Abstract
Unusual DNA conformations including cruciforms play an important role in gene regulation and various DNA transactions. Cruciforms are also the models for Holliday junctions, the transient DNA conformations critically involved in DNA homologous and site-specific recombination, repair and replication. Although the conformations of immobile Holliday junctions in linear DNA molecules have been analyzed with use of various techniques, the role of DNA supercoiling has not been studied systematically. We utilized Atomic Force Microscopy (AFM) to visualize cruciform geometry in plasmid DNA with different superhelical densities at various ionic conditions. Both folded and unfolded conformations of the cruciform were identified, and the data showed that DNA supercoiling shifts the equilibrium between folded and unfolded conformations of the cruciform towards the folded one. In topoisomers with low superhelical density the population of folded conformation is 50 to 80 %, depending on ionic strength of the buffer and a type of cation added, whereas in the sample with high superhelical density, this population is as high as 98-100%. The time-lapse studies in aqueous solutions allowed us to observe the conformational transition of the cruciform directly. The time-dependent dynamics of the cruciform correlates with the structural changes revealed by the ensemble-averaged analysis of dry samples. Altogether, the data obtained show directly that DNA supercoiling is the major factor determining the Holliday junction conformation.
DNA cruciforms play an important role in regulation of biological processes involving DNA. These structures are formed by inverted repeats and require DNA supercoiling for their stable existence (1). Inverted repeats are associated with origin of replication (1-3). Cruciforms are suggested to be involved in the regulation of gene expression (1, 4), and may also play role in nucleosome structure and function (5). In addition, the cruciform is inherent model for Holliday junction, an intermediate in homologous and site-specific recombination (reviewed in (6)). Holliday junctions are also targets for many architectural and regulatory proteins such as histones H1 and H5, topoisomerase IIbeta, HMG proteins, HU, p53, proto-oncogene protein DEK to mention a few (7-15). The biological role for some of these interactions is still unclear. So far, immobile Holliday junctions or four-way DNA junctions were primary experimental models for structural studies of Holliday junctions by the use of variety of methods (electrophoresis, FRET1, X-Ray, thermodynamics, chemical and enzymatic probing (reviewed in (6, 14, 16)). According to these studies, it can be summarized that the Holiday junction adopts two distinct conformations: folded and unfolded. Unfolded conformation, in which adjacent arms are nearly perpendicular to each other and the structure, has a 4-fold symmetry, and exists at low concentration of metal ions. Folded conformation (or stacked, X-type), in which four arms undergo pairwise coaxial stacking, could be modeled with two duplexes exchanging strands at the junction point. This conformation is stabilized by high ionic strength divalent or polyvalent cations in particular (e.g. Mg2+ cations at the concentration more then 100 μM). Sodium cations also shift equilibrium distribution towards folded conformation though much higher concentrations of cations are required (17). Also, folded conformation of cruciform can be parallel or antiparallel. Synthetic immobile DNA junction exists in antiparallel geometry and no parallel geometry was observed (6, 16, 18, 19). However, computer modeling showed that interarm angle can vary in a broad range of values (20). Furthermore, thermodynamic studies on short Holliday junctions tethered to form parallel or antiparallel orientation revealed that there is no significant difference in free energy between these orientations (21).
Atomic Force Microscopy (AFM) was successfully applied for imaging cruciforms in supercoiled DNA (5, 22-24). These early studies showed that the cruciform in supercoiled DNA can adopt folded and unfolded conformations, but in folded conformation parallel rather than antiparallel orientation of the helical strands is observed. It was hypothesized that DNA supercoiling is an important factor in such conformational preference for the cruciform (5, 24). To test this hypothesis, we systematically analyze the structural and dynamic properties of the cruciform in plasmids with different superhelical densities. We employed AFM, including the time-lapse imaging in aqueous solutions, to study cruciform structure in buffers with different concentrations of metal ions. The results presented here indicate that the geometry of cruciform is primarily governed by DNA supercoiling. DNA supercoiling shifts equilibrium between folded and unfolded conformations of cruciform towards the folded one with the parallel orientation of the DNA helical strands rather than the antiparallel configuration typically observed in synthetic Holliday junctions. The effect of DNA supercoiling is enhanced in the presence of cations. These findings provide additional support for studies the role of DNA supercoiling in the structure and dynamics of Holliday junctions (cruciforms).
MATERIALS AND METHODS.
Plasmids. Topoisomer samples of plasmid pFR6 (3396 bp)(a kind gift from V. Potaman, Texas A&M University), are prepared as described earlier (22, 25). Each topoisomer sample contains a set of topoisomers and is characterized by mean value of superhelical density <σ>. Two topoisomer samples: one with low superhelical density denoted as t3low (mean value <σ> = -0.038) and another with high superhelical density denoted as t7high (mean value <σ> = -0.085) are used for analysis of cruciform geometry and dynamics.
Synthetic DNA Holliday junction with two arms of 320 bp and two arms of 64 bp are symmetrically positioned relative to the center of the junction was synthesized by ligation of DNA fragments to the core structure of immobile Holliday junction. The core structure of Holliday junction was obtained by step-wise annealing of four oligonucleotides. The detailed procedure can be found in the Supporting Information section.
AFM procedure was performed as described earlier (22, 23, 25-28) A mica surface was modified for 30 min with 0.5 mM 1-(3-aminopropyl)silatrane (APS; (26)) for experiments in solution and with 0.16 mM APS for imaging of dried samples. The sample was deposited on APS-modified mica for 2-5 min, then rinsed with deionized water and dried with argon (for experiment with dried sample) or applied directly without a rinsing/drying procedure for AFM imaging with hydrated sample.
Time-lapse imaging in hydrated samples was performed as described earlier (24, 27). Briefly, the APS-modified mica with the sample droplet on the top of it was placed on the scanning stage of MultiMode SPM Nanoscope IIIa (Veeco, Santa Barbara, CA). The optical head with installed liquid cell was mounted above the sample, additional buffer was added into the space between the cell and mica surface, and the scanning tip (NP probe, Veeco, Santa Barbara, CA) was approached to the surface and engaged. The buffer was replaced when necessary with two syringes attached to the flow cell without terminating the scanning.
