Abstract
Protein structural features are usually determined by defining regularities in a large population of homogeneous molecules. However, irregular features such as structural variation and flexibility are likely to be missed, despite their vital role for their biological function. In this paper, we report the observation of striking irregularities in the flexibility of the coiled-coil region of the human Rad50 DNA repair protein. Existing methods to quantitatively analyze flexibility are applicable to homogeneous polymers only. Because protein coiled-coils cannot be assumed to be homogeneous, we develop a method to quantify the local flexibility from high-resolution atomic force microscopy images. Indeed, in Rad50 coiled-coils, two positions of increased flexibility are observed. We discuss how this dynamic structural feature is integral to Rad50 function.
The protein complex containing Rad50 and Mre11 (R/M) plays a pivotal role in maintaining genome stability. Its vital importance is underscored by its conservation from bacteriophages to humans (1). Based on amino acid sequence similarities, Rad50 belongs to the structural maintenance of chromosomes (SMC) family of proteins. Their shared amino acid sequence predicts a very distinct structure, including a bipartite globular ATPase domain made up from the N and C termini of the protein separated by an extensive central region predicted to form a coiled-coil (2). Accumulated recent evidence reveals that Rad50 and other SMC proteins are arranged as intramolecular coiled-coils bringing together the N and C termini from one polypeptide to form a functional ATPase at one end of an elongated structure (Fig. 1b and refs. 3–5). The SMC family members all exist as complexes of two such extended proteins. Their diverse functions must all require this striking structure. Many proposals for the mechanism of action of R/M and other SMC complexes require the coiled-coils to be rigid and act as lever arms (6). However, images of R/M show the coiled-coils in a variety of conformations, suggesting a high degree of flexibility (refs. 3 and 7 and Fig. 1a). This finding has been confirmed by time-resolved atomic force microscopy [AFM, also called scanning force microscopy (SFM)] imaging of R/M partially immobilized in buffer, showing that individual coiled-coils are quite flexible (3).
Fig. 1.
High-resolution AFM images of R/M complexes. (a) Overview of R/M complexes deposited on mica and imaged in air, showing a high variety of conformations. Scan range, 500 nm; z range, 3 nm. (b) Schematic representation of the architecture of the human R/M complex. C, Rad50 carboxyl-terminal domain; N, Rad50 amino-terminal domain; M, Mre11; Zn2+-hook in dark blue. (c) Software zoom of a R/M complex, oriented like a; scan width, 125 nm; z range, 3 nm. (d) Cross section through the Rad50 coiled-coils of a R/M complex.
To advance the structural characterization of the Rad50 coiled-coils, we developed a method to characterize their flexibility quantitatively from high-resolution AFM images of R/M. For this analysis, we used R/M complexes with two separate, well-resolved coiled-coils such as shown in Fig. 1c. In Fig. 1d, a cross-section through the coiled-coils is plotted to emphasize the resolution obtained. The high-resolution images allowed detailed analysis of the trajectory of the coiled-coils. In case of a homogeneous polymer, the local flexibility F at every position corresponds directly to the inverse of the persistence length P, which is commonly used to describe the global bending rigidity of polymers. The persistence length of homogeneous biopolymers can be determined from static images by using statistical mechanics. The bending rigidity of DNA, for example, has been quantified directly from AFM images (8). In this method, P is determined from measured end-to-end distance R and contour length L by using the following equation:
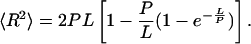 |
[1] |
For polymers like DNA, this method yields an average persistence length that is in good agreement with values obtained by other techniques (8). This method has been modified to describe known segments of different persistence length (9). Another method for the determination of overall flexibility of biopolymers involves measuring of polymer extension as a function of the applied force, with optical tweezers (10) or AFM force spectroscopy (11), and to fit a worm-like-chain model to these data (12). Because structural differences along the polymer chain will be averaged out with these methods, they are applicable to homogeneous polymers only. However, a coiled-coil of two interwound amino acid chains cannot be assumed to be homogeneous. To describe this likely inhomogeneity, we developed a method that allows quantitative analysis of local flexibility along a polymer chain and therefore maps the spatial distribution of the flexibility along the polymer chain.
