Abstract
We demonstrate the detection of nanometer-scale conformational changes of single DNA oligomers through a micromechanical technique. The quantity monitored is the displacement of a micrometer-size bead tethered to a surface by the probe molecule undergoing the conformational change. This technique allows probing of conformational changes within distances beyond the range of fluorescence resonance energy transfer. We apply the method to detect single hybridization events of label-free target oligomers. Hybridization of the target is detected through the conformational change of the probe.
We describe measurements of nanometer-scale conformational changes of single DNA oligonucleotides, 40–90 bases long. The experiments are based on a micromechanical technique, which we have introduced previously (1). By measuring the contour length shortening of a single-probe molecule on formation of the double-stranded (ds) structure, we apply the method to detect hybridization of single unlabeled target oligomers. There are two interesting aspects to this approach. First, the ability to monitor directly certain conformational changes of DNA and RNA provides a tool for investigating processes such as (protein-induced) bending and looping, which have regulatory roles in transcription and splicing. The most detailed information on static conformations is provided by labor-intensive structural studies. At the other end, simple gel-shift assays provide a partial characterization, such as a bending angle. Single-molecule methods, in principle, offer a direct way of studying conformational changes, including dynamics. Recently, the conformational change involved in the catalytic activity of a ribozyme has been studied in detail by single-molecule fluorescence resonance energy transfer (FRET) (2). However, FRET is limited to distances <10 nm, whereas many interesting structures formed by DNA, such as bends and loops, involve larger (10- to 30-nm) scales (10 nm is the contour length of a dsDNA 30 mer). With the method described here, we can detect nanometer-scale conformational changes of single DNA oligomers of length 30–90 bases, thus extending the range covered by FRET.
A second aspect is the possibility of developing sensitive assays based on single-molecule detection. DNA hybridization assays are ubiquitous in genomic analysis, gene expression studies, and, increasingly, diagnostics. The sensitivity and throughput of the assays have recently been improved through the introduction of DNA arrays and the development of several new sensitive detection techniques. These include molecular beacons (3–6), nanoparticle composites (7–10), surface plasmon resonance (11, 12), fiber-optic arrays (13–16), and conductivity/capacitance measurements (17, 18). The most widely used detection methods rely on labeling target DNA, usually by fluorescent dyes. However, the resulting sensitivity limits the range of applications, specifically for DNA arrays where small populations of cells are to be analyzed. Thus improved sensitivity would be valuable.
With our method, we implement a totally different concept, which is to detect hybridization through the conformational change induced in the probe; this eliminates the need to label the target. So far, this strategy has been successfully implemented only with the molecular beacons but not for the detection of single molecules. Here we demonstrate a micromechanical method that exploits a conformational change in a single-probe molecule to detect hybridization of a single target. The method is compatible with microfluidics, thus it can form a platform for sensitive DNA-array-based assays.
A single-stranded (ss)DNA oligomer such as the probe in our experiment (≈60 bases long) is a flexible polymer (persistence length lp ≈ 1.6 nm or three to four bases), which assumes random coil conformations; the corresponding ds molecule, however, is stiff (lp ≈ 50 nm, contour length ≈ 20 nm for a 60 mer). The ss can be stretched well beyond the contour length of the corresponding ds, if a sufficient force is applied at the ends. Such force-extension curves have been measured for single-micrometer-long DNA molecules (19–22); when plotted vs. the extension E relative to the ds contour length, data for DNA of different lengths fall on the same curve. If hairpins cannot form, e.g., under denaturing conditions (21), or in the case of this experiment because of sequence design, at a force of ≈3 pN, the ss is stretched to a relative extension E = 1 (see figure 4 in ref. 21). Consider a ss under tensions >3 pN, therefore E > 1. On hybridization, the end-to-end distance (EED) for this molecule will shorten. On the other hand, if the tension was <3 pN (E < 1), then on hybridization, the EED will increase (Fig. 1a). In the experiments, we observe both scenarios.
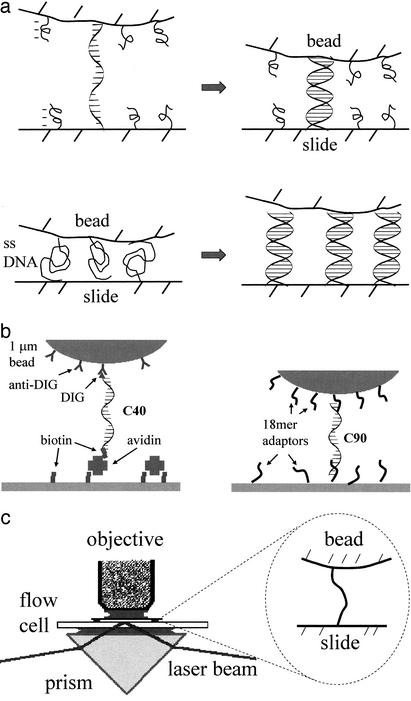
Fig. 1.