Samples t3low and t7high were prepared and imaged in the following buffers: HE buffer (10 mM HEPES, 1 mM EDTA, pH 7.0), HEN buffer (10 mM HEPES, 1 mM EDTA, 200 mM NaCl, pH 7.0) and HM buffer (10 mM HEPES, 10 mM MgCl2, pH 7.0).
Short synthetic Holliday junction were prepared and imaged in TE buffer (10 mM TRIS-HCl, 1mM EDTA, pH 7.0) and TM buffer (10 mM TRIS, 10 mM MgCl2, pH 7.0).
Data analysis was performed using FemtoScan Online software package (version 1.6 (4.4); Moscow State University and Advanced Technologies Center, Moscow, Russia). The angles between DNA helices at junction point were measured as described in our recent paper (28). To measure the angle value lines were drawn over the middle of DNA filaments just near the junction point over the filament sections ca. 10 nm that typically appears as straight lines. The angle between these lines was measured (see Fig. S1 in Supporting Information section for specifics). As the cruciform hairpin arms are not necessary continuations of the plasmid main strands, both the angles between hairpin arm and the angles between DNA main strands were measured. Lengths of cruciform arms were measured as described before (27). A cruciform arm was approximated by a segmented line, and the length of the arm was obtained from the sum of the segments lengths (see Fig. S1 in Supporting Information section for specifics).
RESULTS
The cruciform structure - effect of DNA supercoiling
The schematics of cruciform (Holliday junction) conformations are presented in Figs. 1a-e. The DNA helices are shown as cylinders and the round-shaped caps on them correspond to loops of the hairpin arms of the cruciform. The structure depicted in Fig. 1a corresponds to the unfolded (open) conformation of the cruciform, whereas Figs. 1b-c and 1d-e show two orthogonal views of folded conformations with parallel or antiparallel orientation of helical strands respectively. Projections c and e are obtained by rotation of projections b and d respectively by 90° around the vertical axis.
Figure 1.
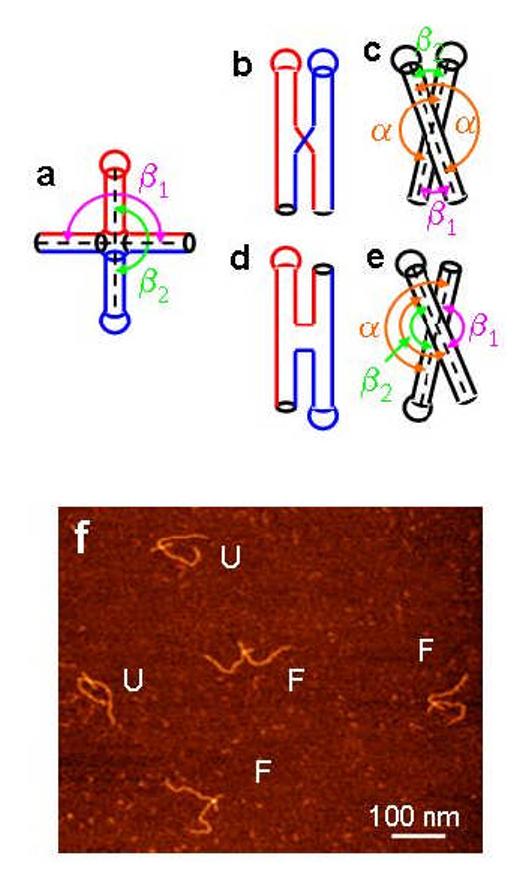
Schematic representation of cruciform conformations: unfolded (a) and folded (b-e) conformations with parallel (b,c) and antiparallel (d,e) orientation of DNA strands. Complementary DNA strands are indicated with red and blue colors. Arches with arrows of different color show “stacking” angle α (brown), angle between main DNA strands β1 (pink) and angle between hairpin arms β2 (green). f. AFM image of dried sample for synthetic Holliday junction deposited from TM buffer. “F” and “U” denote Holliday junctions in folded and unfolded conformations, respectively.
Figure 1f shows the AFM image of the synthetic Holliday junction assembled from 4 linear arms in TEM buffer. Folded and unfolded conformations of the junction on the AFM image could be found and are marked with F and U, respectively. Typical images of supercoiled DNA molecules with the cruciform in unfolded and folded conformations are shown in Figs. 2a (sample t3low, <σ> = -0.038 in HE buffer) and Fig. 2b (sample t7high, <σ> = -0.085 in HE buffer) respectively. The AFM images for the sample with low supercoiling density (t3low) reveal both folded and unfolded conformations of cruciforms, but the cruciform appears in folded conformation for sample t7high (Fig. 2b). To quantify this effect we counted the occurrence of both conformations of Holliday junctions; we found that folded conformation exists in 16%, 48% and 98% of short Holliday junctions and samples t3low and t7high respectively (see Table 1).
Figure 2.
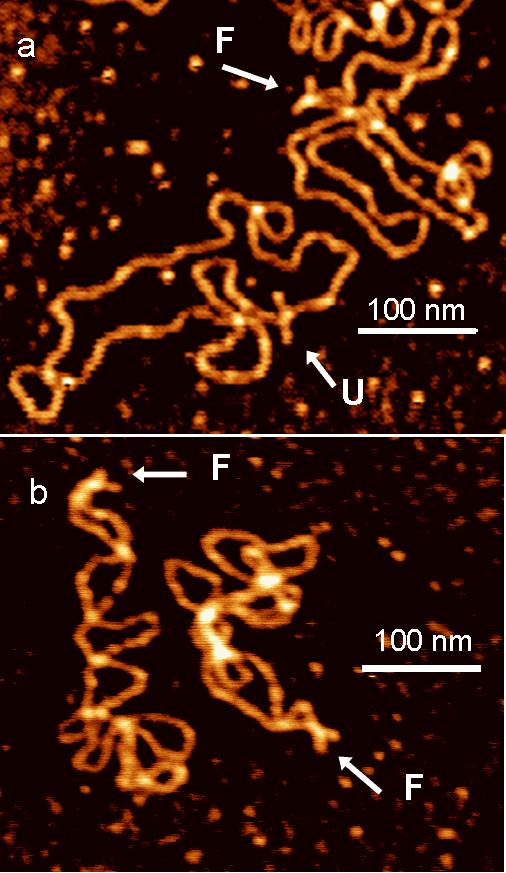
AFM images of dried sample t3low (a) and sample t7high (b) deposited from HE buffer. “F” and “U” denote cruciform in folded and unfolded conformations, respectively. Arrow shows the position of cruciform in DNA molecule.