To locally determine flexibility in a polymer, we used and extended a method derived from quantifying bending angles of protein–DNA complexes (13). A slightly different method has been used to identify subtle sequence-dependent differences in flexibility in DNA molecules (14). In our analysis, the local curvature κ of a specific polymer at a certain position is defined as the inverse radius of a circle following the trajectory along the molecule at that position. The elastic energy ΔU necessary to deflect the polymer can be expanded around the intrinsic curvature 〈κ〉 by a Taylor series as:
 |
[2] |
Neglecting higher-order terms, the elastic bending energy of the polymer can thus be described as a harmonic oscillator:
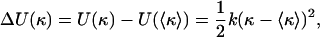 |
[3] |
using a spring constant
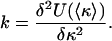 |
[4] |
The local flexibility F can now be related to this spring constant as:
 |
[5] |
where kB = Boltzmann's constant.
Visualization of R/M complexes by AFM reveals the Rad50 coiled-coils in a variety of conformations (Fig. 1a and ref. 3) imposed by thermal equilibrium that will follow Boltzmann's law. Thus, by using Eq. 3, the observed curvatures will be distributed according to
 |
[6] |
giving a normal distribution around average curvature 〈κ〉, with a standard deviation of 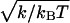 . Fitting Eq. 3 to the natural logarithm of this observed curvature distribution directly yields a value for the spring constant k, and thus, via Eq. 5, also the flexibility F.
. Fitting Eq. 3 to the natural logarithm of this observed curvature distribution directly yields a value for the spring constant k, and thus, via Eq. 5, also the flexibility F.
Materials and Methods
DNA samples were prepared for AFM imaging by depositing 1 ng/μl 1,000-bp DNA (DNA Quantitation Standard Kit, Eurogentec, Seraing, Belgium) in 5 mM MgCl2 and 10 mM Hepes-KOH (pH 7.5) onto freshly cleaved mica. The human R/M complex was purified as described (3). For AFM analysis, 20 μl containing 400 ng of the protein complex in buffer (150 mM KCl/25 mM Tris·HCl, pH 7.8/10% glycerol) was deposited onto freshly cleaved mica. After ≈20 s, the mica was washed with MilliQ-filtered water and blown dry in a stream of nitrogen gas. Samples were imaged in air at room temperature and humidity with a NanoScope IIIa (Digital Instruments, Santa Barbara, CA), operating in tapping mode with a type E scanner. Tapping mode silicon tips OMCL-AC160TS-W2 (Olympus, Melville, NY) were used for imaging DNA and SuperSharpSilicon tips, type SSS–NCH-8 (NanoSensors, Neuchatel, Switzerland) were used for imaging R/M complexes. Image processing was done by using Interactive Data Language (IDL; Research Systems, Inc., Boulder, CO) and consisted only of a background correction. The coordinates of the molecular trajectories were traced manually by using a similar routine as described by Rivetti and Codeluppi (15). Further data analysis and simulations were also implemented in IDL.
Analysis of DNA Flexibility with the Local Curvature Method. First, we tested the local curvature method by analysis of 1,000-bp DNA molecules. DNA can be considered homogeneous in terms of flexibility, although subtle sequence-dependent differences have been reported (14). One hundred DNA molecules were imaged by AFM (Fig. 2a) and traced. Applying Eq. 1 yielded an average persistence length of 45 nm, well within the range of previously reported values (8).
Fig. 2.
Validation of the local curvature method. (a) Two 1,000-bp DNA molecules imaged by tapping mode AFM at a scan range of 500 nm and a z range of 3 nm. (b) Result of simulation of two 1,000-bp DNA molecules with a persistence length (P) of 45 nm at a tip radius of 5 nm and a DNA height of 1 nm, which corresponds to the height observed in a. Image settings are as shown in a.(c) Experimentally measured curvature distribution of DNA based on 100 molecules. Curvature was calculated by using three 5-nm-spaced points along the chain, and iterated every 1 nm, resulting in ≈290 data points per DNA molecule. (d) Calculated bending energy ΔU for the curvature distribution shown in c. Solid line shows a parabolic fit to energy values below 1.5 kBT. Dotted line shows the expected bending energy curve based on a P = 45 nm. (e) Persistence length Pout as determined for the simulated homogeneous polymers. Filled squares show persistence length calculated with the end-to-end method. Open circles are measured with the curvature analysis method at a tip radius of 5 nm, whereas open squares represent data obtained with tip radius 10 nm. (Inset) A zoom for small persistence lengths, in the range of the tip size. The observed persistence length increases monotonously with the inputted persistence length.