(a Upper) Detection of single hybridization events. A single tether is kept under tension by the repulsive bead–slide interaction due to the negatively charged polymers at the surfaces; on hybridization, the tether shortens. (Lower) The opposite case of many tethers, each under weak tension and therefore closer to a coiled state. On hybridization, the-end-to-end distance of the tethers increases. (b) Two schemes used to tether 1-μm-diameter beads to the slide surface through a probe oligomer. There is also a BSA monolayer on the surfaces (not shown). For some experiements, we hybridize to the free adaptors on the bead a ss 75 mer (not shown) unrelated to C90, to increase repulsion. (c) Schematics of the optical setup: the He-Ne laser beam is guided through the prism to create an evanescent wave at the bottom of the flow cell; light scattered by a single bead is collected through the objective.
A micrometer-size polystyrene bead is tethered to the surface of a microscope slide by a single DNA oligomer (the probe). A repulsive force between bead and slide stretches the tether; the origin of the force is negatively charged polymers on the surfaces. Hybridization of the target to the probe shortens the molecular tether, pulling the bead closer to the slide (Fig. 1a Upper). The bead-slide separation is monitored with subnanometer resolution by evanescent wave scattering, so the hybridization event can be detected.
With many (>10) DNA tethers holding the bead, the bead-slide repulsive force is not strong enough to stretch the ss tethers. Then hybridization with the target lengthens the molecular tethers, pushing the bead away from the slide (Fig. 1a Lower).
Our results demonstrate the label-free detection of single-hybridization events. We have detected a specific unlabeled target sequence at a concentration of 2 nM, in a total volume of 80 μl, and in the presence of 50-fold excess concentration of unrelated oligomers. Because the signal is inherently independent of target concentration and amount, very low detection limits seem possible. In addition, we believe the method can be used to monitor other kinds of conformational changes in DNA, such as are induced by protein binding.
Materials and Methods
Flow cells were constructed with a microscope slide and cover glass separated by 75-μm-thick spacers and glued together; typical cell volume was 80 μl. Slides were washed with soap and water in an ultrasound bath, rinsed, cleaned with ``piranha solution'' (5 parts water/1 part H2O2/1 part H2SO4) at 60°C for 15 min, rinsed, silanized with AquaSil (Pierce) for 15 min, rinsed, and baked for at least 30 min at 100°C. Some experiments were also performed with nonsilanized slides.
Preparation of Tethered Beads. DNA oligonucleotides were purchased from Operon Technologies (Alameda, CA), HPLC purified. The experiments were performed on beads tethered to the bottom of the flow cell (formed by the upper surface of the slide). In one scheme (I) (Fig. 1b Left), the probe was a 40 mer (C40) modified with digoxigenin (DIG) at one end and biotin at the other end. Amino-modified 1-μm-diameter polystyrene beads (Polysciences) were functionalized with anti-DIG by incubation in a 8% solution of gluteraldehyde (in PBS) followed by coupling of anti-DIG (Fab fragment, Roche Applied Sciences), blocking by BSA, and coupling to C40.
In scheme II (Fig. 1b Right), the 90-mer probe C90 was attached to the bead and slide through adaptor oligomers 18BIOT-B and -G. One-micrometer-diameter polystyrene beads functionalized with streptavidin (Sigma) were incubated with 18BIOT-B (0.1 pmol/microliter in PBS) overnight. The batch was then divided into several aliquots; for multiple-tether studies, C90 was added in the ratio of 103 oligos per bead; for single-tether studies, the ratio was 5 oligos per bead, or alternatively a mixture in the ratio 1:100 of C90 and an unrelated 75 mer lacking the part complementary to the adaptor oligomer on the slide. The result is a charged polymer layer providing a repulsive force between bead and slide. Finally, beads were blocked with excess biotin.
The surface of the flow cell was functionalized by incubating with the following solutions: biotinylated BSA (Sigma) and BSA (fatty-acid free, Sigma) in the ratio 1:100, (BSA) = 5 mg/ml, in PBS, pH 6, overnight; neutravidin (Pierce) 0.1 mg/ml for >4 h. For scheme II, biotinylated adaptor oligomer 18BIOT-G was introduced (0.1 pmol/microliter, >4 h) after the neutravidin step.
