detectable activity against tyrosine-phosphorylated substrates in vitro, our results suggest that the biologically important activity of PTPRQ is as a PIPase rather than as a PTPase. In this regard, PTPRQ resembles certain other PTPase superfamily members, including PTEN, TPIP, and myotubularin, that have been shown to be active against PIPs. However, PTPRQ has a broader substrate specificity than those phosphatases, and its activity against PIPs depends on different amino acid residues in the PTPase catalytic domain.
PTEN and TPIP are related PTPase-like proteins that differ from consensus active PTPases in having two lysines in the catalytic site. These lysines have been shown to be important for PTEN activity against PI(3,4,5)P3 (21) but are not present in PTPRQ. Instead, the activity of PTPRQ against PIPs depends on the presence of Glu in place of Asp in the WPD motif that is characteristic of consensus active PTPases, including other receptor type III phosphatases. This carboxylic residue plays a key role in catalysis by protonating the phenolic oxygen of tyrosine, which allows efficient dissociation of the dephosphorylated substrate from the enzyme. When the Asp in RPTPα was mutated to Ala, catalytic activity against phosphotyrosine was completely eliminated (22). Activity was reduced by at least 2 orders of magnitude when the Asp was replaced by Glu, which differs from Asp only in having one additional carbon in the side-chain spacer (22, 23). Consistent with this well-documented role of Asp in catalysis in active PTPases, we found that the activity of recombinant cytoPTPRQ against para-nitrophenyl phosphate, a widely used PTPase substrate, was greatly increased when the Glu (E2171) in WT cytoPTPRQ was substituted by the consensus Asp of a standard active PTPase. However, the presence of Glu instead of Asp-2171 cannot be the only factor that makes cytoPTPRQ a poor PTPase, because cytoPTPRQ(E2171D) did not show consistently higher activity than WT cytoPTPRQ when assayed with tyrosine-phosphorylated peptides or protein substrates. In contrast, substitution of Glu with Asp completely eliminated the activity of cytoPTPRQ against PI(3,4,5)P3 and other PIP substrates and eliminated the ability of cytoPTPRQ overexpression to induce apoptosis and inhibit cell cycle progression. This finding suggests that PTPRQ has potential growth-regulatory functions that depend on activity against PIPs rather than against phosphotyrosine.
A second subfamily of PTPase-like proteins that have activity against PIPs is represented by myotubularin. Like PTEN, myotubularin has low activity against para-nitrophenyl phosphate and was reported to have dual-specificity protein phosphatase activity before its activity against phosphatidylinositol 3-phosphate was recognized. Members of the myotubularin family of PTPase-like proteins, which includes at least 10 human genes, all appear to be specific for phosphatidylinositol 3-phosphate rather than for PI(3,4,5)P3 (4, 5). All are unusual in having two Asp residues in the Cx5R catalytic site (1, 24). PTPRQ does not have these two Asp residues.
Unlike PTEN or myotubularin family members, PTPRQ can also hydrolyze the D5 and D4 phosphates of PIPs. Type I 5-phosphatases (active against inositol polyphosphates) and type II 5-phosphatases (active against inositolpolyphosphates and PIPs) both contain the two catalytic domain signature motifs, (F/I)WxGDxN(F/Y)R and (R/N)xP(S/A)(W/Y)(C/T)DR(I/V)(L/I) (2). PTPRQ does not contain these sequence motifs. To suggest a provisional model of how the structure of the PTPRQ substrate pocket could permit activity as a 5-phosphatase and a 3-phosphatase, we used a homology modeling program to predict the 3D structure of the PTPRQ catalytic domain based on the known structures of PTEN (14) and the standard active PTPase PTPα (25). The model (Fig. 9, which is published as supporting information on the PNAS web site) suggested that, like PTEN, PTPRQ has a much larger catalytic pocket than PTPα, allowing it to accommodate PI(3,4,5)P3, which is larger than phospho-tyrosine, and could not enter the smaller catalytic pocket of a standard PTPase (Fig. 9B). The sequence of the catalytic motif of PTEN contains two invariant Lys residues (Lys-125 and Lys-128 in human PTEN) that are found only among PTEN family members. Mutation analysis and crystal structure determination suggests that Lys-125 is involved in binding the D1 phosphate and Lys-128 is involved in binding the D5 phosphate. PTPRQ does not have either of these lysines (or other positively charged groups) in these positions. This may permit PTPRQ more flexibility in accommodating and orienting different inositol phospholipids and allow it to cleave D5 and D4, as well as D3, phosphates.