By using three points at a 5-nm spacing along the trajectory of the DNA, iterated at 1-nm intervals, the local curvature was measured. The resulting curvature distribution was found to be constant along the DNA chain, as expected for a homogeneous polymer. The curvature distribution and its calculated bending energy are plotted in Fig. 2 c and d, respectively. In Fig. 2d, the dotted line shows the calculated bending energy curve for a polymer with a persistence length of 45 nm. For very small curvatures, the data closely matched the calculated energy curvature, validating the method.
However, for curvatures exceeding 0.02 nm–1 the experimental data deviate substantially from the theoretical curve. Because of resolution limits of AFM imaging, high-curvature features are significantly obscured. This effect is enhanced by our tracing method, which moderately filters the coordinates of trajectory of the polymer. Furthermore, to determine the local curvature, we need three points along the trajectory of the molecule, which may average sharp bends within this range. As a consequence of these three effects, sharp kinks will be recorded having a too small curvature, effectively squeezing the tails of the curvature distribution to the center and thus broadening the ΔU(κ) curve. Fitting a broader range of curvatures to these data points resulted in an erroneous persistence length of 204 nm.
To quantify the combined effects of tip-sample convolution and image processing, we simulated AFM images of homogenous DNA-like polymers. The simulated polymers had the same dimensions as DNA, but varied in persistence length. The effect of tip-convolution was generated by using a parabolic tip, similar to the procedure described by Rivetti and Codeluppi (15). Fig. 2b shows a simulated image of two 1,000-bp DNA molecules with a persistence length of 45 nm, similar to the molecules measured in Fig. 2a. By using the end-to-end distance method, the persistence length of the molecules in the simulated images closely matched the input values, confirming the validity of the simulation.
Curvature distributions were generated after tracing 100 molecules in these simulated images. By using the procedure described above, the local flexibility was measured. In Fig. 2e, the resulting values for the persistence length, which correspond to 1/F, are plotted as a function of the input values. For finite tip radii, the measured persistence length seems to be overestimated by a factor up to 10. Reversibly, based on Fig. 2e and the experimentally determined persistence length of DNA according to Eq. 1, we can estimate the tip radius used to produce the image in Fig. 2a to be ≈6 nm. This finding fits well with the observed width of the DNA. The measured values converge to the input numbers at persistence lengths beyond 600 nm. Thus, our local curvature method overestimates the persistence length if it is of the same order of magnitude as the image resolution, which is limited in AFM imaging by the finite size of the tip. Despite this overshoot in the absolute value of the calculated persistence length, however, a monotonously increasing observed persistence length was found for small values of the input persistence length (Fig. 2e Inset). Thus, our simulations show that a relative flexibility can be obtained sensitively and reliably, even though sharp bends are obscured by tip convolution limitations.