Table 1.
Occurrence (percent) of folded and unfolded conformations in synthetic Holliday junction and in topoisomer samples t3low and t7high. The number of molecules measured for specific conformation is shown in parentheses.
| Buffer | HE | HEN | HM | |||
|---|---|---|---|---|---|---|
| Folded | Unfolded | Folded | Unfolded | Folded | Unfolded | |
| Synthetic Holliday junction | 16a (37) | 84a (195) | 48b (98) | 52b (107) | ||
| t3low | 48 (125) | 52 (136) | 52 (127) | 48 (115) | 81 (182) | 19 (42) |
| t7high | 98(288) | 2(5) | 99 (148) | 1(1) | 100 (150) | 0(0) |
in TE buffer
in TM buffer
Effect of ionic conditions on conformational transitions of cruciform
Ionic conditions have a profound effect on the conformations of immobile Holliday junctions. What is effect of ionic strength and divalent cations on conformations of Holliday junctions under the negative supercoiling stress? To answer this question, we performed imaging of t3low and t7high samples as well as synthetic Holliday junctions at various ionic conditions. Images presenting folded and unfolded conformations for different samples are shown in Fig. 1f and Fig. 2. First of all, we analyzed the partition of the Holliday junctions between the unfolded and folded conformations as a function of the buffer composition and DNA supercoiling. Results of this analysis are summarized in Table 1. The cruciform in sample t7high predominantly (98%) adopts a folded conformation, even in low-ionic strength buffer, so the distribution between two conformations very slightly depends on ion concentration. Quite different results were obtained for sample t3low and synthetic Holliday junctions. The population of both conformations for the sample t3low remains nearly equal when buffer with low ionic strength is used, whereas 10 mM Mg2+ significantly shifts equilibrium between folded and unfolded conformation of the cruciform towards the folded one. Synthetic Holliday junctions adopt unfolded conformation in HE buffer but addition of 10 mM Mg2+ also shifts equilibrium towards the folded one. These data are in a qualitative agreement with corresponding data in solution (6, 18, 19).
Geometry of cruciform in folded conformation.
The AFM data allow us to characterize conformations of the cruciforms. According to the schematics for Holliday junction (Figs. 1b-e), angle α between the hairpin arm and its continuation in the main DNA strands when equal to 180°, indicates the stacking of these arms. At the same time, angle β2, between the hairpin arms or angle β1 between long DNA strands allows to distinguish between parallel conformation (scheme b, c in Fig. 1, acute angles β1) and antiparallel conformation (scheme d, e in Fig 1, obtuse angles β1). To retrieve this information, we performed angle measurements between all arms of the cruciform.
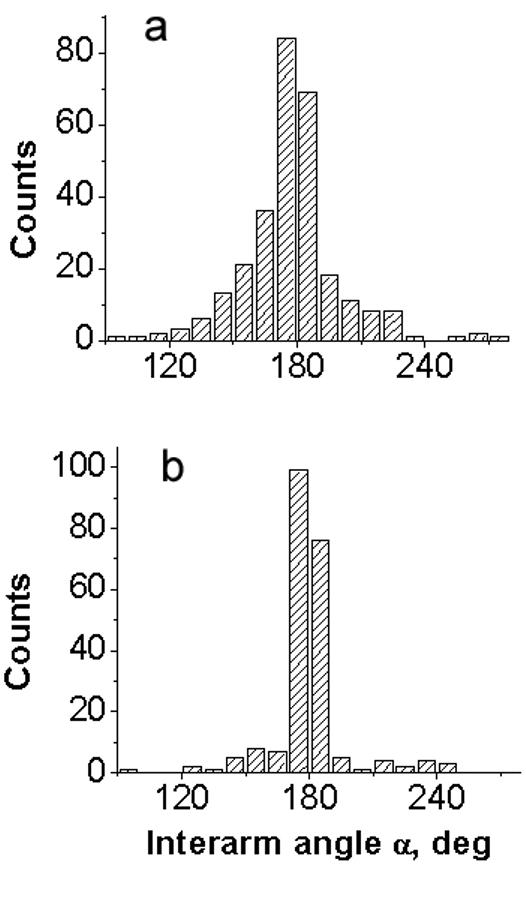
A set of the large number of images (between 86 and 150 frames) was obtained for each sample in different ionic conditions (HE, HEN and HM buffer). Figure 3 shows the results for the measurements of the interarm angle α (see Fig. 1c, e for the explanations). The histograms for the distribution of angle α values obtained for sample t7high in HM buffer (Fig. 3a) and for sample t3low (Fig. 3b) in HE buffer are centered around 180° value. The distributions are quite narrow, suggesting that the hairpin arm is a continuation of the corresponding main strand, or in other words, the sections of the cruciform are stacked. Similar results were obtained for sample t3low in HEN and HM buffers as well as for sample t7high in HE and HEN buffers (data not shown).
Figure 3.
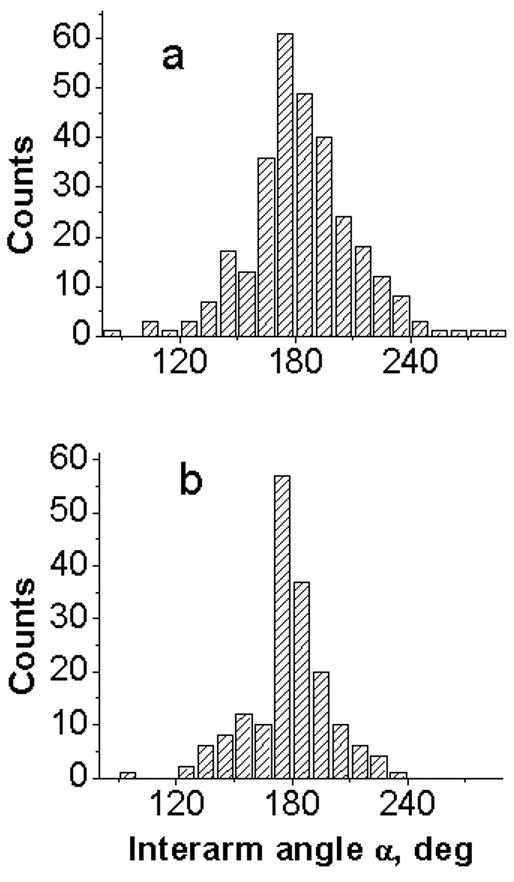
“Stacking” angle α (see schematics in Fig. 1c,e) measured for dried samples: (a) sample t7high deposited in HM buffer and (b) sample t3low deposited in HE buffer.