The PI(3,4,5)P3 3-phosphatase activity of PTEN is required for its inhibitory effect on Akt/PKB phosphorylation and its ability to function as a tumor suppressor. Some of the PTEN mutations (e.g., G129E) identified in patients, which cause deregulated cell proliferation, eliminate the PIPase activity without significantly decreasing PTPase activity (26–28). This finding supports the hypothesis that it is the PIPase activity of PTEN, rather than its weak dual-specificity phosphatase activity, that is critical for its activity as a tumor suppressor. An analogous argument supports the hypothesis that it is the PIPase activity of PTPRQ, rather than its PTPase activity, that is essential for its proapoptotic and antiproliferative activity of PTPRQ in cultured cells. The cytoPTPRQ(E2171D) mutant retains PTPase activity but loses PIPase activity and does not inhibit cell proliferation or induce apoptosis. The 5-phosphatase and 3-phosphatase activities of PTPRQ may play complementary roles in inhibiting signaling via PI(3,4,5)P3 produced by activated PI3-kinase. PI(3,4)P2 (the product of 5-phosphatase activity) and PI(4,5)P2 (the product of 3-phosphatase activity) are both less active in activating Akt/PKB phosphorylation than is PI(3,4,5)P3 (17). Thus, the 5-phosphatase p150Ship inhibits proliferation stimulated by macrophage colony-stimulating factor (29) and the related inositol 5-phosphatase SHIP2 is a negative regulator of signaling downstream of the insulin receptor (30). Phosphorylation of Akt/PKB is reduced by 5-phosphatase IV in parallel with reduced intracellular levels of PI(3,4,5)P3, which increases cell susceptibility to apoptotic stimuli (31). Phosphatidylinositol 5-phosphatase activity can also participate in regulation of other pathways. For example, synaptojanin is a nerve terminal protein with 5-phosphatase activity, which appears to play a role in regulating synaptic vesicle recycling (32). Interestingly, many of the ``inactive'' D2 domains of double-domain receptor-like PTPases also have Glu in place of Asp in this position. It will be interesting to determine whether some D2 domains may have inositol phosphatase activity, which would give these receptor-like proteins significant activity against both phosphotyrosine, via their D1 domains, and against phosphatidylinositol, via their D2 domains.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Aprikyan (University of Washington) for his help with the flow cytometry analysis. This research was supported by National Institutes of Health Grant DK54857 (to D.F.B.-P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PTPase, protein tyrosine phosphatase; PTPRQ, PTPase RQ; cytoPTPRQ, PT-PRQ cytoplasmic domain; EGFP, enhanced GFP; PDGFR, platelet-derived growth factor receptor; PI3-kinase, phosphoinositide 3-kinase; PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate; PIP, phosphatidylinositol phosphate; PIPase, phosphatidylinositol phosphatase; PKB, protein kinase B.
References
- 1.Maehama, T., Taylor, G. S. & Dixon, J. E. (2001) Annu. Rev. Biochem. 70, 247–279. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck, B., Leevers, S. J., Ahmadi, K., Timms, J., Katso, R., Driscoll, P. C., Woscholski, R., Parker, P. J. & Waterfield, M. D. (2001) Annu. Rev. Biochem. 70, 535–602. [DOI] [PubMed] [Google Scholar]
- 3.Tamura, M., Gu, J., Matsumoto, K., Aota, S., Parsons, R. & Yamada, K. M. (1998) Science 280, 1614–1617. [DOI] [PubMed] [Google Scholar]
- 4.Kim, S. A., Taylor, G. S., Torgersen, K. M. & Dixon, J. E. (2002) J. Biol. Chem. 277, 4526–4531. [DOI] [PubMed] [Google Scholar]
- 5.Laporte, J., Blondeau, F., Buj-Bello, A. & Mandel, J. L. (2001) Trends Genet. 17, 221–228. [DOI] [PubMed] [Google Scholar]
- 6.Wright, M. B., Hugo, C., Seifert, R., Disteche, C. M. & Bowen-Pope, D. F. (1998) J. Biol. Chem. 273, 23929–23937. [DOI] [PubMed] [Google Scholar]