Results
Quantitative Analysis of Flexibility Along the Rad50 Coiled-Coil. To determine the flexibility of the Rad50 coiled-coil, the contours of 100 R/M complexes were traced. Three typical R/M complexes with their traced coordinates are shown in Fig. 3 a–c. From the traced coordinates, we directly obtained an average contour length of 47 nm for a single Rad50 coiled-coil and an average end-to-end distance of 42 nm. Naively, neglecting in-homogeneities in the amino acid chains, Eq. 1 results in a global persistence length of 30 nm for a single Rad50 coiled-coil. Next, the traced coordinates of the R/M complexes were evaluated by using our local curvature method. We defined positive curvature as bending inward forming a circular R/M complex configuration, whereas outward bends were assigned a negative curvature (Fig. 3d). The curvature of all traces was analyzed by using three 5-nm-spaced points. This procedure was iterated, following the trajectory of the molecule with 1-nm intervals. The curvatures along 200 Rad50 coiled-coils were plotted in equally spaced segments according to the distance from the center of the globular domain. Fig. 4 a, c, and e, for example, shows three histograms of the local curvature at 37, 40, and 44 nm from the globular domain, respectively. At 40 nm, a significantly broader curvature distribution was observed compared with its neighboring segments. The curvature distribution represented in terms of the bending energy is plotted in Fig. 4 b, d, and f. A parabola describing Eq. 3 was fitted to all data points, with a ΔU <1.5 kBT. By adopting this threshold, very high curvatures were omitted, which is justified because the number of these occurrences is underestimated, as described above.
Fig. 3.
Tracing of R/M complexes. (a–c) The trajectory of the Rad50 molecules within the R/M complexes as determined by tracing; scan range, 125 nm; z range, 3 nm. (d) Schematic representation of a R/M complex with + and – signs denoting our curvature sign convention.
Fig. 4.
Curvature distributions and corresponding energy plots of Rad50 coiled-coils. (a, c, and e) Curvature distributions of specific segments of the Rad50 molecule 37, 40, and 44 nm from the center of the R/M complex globular domain, respectively. (b, d, and f) Corresponding calculated bending energies. Lines represent parabolic fits to values <1.5 kBT.
A major result of this report is that the observed flexibility of the Rad50 coiled-coils seems to vary along its length, as shown in Fig. 5a. Two sections of clearly increased flexibility could be distinguished at 27 and 40 nm. These two sections of increased flexibility appeared in all curvature distributions, independent of the chosen bin size. Whereas the flexibility of the polymer segments changed along the Rad50 coiled-coil, the intrinsic curvature along the coiled-coil, deduced from the minimum position of the elastic energy, only moderately varied (Fig. 5b), with all sections having a positive value, i.e., bending toward the center of the complex. The corresponding radii of curvature, 46 to 86 nm, were all of the same magnitude or larger than the total length of the molecule, giving the molecule a moderately bent average conformation. Thus, the Rad50 coiled-coil does not contain segments with locally increased intrinsic curvature, and the large variety of coiled-coil conformations originates mainly from its two relatively flexible segments.
Fig. 5.
The Rad50 coiled-coil contains specific segments of increased flexibility. (a) Flexibility distribution along the Rad50 coiled-coil. Segments within 10 nm of the center of the globular R/M complex domain, marked I, are indicated by white bars. These regions do not contribute to the Rad50 coiled-coil. The last bin is also displayed in white because end effects distort its value. Gray bars represent segments along the coiled-coil. Two segments displaying increased flexibility are marked II and III. (b) Intrinsic curvature distribution along the Rad50 coiled-coil. The first bins, indicated in white and corresponding to the R/M complex globular domain, are not relevant with respect to the curvature analysis of the Rad50 coiled-coil; the last bin is affected by end effects. (c) Schematic cartoon of Rad50 in ribbon representation derived from the crystal structures of the ATPase domain (left) and the zinc hook (right) the Rad50 homologue in Pyrococcus furiosus (5, 20). The structure of the coiled-coil that connects these two features has not been determined and is indicated by the dotted line. (d) Probability of coiled-coil formation along the Rad50 amino acid chain is indicated by using a gray scale with black and white representing the lowest and highest probability of forming a coiled-coil, respectively. The amino acid chain is folded back onto itself at the position of the zinc hook, marked IV. The globular domain is marked I. Two positions, marked II and III, appear in the coiled-coil where both amino acid chains have a predicted decreased probability in coiled–coil formation. These positions colocalize with increased flexibility segments II and III in a.