Another set of angles provides the information on the type of the folded conformation, parallel or antiparallel. These are the angle between the hairpin arms and the main strands β1 (see Figs. 1c, e and Materials and Methods for specifics). If the angle is less than 90°, the conformation of the cruciform is referred to as the parallel conformation; if the angle is more than 90°, the helices are in antiparallel orientation (24). Two typical histograms of the angle distributions for β1 angles are shown on Figs. 4a and 4b for sample t7high in HM buffer and for sample t3low in HE buffer, respectively. A dashed vertical line is drawn at 90°, so the points to the left of this line correspond to the parallel orientation of the cruciform, whereas all points to the right of these lines correspond to the antiparallel one. Fig. 4a shows that the cruciform for the t7high sample adopts a folded conformation with a parallel orientation of the arms. At lower supercoiling density (sample t3low, Fig. 4b), the preference to the parallel configuration remains, although the entire distribution is shifted to greater angle values. The mean values of β1 angles and their standard deviations calculated from similar measurements are summarized in Table 2. The data show that cruciforms in the two samples at different ionic conditions adopt parallel orientation, whereas synthetic Holliday junction at 10 mM MgCl2 predominantly adopts antiparallel one. The data on orientation of DNA arms in synthetic Holliday junctions is in qualitative agreement with corresponding results obtained in solution (6, 17). It is should also be noted that Mg2+ folds the cruciform even more tightly, increasing the effect of the supercoiling. Monovalent cations work in the same way, but the effect is considerably less pronounced than that of the divalent cations.
Figure 4.
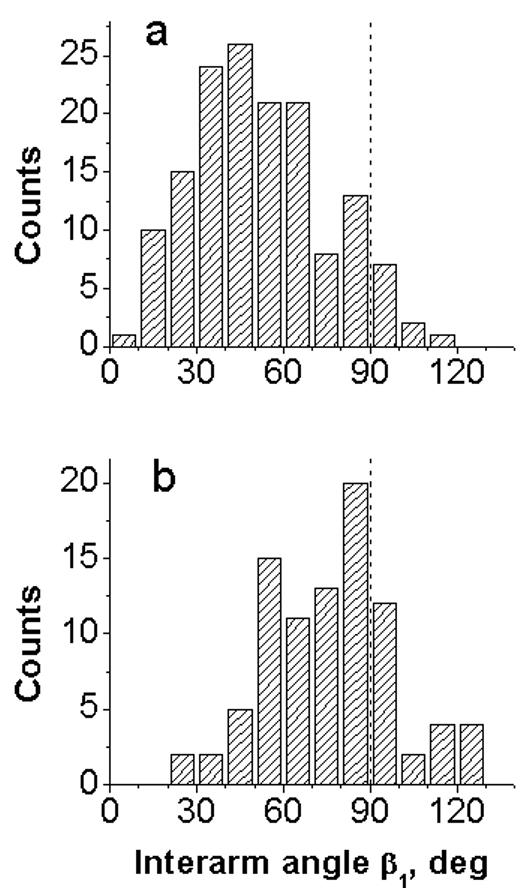
Angle β1 between main DNA strands for cruciform in folded conformation (see schematics in Fig. 1 (c,e) measured for dried samples: (a) sample t7high deposited from HM buffer and (b) sample t3low deposited from HE buffer.
Table 2.
Angle β1(see schematics in Fig. 1cande)measured for cruciform in folded conformation for synthetic Holliday junction and for topoisomer samples t3low and t7high in different buffers. Number of measured molecules is shown in parentheses.
| Buffer | HE | HEN | HM |
|---|---|---|---|
| Synthetic Holliday junction | N/A | 95 ± 40 (205)a | |
| t3low | 75 ± 21 (90) | 67 ± 23 (92) | 68 ± 29 (106) |
| t7high | 76 ± 22 (86) | 64 ± 23 (148) | 52 ± 24 (150) |
in TM buffer
Geometry of the cruciform in unfolded conformation.
The set of data obtained allows us to analyze the geometry of the unfolded conformation of the cruciform, which could be found in t3low sample at low ionic strength conditions. We measured all four angles between adjacent arms to characterize this conformation. These angles are 85-94 ± 27°, suggesting that unfolded conformation of the cruciform is close to a planar geometry, and has 4-fold symmetry. These values correlate well with data obtained by other methods (29).
Time-lapse observations of the cruciform conformational dynamics
To study the dynamics of the cruciform, we performed time-lapse imaging, and captured large series of consecutive images for the same molecules in aqueous solutions without drying the sample (24, 27).
Selected time-lapse images illustrating the dynamics of the cruciform for sample t3low and t7high in HE buffer are presented in Figure 5. The cruciform in sample t3low (Fig. 5 a-f) shows a high conformational dynamics. It changes conformation several times and flips from folded to unfolded and back to folded conformation several times during the time of the experiment. Figs. 5g-k in which traces of the cruciform only are shown summarizes these conformational changes (these images are made by placing traces of molecules on black background as described earlier (27)). The full set of data that comprises of 91 frames assembled as a movie file can be viewed in the Supporting Information section. File S1A was assembled from unprocessed AFM images of the molecule sections containing the cruciform; file S1B, was assembled from the images of the cruciforms traces placed on black background and centered at the junction. Similar data were obtained for other molecules in separate time-lapse experiments. Interestingly, transitions from folded to unfolded states and vice a versa appeared as two state flips, so we were unable to observe transient states between the two conformations suggesting that the lifetime of these states is much shorter than the image acquisition time.