- 7.Brady-Kalnay, S. M. & Tonks, N. K. (1995) Curr. Opin. Cell Biol. 7, 650–657. [DOI] [PubMed] [Google Scholar]
- 8.Andersen, J. N., Mortensen, O. H., Peters, G. H., Drake, P. G., Iversen, L. F., Olsen, O. H., Jansen, P. G., Andersen, H. S., Tonks, N. K. & Moller, N. P. (2001) Mol. Cell. Biol. 21, 7117–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith, D. B. & Johnson, K. S. (1988) Gene 67, 31–40. [DOI] [PubMed] [Google Scholar]
- 10.Taylor, G. S. & Dixon, J. E. (2001) Anal. Biochem. 295, 122–126. [DOI] [PubMed] [Google Scholar]
- 11.Hart, C. E., Seifert, R. A., Ross, R. & Bowen-Pope, D. F. (1987) J. Biol. Chem. 262, 10780–10785. [PubMed] [Google Scholar]
- 12.Rosato, M. T., Jabbour, A. J., Ponce, R. A., Kavanagh, T. J., Takaro, T. K., Hill, J. P., Poot, M., Rabinovitch, P. S. & Faustman, E. M. (2001) J. Immunol. Methods 256, 35–46. [DOI] [PubMed] [Google Scholar]
- 13.Poot, M. & Pierce, R. H. (1999) Cytometry 35, 311–317. [DOI] [PubMed] [Google Scholar]
- 14.Lee, J. O., Yang, H., Georgescu, M. M., Di Cristofano, A., Maehama, T., Shi, Y., Dixon, J. E., Pandolfi, P. & Pavletich, N. P. (1999) Cell 99, 323–334. [DOI] [PubMed] [Google Scholar]
- 15.Myers, M. P., Stolarov, J. P., Eng, C., Li, J., Wang, S. I., Wigler, M. H., Parsons, R. & Tonks, N. K. (1997) Proc. Natl. Acad. Sci. USA 94, 9052–9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper, J. A., Bowen-Pope, D. F., Raines, E., Ross, R. & Hunter, T. (1982) Cell 31, 263–273. [DOI] [PubMed] [Google Scholar]
- 17.Kandel, E. S. & Hay, N. (1999) Exp. Cell Res. 253, 210–229. [DOI] [PubMed] [Google Scholar]
- 18.Li, D. M. & Sun, H. (1998) Proc. Natl. Acad. Sci. USA 95, 15406–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morimoto, A. M., Berson, A. E., Fujii, G. H., Teng, D. H., Tavtigian, S. V., Bookstein, R., Steck, P. A. & Bolen, J. B. (1999) Oncogene 18, 1261–1266. [DOI] [PubMed] [Google Scholar]
- 20.Rameh, L. E., Chen, C. S. & Cantley, L. C. (1995) Cell 83, 821–830. [DOI] [PubMed] [Google Scholar]
- 21.Maehama, T. & Dixon, J. E. (1999) Trends Cell Biol. 9, 125–128. [DOI] [PubMed] [Google Scholar]
- 22.Wu, L., Buist, A., den Hertog, J. & Zhang, Z. Y. (1997) J. Biol. Chem. 272, 6994–7002. [DOI] [PubMed] [Google Scholar]
- 23.Flint, A. J., Tiganis, T., Barford, D. & Tonks, N. K. (1997) Proc. Natl. Acad. Sci. USA 94, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor, G. S., Maehama, T. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilwes, A. M., den Hertog, J., Hunter, T. & Noel, J. P. (1996) Nature 382, 555–559. [DOI] [PubMed] [Google Scholar]
- 26.Furnari, F. B., Huang, H. J. & Cavenee, W. K. (1998) Cancer Res. 58, 5002–5008. [PubMed] [Google Scholar]
- 27.Ramaswamy, S., Nakamura, N., Vazquez, F., Batt, D. B., Perera, S., Roberts, T. M. & Sellers, W. R. (1999) Proc. Natl. Acad. Sci. USA 96, 2110–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers, M. P., Pass, I., Batty, I. H., Van der Kaay, J., Stolarov, J. P., Hemmings, B. A., Wigler, M. H., Downes, C. P. & Tonks, N. K. (1998) Proc. Natl. Acad. Sci. USA 95, 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lioubin, M. N., Algate, P. A., Tsai, S., Carlberg, K., Aebersold, A. & Rohrschneider, L. R. (1996) Genes Dev. 10, 1084–1095. [DOI] [PubMed] [Google Scholar]
- 30.Clement, S., Krause, U., Desmedt, F., Tanti, J. F., Behrends, J., Pesesse, X., Sasaki, T., Penninger, J., Doherty, M., Malaisse, W., et al. (2001) Nature 409, 92–97. [DOI] [PubMed] [Google Scholar]
- 31.Kisseleva, M. V., Cao, L. & Majerus, P. W. (2002) J. Biol. Chem. 277, 6266–6272. [DOI] [PubMed] [Google Scholar]
- 32.McPherson, P. S., Garcia, E. P., Slepnev, V. I., David, C., Zhang, X., Grabs, D., Sossin, W. S., Bauerfeind, R., Nemoto, Y. & De Camilli, P. (1996) Nature 379, 353–357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.