Several controls were performed on various aspects of these constructions. Hybridization properties of the oligomers were checked by gel electrophoresis. The specific coupling of the adaptor oligomers to the surfaces was checked by fluorescence microscopy. Specific attachment of the beads through the DNA tethers was checked with control beads lacking the tethers and by cutting off tethered beads using a restriction enzyme.
Optical Setup. The principle of the measurement is to create an evanescent optical wave at the glass–solution interface where the beads are tethered. A bead illuminated by this evanescent wave scatters some light. Because the intensity of the evanescent wave decreases (exponentially) with the distance from the interface, the closer a bead is to the interface, the higher the scattered intensity. Thus measuring the scattered intensity yields a measurement of the distance between the bead and the interface: I = Ic exp(–h/δ), where I is the scattered intensity, Ic the intensity at contact, h the separation between the bead and the slide, and δ the penetration depth of the evanescent wave (δ = 86 nm in our setup). Therefore a displacement of the bead can be measured as: h2–h1 = Δh = δ ln (I2/I1) (1, 23–25). This relation has been shown experimentally to remain valid down to contact (24). The penetration depth δ is calculated from the incidence angle and the refractive indexes; however, we also perform an independent calibration of the displacement measurements (Δh) by observing the vertical motion of a free (untethered) bead and comparing the gravitational potential thus obtained to the weight of the bead (23, 26, 27). The absolute bead-slide separation can also be calibrated by collapsing the bead on the slide surface at the end of the measurement to obtain the contact intensity.
The optical setup is simple. The flow cell is optically coupled to a Dove prism through immersion oil (Fig. 1c). The beam from a 20 mW He-Ne laser is steered through the prism to create an evanescent wave at the bottom of the flow chamber. Light scattered by a single bead is collected through a microscope objective (×100, numerical aperture 1.3, oil immersed, Leitz) and focused on a photodiode mounted on a trinocular tube. The signal is recovered through phase-sensitive detection: before entering the prism, the beam is chopped (≈1 kHz) and a portion split into a reference detector. Signal and reference are mixed in a lock-in amplifier (Stanford Research, Sunnyvale, CA) and the output acquired by a computer.
Experimental Procedure. A suspension of beads in buffer TST100 (Tris 20 mM/NaCl 100 mM/Tween 20 μM, pH 8) is introduced in the flow cell. After ≈1 h, some beads have tethered to the bottom and are visible with evanescent wave illumination. A single bead (which appears as a bright diffraction pattern against a dark background) is brought into the field of view of the photodiode. The vertical fluctuations of the bead are monitored for some time, then the hybridization buffer (TST100 for most experiments) containing, as a control, an unrelated 60 mer at a concentration of 100 nM is introduced. After some time, the same solution with the added target oligomers is introduced.
Results
Preparation of Tethered Beads. A colloidal particle near a surface experiences forces that are described by the so-called Derjaguin–Landau–Verwey–Overbeek interaction potential, a sum of Van der Waals attraction and electrostatic repulsion, if the surfaces have same sign charge. This interaction potential has a minimum at contact, but there is an electrostatic barrier that may prevent the particle from sticking to the surface. Depending on the surface charge and ionic strength, the potential may have a secondary minimum beyond the barrier, typically at a separation of some ≈10 nm, or it may be completely repulsive (28).
Previously we have shown (1, 25) that it is possible to attach a micrometer-size bead to a microscope slide through a single-nanometer-size molecular contact (e.g., a globular protein), while the bead is effectively separated from the surface by the potential barrier. Furthermore, from the thermal motion of the bead, one can reconstruct the potential and thus the forces on the bead and the molecular contact. In the present case, the Van der Waals attractive force is small [e.g., for a 1-μm-diameter bead, FVdW ≈ 0.7 pN at a separation h = 20 nm, using the Hamaker constant measured in (27): A = 1.0 × 10–14 ergs] compared with the elastic response of the DNA tether (Fel ≈ 3 pN for a ss 60 mer at a relative extension of 1; see below); thus, the restoring force that keeps the bead close to the surface is characteristic of the tether. We exploit this circumstance in the data analysis (Fig. 3 b and c).