Figure 5.
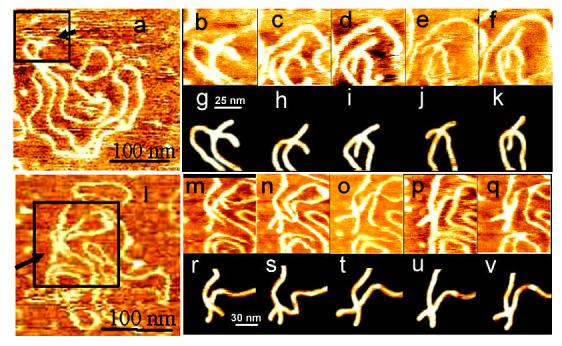
Dynamics of the cruciform observed with time-lapse AFM for hydrated sample t3low (frames a-k) and sample t7high (frames l-v) in HE buffer.
Initial image of the molecule with cruciform is shown in frame a (sample t3low) and frame l (sample t7high). Arrow is pointed to the cruciform. Images of enlarged area specified by the square in initial image, show time progress of the cruciform structure (frames b-f for sample t3low and frames m-q for sample t7high). Traces of the same molecule on black background are shown in frames g-k (sample t3low) and frames r-v (sample t7high). The capture time for images are 0 min (a), 11 min (b), 44 min (c), 60 min (d), 134 min (e), 155 min (f) (sample t3low) and 0 min (l), 13 min (m), 22 min (n), 71 min (o), 233 min (p), 363 min (q) (sample t7high).
The results for the cruciform dynamics within the plasmid with high supercoiling density (sample t7high) obtained in one the time-lapse experiment are shown in Figs. 5l - q. This cruciform remains in a folded conformation even over an observation period of 9.5 hours. Other observations with the same t7high sample were consistent with this long one we were unable to detect the transition between folded and unfolded conformations. The only change was the variation of the angle between the arms. These dynamics are summarized in Figs. 5r-v and the full set of data is placed as movie files in Supporting Information. File S2A is assembled from unprocessed AFM images of the molecules sections containing the cruciform. Movie file S2B was assembled from the images of the cruciforms traces placed on black background and centered at the junction. Overall, the time-lapse observations showed that the cruciform undergoes conformational transitions if the supercoiling density is low (sample t3low), but the conformation does not change if the supercoiling density is high. This conclusion is consistent with the data obtained in the studies of dried samples when the analysis was performed by averaging over ensemble of molecules (see Table 1). These data show that the populations of folded and unfolded conformations of the cruciform were nearly equal for t3low sample, whereas equilibrium distribution between these conformations in t7high sample is shifted toward the folded one.
To quantify the data on dynamics, we measured the “stacking” of α angles in folded conformation of the cruciform in the same way as for dried samples (for definition see Figs. 1c, e). We assembled the data from several molecules. Two representative histograms of distributions are presented in Fig. 6a (sample t7high in HM buffer) and Fig. 6b (sample t3low in HE buffer). Similar results were obtained in the case of sample t3low in HEN and HM buffer as well as for sample t7high in HE and HEN buffer (data not shown). Similar to dried samples, the maxima of these peaks are centered at ∼180° and the widths are quite narrow.
Figure 6.
“Stacking” angle α (see schematics in Fig. 1c,e) measured in hydrated sample: (a) sample t7high in HM buffer and (b) sample t3low in HE buffer.
We also plotted the interarm angles (see Fig. 1) as a function of time. The graphs for the time-lapse experiments for two molecules of t3low and t7high samples in HE buffer are shown as time trajectories in Figs. 7a, b, respectively (the same molecules are presented in Fig. 5 and as movie files in Supporting Information). The data for both angles are indicated in pink and green colors for β1 and β2, respectively (the same colors as in Figs. 1a, c, e). Vertical dashed lines in Fig. 7a separate regions between different conformations of cruciform (folded or unfolded). The changes of conformation of cruciforms are clearly seen from the Fig. 7a. The cruciform is initially in folded conformation and both β1 and β2 are ∼70°, then the conformation of cruciform changes to the unfolded one, and this transition is accompanied by a significant change in the angle value (from ∼70° to ∼180°). The cruciform remains in unfolded conformation for some time and β2 angle fluctuates around 180°, whereas β angle varies between ∼100° and ∼180°, then conformation changes to the folded one and both β1 and β2 values drop to ∼100°. Finally, the cruciform changes conformation to the unfolded one and both angles becoming closer to 180°. Similar experiments with sample t7high did not reveal any transition between the conformations of the cruciform in the sample t7high (see Fig 7b); values for β1 and β2 angles vary between ∼40 and ∼140 degrees, although some dynamics of the cruciform within the family of the folded conformation was observed. For example, the interarm angles β2 and β1 are >90° for frames between 8 and 25 corresponding to antiparallel orientation of cruciform arms (see Fig. 1), whereas at the rest of this graph, angle values are <90° so this orientation is parallel. As it can also be seen from this figure, both β1 and β2 angles change in a coordinated manner. Such correlated mobility confirms the conclusion made earlier that the hairpin arms and the main strands of the cruciforms are stacked. To find the mean value of angle β1, we assembled data from several molecules, and the total number of images analyzed was not less than 100 in each cases. We found the mean values of angle β1 of cruciforms in fully hydrated samples are quite similar to corresponding values found for dried samples (see Table 3). The highest difference between the data for dried samples and in solution is 8 degrees (standard deviation 23 degrees) for sample t7high in HM buffer, so it can be concluded that data in dried sample are consistent with the time-lapse data obtained in buffer.
Figure 7.