Mapping Rad50 Regions of Increased Flexibility on Its Amino Acid Sequence. What is the structural origin of the variation in local flexibility? Crystallographic studies have provided atomic resolution structures for the N and C termini of Rad50 (16), as well as the region in the middle of the predicted coiled-coil (5). The N and C termini juxtapose to form a globular ATPase domain. The middle of the predicted coiled-coil forms a structure, referred to as a zinc hook, where the amino acid chain makes a sharp U-turn. However, no high-resolution structural data are available for the predicted coiled-coil that connects the globular ATPase domain to the zinc hook (Fig. 5c). The probability of forming a coiled-coil along the Rad50 amino acid chain was determined by using the program coils (17). The result is graphically displayed in Fig. 5d. Interestingly, in addition to the zinc hook (denoted IV) and the ATPase domain (denoted I), two positions, denoted II and III, were revealed in which both strands of the coiled-coil had a decreased probability of adopting a coiled-coil structure. Positions II and III are located ≈110 and 70 aa from the zinc hook, respectively. Assuming a rise of 1.5 Å per amino acid residue and taking into account the broadening effect of tip-convolution which is ≈4 nm, positions II and III of decreased coiled-coil probability (Fig. 5d) mapped to segments II and III of locally increased flexibility (Fig. 5a). Our results suggest that the observed flexibility of the Rad50 coiled-coil resides in segments in which the coiled-coil structure is disrupted.
Discussion
Our detailed analysis of AFM images of a wide variety of molecular conformations has resulted in high-resolution data on the local molecular flexibility. By analysis of high-resolution AFM images of the R/M complex, we have revealed structural variations in the Rad50 protein that are spaced 12 nm apart. Although other techniques have been used to quantify polymer flexibility, the method presented here makes it possible to reveal differences in flexibility within a molecule. The global persistence length of a single disrupted amino acid chain, determined from force-distance measurements, is 2.0 nm (18). Quantitatively we observed the measured flexibility of the Rad50 coiled-coils to vary from 0.007 nm–1 for the stiff segments to 0.016 nm–1 for the more flexible segments. By using a correction based on Fig. 1e for a tip radius of 5 nm, these flexibilities correspond to apparent persistence lengths of 17 and 6.0 nm, respectively. This result indicates that the flexible segments approach the flexibility of two non-ordered amino acid chains.
The remarkable architecture of the R/M complex has led to speculation about its mechanism of action. The flexibility map that we obtained for Rad50 highlights the dynamic nature of its structure and also rules out some mechanistic models. Because the coiled-coils are not stiff, they cannot simply transmit changes in conformation of the globular domain to changes in relative orientation of the ends of the coiled-coil (19). However, the flexibility we observe is a distinct advantage for other proposed mechanisms for R/M function. We have observed tethering of DNA via apparent interactions between the coiled-coil ends of multiple DNA-bound R/M complexes (3). The atomic level structure of the end of an archaeal Rad50 coiled-coil has revealed a hook-like structure with a conserved CXXC-motif such that two coiled-coil ends can dimerize by coordination of Zn2+ (5). Human Rad50 has a similar CXXC-motif and ability to dimerize in the presence of Zn2+ (5). This mode of dimerization requires the two coiled-coil ends to assume a specific orientation relative to each other. The multiple R/M complexes bound to DNA do not seem to be in any regular arrangement (3). The flexibility of the coiled-coils that we demonstrate here would allow the hook ends of multiple DNA-bound complexes to form correctly oriented intermolecular Zn2+-fingers independent of the orientation of their DNA-bound globular domains.
The sequence similarity among SMC family members defines a common architecture that likely includes flexible coiled-coils. Interestingly, a flexible structure would be advantageous for another set of SMC proteins in the recently proposed mechanism of sister chromatin cohesion by the Saccharomyces cerevisiae SMC1/3 cohesin complex. The SMC1 and -3 proteins also have intramolecular coiled-coils and dimerize via a globular domain at the apex of the coiled-coils (4). The ATPase domains of SMC1 and -3 can be bridged by a third cohesin subunit to make a large protein ring. Nasmyth and colleagues (4) have proposed that this protein ring could encircle a chromosome before replication, allowing a replication fork to pass through and thus form a topological link around the two resulting daughter chromosomes. The size of the ring is large enough to allow passage of a replication fork and encircle two 10-nm chromatin fibers. A flexible ring would more easily allow closure in the first place and accommodate replication fork passage, which itself has a dynamic and irregular shape. The SMC proteins function in diverse cellular pathways and likely have diverse mechanistic roles. The flexibility we describe here for human Rad50 coiled-coils may be a defining structural feature with exceptional mechanistic versatility.