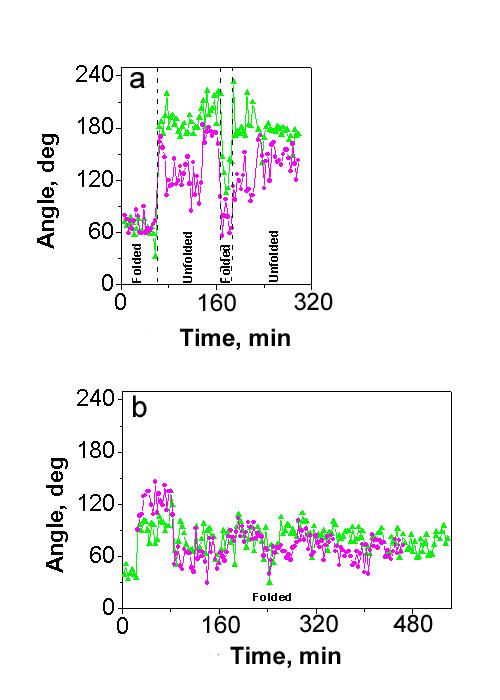
Dynamics of time-dependent variation of angle β1 (pink line) and angle β2 (green line) (see schematics in Fig. 1a,c and e) for samples t3low (a) and t7high (b) in HE buffer. The vertical dashed lines indicate the change in cruciform conformation as named on the bottom of the graph. The time difference between two consecutive frames is ∼3 min.
Table 3.
Angle b1 (see schematics in Fig. 1c and e) measured for cruciform in folded conformation for dried and hydrated samples t3low and t7high.
| Buffer | HE | HM | ||
|---|---|---|---|---|
| Dry sample | In buffer | Dry | In buffer | |
| t3low | 75 ± 21 | 74 ± 18 | 68 ± 29 | 66 ± 24 |
| t7high | 76 ± 22 | 70 ± 24 | 52 ± 24 | 44 ± 23 |
Branch migration.
We measured the lengths of the hairpin arms for the time-lapsed images to evaluate the effect of the supercoiling on branch migration. A similar approach was applied in our paper (27) for analysis of branch migration in synthetic Holliday junctions. We plotted time-dependent variation of the lengths of cruciform hairpin arms of t3low sample in unfolded (Fig. 8a) and folded (Fig.8b) conformations in HE buffer assembling the results for various experiments that are separated by vertical dashed lines. The data for two hairpin arms are indicated with different colors. Some sections of the time trajectories for unfolded cruciforms (Fig. 8a, molecules III and IV) show a gradual change in the hairpins lengths in which both hairpins increase their length (molecule III) and decrease their length (molecule IV), and these changes exceed the noise-level fluctuations of the arms lengths (these change are marked with arrows). Importantly, both hairpins change their length in coordinated manner. Such a coordinated change in the hairpin arms length was not observed for folded cruciforms (Fig. 8b). This finding is consistent with the model in which unfolding of Holliday junction is required for branch migration (19, 27, 30, 31).
Figure 8.
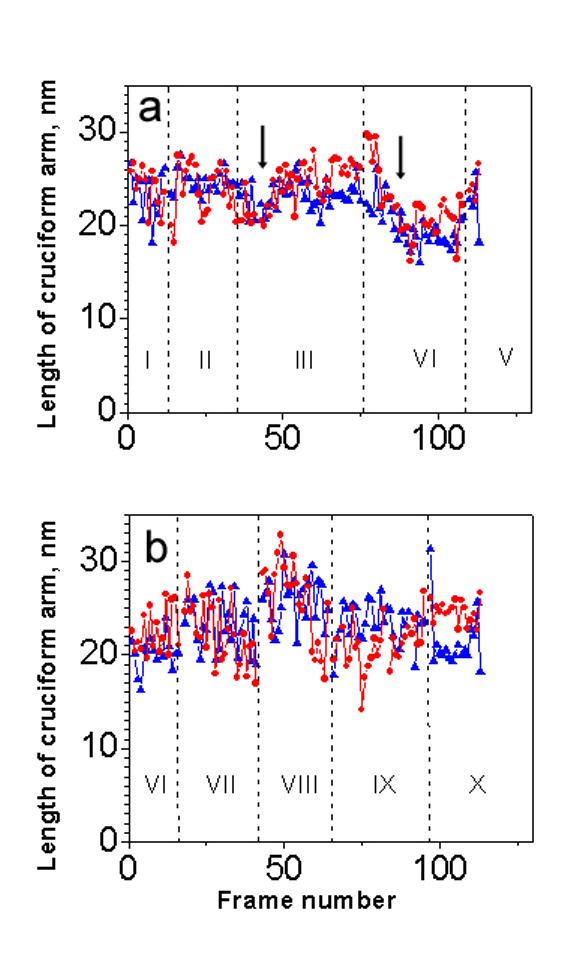
Time-dependent variation of the length for hairpin arms measured in selected molecules with cruciform in unfolded conformation (a, molecules I-V) and in folded conformation (b, molecules VI-X) for sample t3low in HE buffer. Vertical dashed lines separate data for different molecules. Arrows indicate the area when the arm length starts to change significantly (see text for details).
DISCUSSION.
The interplay between the local DNA structures and the global DNA conformations (topology) plays a significant role on DNA functions. The cruciforms formed by inverted repeats in supercoiled DNA are alternative DNA structures that critically involved in various DNA transactions. Cruciforms adopt different conformations. This conformational transition can have a strong effect on global DNA structure and dynamics playing role of a molecular switch, influencing the search of homologous regions within the supercoiled DNA molecule or in a topological domain (5). In our early paper, (24) we applied for the first time AFM for direct imaging the cruciform dynamics; we were able to characterize the transition between folded and unfolded conformations of the cruciforms. These data suggested that DNA supercoiling can be a critical factor determining the conformation of the cruciform. The results obtained in this paper not only support this hypothesis, but also provide a number of important characteristics to the structure and dynamics of the cruciform depending on the DNA supercoiling and ionic conditions. Importantly, these studies revealed some similarities while also revealing drastic differences between the four-way junction with free ends (synthetic Holliday junctions) and the cruciforms, the mobility of which is limited by the DNA topology.
Both synthetic junctions and cruciforms are capable of forming unfolded and folded states, but in contrast to synthetic junctions the conformation of which is determined by ionic conditions, primarily by the presence of divalent cations, the DNA supercoiling is the most critical factor in determining the cruciform conformation. Therefore, the cruciform in sample t7high (high supercoiling density) adopts predominately folded conformation even at low ionic strength and the absence of Mg2+ cations, although these cations further shift the equilibrium between the folded and unfolded conformations towards the folded one. The cruciforms in sample t7high in the presence of Mg2+ cations fold tighter that makes a hairpin arms to approach very closely to each other.
The most dramatic difference between the structural features of cruciforms and synthetic Holliday junctions is the geometry of folded conformation. It was shown that in the absence of the topological constraints and in the presence of divalent cations, Holliday junctions fold to the conformation with antiparallel orientation of the exchanging arms (6, 16, 18, 19). Our data show that the cruciform adopts parallel configuration of exchanging arms even in the absence of divalent cations. The cations further enforce this conformation of the cruciform. These findings suggest that negative DNA supercoiling stabilizes the folded conformation with parallel orientation of the arms. What is the energetic difference in these two conformations of the junction? Does the DNA supercoiling provide sufficient energy to stabilize an alternative conformation? Thermodynamic analysis of the stability between the parallel and antiparallel conformation performed on tethered junctions (21) revealed rather small differences in free energies for these two states (ca. 1.5 kcal/mol) in favor of antiparallel geometry. The molecular modeling analyses (20) came to qualitatively similar conclusions, although the free energy for antiparallel conformation was estimated at 18 kcal/mol lower than for the parallel conformation. It is instructive to compare these values with the free energy of DNA supercoiling. According to the a well-known expression for the free energy of DNA supercoiling, ▵G= 1100 RT (▵Lk)2/N, where R - the gas constant, T-temperature (in K), N-the length of plasmid in base pairs and ▵Lk-linking number difference that is proportional supercoiling density σ value (▵Lk = σN/γ, where γ-number of base pairs per turn in linear DNA (1, 32)). For the plasmid used in this work (N=3396 bp), the free energy increment per one supercoiling turn estimated for the supercoiling density close to the transition value (▵Lk=13, (22)) is ca. 8.4 RT or 5 kcal/mol. This estimate shows that supercoiling energy is in the range sufficient for stabilizing the unfavorable folded conformation of the junction. The stabilization of unfavorable DNA conformations is a ubiquitous property of negative DNA supercoiling, facilitating dramatically different dynamics of DNA required for its functioning. The fact that unwinding stress is needed for folding of the junction into parallel conformation can be useful in modeling of folded conformations of the Holliday junctions.
The measurements of the angles between the arms of the cruciforms showed that the hairpin arm and the one of the arms of the main DNA strand form almost continuous helix. Moreover, our time-lapse experiments show that these arms move in a coordinated manner. These findings suggest that the helices interact within the joint point probably due to stacking between the helices. A similar stacking interaction was observed for the antiparallel conformation of the Holliday junction recently confirmed by x-ray crystallographic analyses (14, 16). Recent single molecule three-color FRET studies of synthetic Holliday junction showed a coordinated movement the arms during the junction dynamics between the two folded states (33). Altogether, these data show that the interhelical interaction at the joint is a common feature for such branched DNA structures, and the most favorable configuration for the helices arrangement dictates by the DNA topology and environmental conditions such as ionic strength. The time lapse AFM studies were critical for direct imaging conformational transitions of the cruciforms, however the time scale of these observations does not relate to the characteristic times for transitions of cruciforms between the various conformations (24, 27, 34). In AFM the majority of time the molecules remains bound to the surface allowing their reliable imaging by the scanning tip; therefore the appearance of the cruciform in a particular conformation is the is determined by how long the cruciform in this configuration stays at the surface. Therefore, time lapse AFM studies provide the propensity of a molecule to adopt a particular conformation or shape rather than the life time of the conformation.
There are a number of interesting biological implications for the results obtained in this paper. The fact that supercoiling shifts the equilibrium between two conformations towards folded conformation affects the dynamics of juxtaposition of two sites in DNA required for such genetic processes as DNA site-specific recombination and transposition. Folded conformation of cruciform restricts the slithering of the DNA domains relative to each other, whereas unfolded one does not (5). There is a plethora of architectural and regulatory proteins that target DNA junctions, four-way junctions in particular (10, 12, 14, 35). The finding that the geometry of the junctions is affected by DNA supercoiling suggests an additional mechanism for involvement of DNA supercoiling into regulatory processes mediated by cruciforms. At the same time, within cells, DNA is supercoiled or arranged in topological domains; therefore cruciforms are in fact natural models for Holliday junctions that are transient forms for DNA during recombination (homologous or site-specific) and aberrant pathways for DNA replication. Branch migration, either protein mediated or spontaneous, is a critical step during these DNA transactions that require unfolding the junction (19, 27, 30, 31). At the same time, folding of the junction is needed for such processes as junctions resolution (14). DNA topoisomerases are indispensable components of the DNA replication and recombination machineries (1, 36, 37), therefore, controlling of the Holliday junction geometry via changing the DNA supercoiling by topoisomerases is another potential regulatory pathway explaining critical involvement of these enzymes in all DNA transactions.
Supplementary Material
ACKNOWLEDGMENT
We thank V. Potaman, Texas A&M University, for providing us with the topoisomer samples of plasmid DNA, and M. Karymov and L. Shlyakhtenko for stimulating discussion of the data and the critical comments and Alex Portillo for the help in editing of the manuscript. .
Footnotes
This research was supported by Grant GM 062235 (Y.L.L.) from the National Institutes of Health.