Acknowledgments
This work was supported by grants from the Dutch Cancer Society and the Netherlands Organization for Scientific Research and Foundation for Fundamental Research on Matter–Earth and Life Sciences.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: R/M, Rad50 and Mre11; SMC, structural maintenance of chromosome; AFM, atomic force microscopy.
References
- 1.Cromie, G. A., Connelly, J. C. & Leach, D. R. (2001) Mol. Cell 8, 1163–1174. [DOI] [PubMed] [Google Scholar]
- 2.Wyman, C. & Kanaar, R. (2002) Curr. Biol. 12, R446–R448. [DOI] [PubMed] [Google Scholar]
- 3.De Jager, M., Van Noort, J., Van Gent, D. C., Dekker, C., Kanaar, R. & Wyman, C. (2001) Mol. Cell 8, 1129–1135. [DOI] [PubMed] [Google Scholar]
- 4.Haering, C. H., Lowe, J., Hochwagen, A. & Nasmyth, K. (2002) Mol. Cell 9, 773–788. [DOI] [PubMed] [Google Scholar]
- 5.Hopfner, K. P., Craig, L., Moncalian, G., Zinkel, R. A., Usui, T., Owen, B. A. L., Karcher, A., Henderson, B., Bodmer, J. L., McMurray, C. T., et al. (2002) Nature 418, 562–566. [DOI] [PubMed] [Google Scholar]
- 6.Hirano, T. (1999) Genes Dev. 13, 11–19. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, D. E., Trujillo, K. M., Sung, P. & Erickson, H. P. (2001) J. Biol. Chem. 276, 37027–37033. [DOI] [PubMed] [Google Scholar]
- 8.Rivetti, C., Guthold, M. & Bustamante, C. (1996) J. Mol. Biol. 64, 919–932. [DOI] [PubMed] [Google Scholar]
- 9.Rivetti, C., Walker, C. & Bustamante, C. (1998) J. Mol. Biol. 280, 41–59. [DOI] [PubMed] [Google Scholar]
- 10.Bustamante, C., Smith, S. B., Liphardt, J. & Smith, D. (2000) Curr. Opin. Struct. Biol. 10, 279–285. [DOI] [PubMed] [Google Scholar]
- 11.Strunz, T., Oroszlan, K., Schafer, R. & Guntherodt, H.-J. (1999) Proc. Natl. Acad. Sci. USA 96, 11277–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. (1994) Science 265, 1599–1600. [DOI] [PubMed] [Google Scholar]
- 13.Van Noort, J., Orsini, F., Eker, A., Wyman, C., De Grooth, B. & Greve, J. (1999) Nucleic Acids Res. 27, 3875–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuccheri, G., Scipioni, A., Cavaliere, V., Gargiulo, G., De Santis, P. & Samori, B. (2001) Proc. Natl. Acad. Sci. USA 98, 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivetti, C. & Codeluppi, S. (2001) Ultramicroscopy 87, 55–66. [DOI] [PubMed] [Google Scholar]
- 16.Hopfner, K. P., Karcher, A., Craig, L., Woo, T. T., Carney, J. P. & Tainer, J. A. (2001) Cell 105, 473–485. [DOI] [PubMed] [Google Scholar]
- 17.Lupas, A., Vandyke, M. & Stock, J. (1991) Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 18.Kellermayer, M. S. Z. & Bustamante, C. (1997) Science 277, 1117.9289852 [Google Scholar]
- 19.Strunnikov, A. V. & Jessberger, R. (1999) Eur. J. Biochem. 263, 6–13. [DOI] [PubMed] [Google Scholar]
- 20.Hopfner, K. P., Karcher, A., Shin, D. S., Craig, L., Arthur, L. M., Carney, J. P. & Tainer, J. A. (2000) Cell 101, 789–800. [DOI] [PubMed] [Google Scholar]