- AFM
- atomic force microscopy
- APS 1-(3-aminopropyl)silatrane
- FRET
- fluorescent resonance energy transfer
SUPPORTING INFORMATION
The section contains the descriptions for procedures for the preparation of synthetic Holliday junction, the measurements of interarm angle values and cruciform arm lengths and the movie files demonstrating dynamics of cruciform. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES:
- 1.Sinden RR. DNA Structure and Function. Academic press; San Diego: 1994. [Google Scholar]
- 2.Kornberg A, Baker TA. DNA Replication. 2nd edition Freeman; New York: 1992. [Google Scholar]
- 3.Jin R, Fernandez-Beros ME, Novick RP. Why is the initiation nick site of an AT-rich rolling circle plasmid at the tip of a GC-rich cruciform? Embo J. 1997;16:4456–66. doi: 10.1093/emboj/16.14.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai X, Greizerstein MB, Nadas-Chinni K, Rothman-Denes LB. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc Natl Acad Sci U S A. 1997;94:2174–9. doi: 10.1073/pnas.94.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shlyakhtenko LS, Hsieh P, Grigoriev M, Potaman VN, Sinden RR, Lyubchenko YL. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J Mol Biol. 2000;296:1169–73. doi: 10.1006/jmbi.2000.3542. [DOI] [PubMed] [Google Scholar]
- 6.Lilley DM. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–59. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 7.Varga-Weisz P, van Holde K, Zlatanova J. Preferential binding of histone H1 to four-way helical junction DNA. J Biol Chem. 1993;268:20699–700. [PubMed] [Google Scholar]
- 8.Varga-Weisz P, Zlatanova J, Leuba SH, Schroth GP, van Holde K. Binding of histones H1 and H5 and their globular domains to four-way junction DNA. Proc Natl Acad Sci U S A. 1994;91:3525–9. doi: 10.1073/pnas.91.9.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West KL, Austin CA. Human DNA topoisomerase IIbeta binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 1999;27:984–92. doi: 10.1093/nar/27.4.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pohler J, Norman DG, Bramham J, Bianchi ME, Lilley DM. HMG box proteins bind to four-way DNA junctions in their open conformation. Embo J. 1998;17:817–26. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. Embo J. 2000;19:6527–35. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Cavallo L, Griffith J. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J Biol Chem. 1997;272:7532–9. doi: 10.1074/jbc.272.11.7532. [DOI] [PubMed] [Google Scholar]
- 13.Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–10. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khuu PA, Voth AR, Hays FA, Ho PS. The stacked-X DNA Holliday junction and protein recognition. J Mol Recognit. 2006;19:234–242. doi: 10.1002/jmr.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zlatanova J, van Holde K. Binding to four-way junction DNA: a common property of architectural proteins? Faseb J. 1998;12:421–31. [PubMed] [Google Scholar]
- 16.Hays FA, Watson J, Ho PS. Caution! DNA crossing: crystal structures of Holliday junctions. J Biol Chem. 2003;278:49663–6. doi: 10.1074/jbc.R300033200. [DOI] [PubMed] [Google Scholar]
- 17.Clegg RM, Murchie AI, Zechel A, Carlberg C, Diekmann S, Lilley DM. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry. 1992;31:4846–56. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]
- 18.Joo C, McKinney SA, Lilley DM, Ha T. Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J Mol Biol. 2004;341:739–51. doi: 10.1016/j.jmb.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 19.McKinney SA, Freeman AD, Lilley DM, Ha T. Observing spontaneous branch migration of Holliday junctions one step at a time. Proc Natl Acad Sci U S A. 2005;102:5715–20. doi: 10.1073/pnas.0409328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srinivasan AR, Olson WK. Computer models of DNA four-way junctions. Biochemistry. 1994;33:9389–404. doi: 10.1021/bi00198a004. [DOI] [PubMed] [Google Scholar]
- 21.Lu M, Guo Q, Seeman NC, Kallenbach NR. Parallel and antiparallel Holliday junctions differ in structure and stability. J Mol Biol. 1991;221:1419–32. doi: 10.1016/0022-2836(91)90942-y. [DOI] [PubMed] [Google Scholar]
- 22.Oussatcheva EA, Pavlicek J, Sankey OF, Sinden RR, Lyubchenko YL, Potaman VN. Influence of global DNA topology on cruciform formation in supercoiled DNA. J Mol Biol. 2004;338:735–43. doi: 10.1016/j.jmb.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 23.Pavlicek JW, Oussatcheva EA, Sinden RR, Potaman VN, Sankey OF, Lyubchenko YL. Supercoiling-induced DNA bending. Biochemistry. 2004;43:10664–8. doi: 10.1021/bi0362572. [DOI] [PubMed] [Google Scholar]
- 24.Shlyakhtenko LS, Potaman VN, Sinden RR, Lyubchenko YL. Structure and dynamics of supercoil-stabilized DNA cruciforms. J Mol Biol. 1998;280:61–72. doi: 10.1006/jmbi.1998.1855. [DOI] [PubMed] [Google Scholar]
- 25.Lushnikov AY, Brown BA, 2nd, Oussatcheva EA, Potaman VN, Sinden RR, Lyubchenko YL. Interaction of the Zalpha domain of human ADAR1 with a negatively supercoiled plasmid visualized by atomic force microscopy. Nucleic Acids Res. 2004;32:4704–12. doi: 10.1093/nar/gkh810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA. Ultramicroscopy. 2003;97:279–87. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 27.Lushnikov AY, Bogdanov A, Lyubchenko YL. DNA recombination: holliday junctions dynamics and branch migration. J Biol Chem. 2003;278:43130–4. doi: 10.1074/jbc.M308228200. [DOI] [PubMed] [Google Scholar]
- 28.Lushnikov AY, Potaman VN, Oussatcheva EA, Sinden RR, Lyubchenko YL. DNA strand arrangement within the SfiI-DNA complex: atomic force microscopy analysis. Biochemistry. 2006;45:152–8. doi: 10.1021/bi051767c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nollmann M, Stark WM, Byron O. Low-resolution reconstruction of a synthetic DNA holliday junction. Biophys J. 2004;86:3060–9. doi: 10.1016/s0006-3495(04)74355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panyutin IG, Hsieh P. The kinetics of spontaneous DNA branch migration. Proc Natl Acad Sci U S A. 1994;91:2021–5. doi: 10.1073/pnas.91.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karymov M, Daniel D, Sankey OF, Lyubchenko YL. Holliday junction dynamics and branch migration: single-molecule analysis. Proc Natl Acad Sci U S A. 2005;102:8186–91. doi: 10.1073/pnas.0407210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vologodskii A. OnLine Biophysical Chemistry Textbook. Biophysical society; 1999. [Google Scholar]
- 33.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–37. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyubchenko YL, Shlyakhtenko LS. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc Natl Acad Sci U S A. 1997;94:496–501. doi: 10.1073/pnas.94.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsons CA, Stasiak A, Bennett RJ, West SC. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–8. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 36.Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu Rev Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 37.Champoux JJ. DNA topoisomerases: structure. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