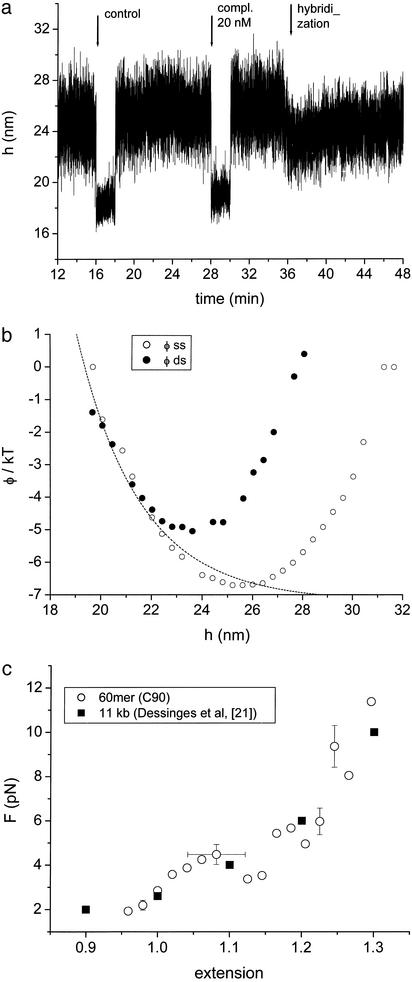
Fig. 3.
(a) Signature of a single hybridization event (t ≈ 36). The bead is tethered by a 90 mer. For 16 < t < 18 and 28 < t < 30, a flow is switched on to introduce first a control (60 mer unrelated to the probe, 100 nM) and then the target (60 mer complementary to the probe, 20 nM); during the flows, the bead is pushed closer to the slide by the drag force. Some time after introducing the target, a single hybridization event (t ≈ 36 min) pulls the bead toward the surface. (b) Interaction potentials obtained from the time trace in a; ϕss (open circles) is obtained by using part of the time series before the hybridization event (t < 36 in a, and it represents the bead held by a ss tether; ϕds is calculated from the time trace after hybridization (t > 36), representing the bead held by a ds tether. The line is an exponential function describing the repulsive part of the potentials. (c) Force-extension curve obtained from b (and including data from two more experiments): F = dϕss/dh, E = relative extension. The errors on F are statistical and of order 10%; the determination of E for each data set is subject to a systematic error indicated by the error bars. The filled squares are data from figure 4 of Dessinges et al. (21) obtained with DNA, which is 200 times longer.
We successfully used different strategies to tether 1-μm-diameter beads to the slide through a probe oligomer (see Materials and Methods). In the first strategy, a 40-mer probe (C40, Fig. 1b) was coupled through DIG–antiDIG and biotin–avidin links; negatively charged BSA layers on the surfaces provide the repulsive force that stretches the single tether.
In a second strategy, 18-mer ``adaptors'' were used to couple the 90-mer probe (C90), which had a sequence of 15 bases at the two ends complementary to the two adaptors (Fig. 1b). For the single-tether measurements, we used an unrelated 78-mer coupled to the bead to form a charged polymer layer providing a repulsive force between bead and slide, keeping the tether under tension (Fig. 1a Upper). To have a high probability of single-tether attachment, we use low probe concentration on the beads (nominally ≈10 molecules/bead).
To obtain the opposite case, where the tethers are in a random coil conformation (low tension), we prepare beads attached by many tethers (nominally ≈103 probe molecules/bead, resulting in an estimated number of tethers >10), and we do not add the 78-mer polymer ``brush'' on the bead (Fig. 1a Lower).
The slide forms the bottom of a flow cell, placed in an evanescent wave-scattering apparatus (Fig. 1c). The intensity of light scattered by a single bead provides a measurement of the displacement of the bead relative to the slide with subnanometer resolution (1, 25). The absolute bead-slide separation is obtained by measuring the contact intensity, Ic. This was done for the data of Fig. 5, whereas for Figs. 2,3,4, we used an average value of Ic to set the absolute vertical scale.
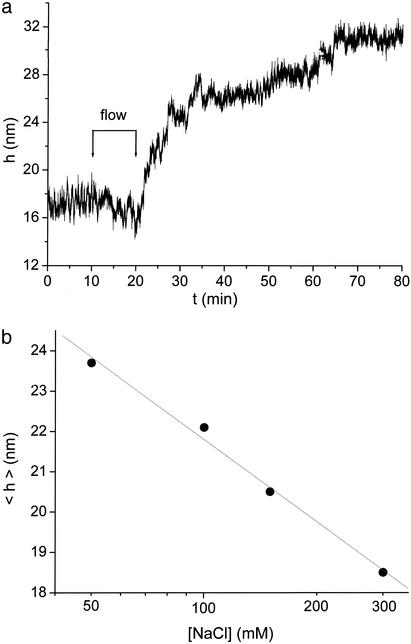
Fig. 5.
(a) Effect of surface charges; the displacement of a bead when charges are added to its surface. The streptavidin-functionalized bead is held by several ssDNA tethers; when biotinylated DNA oligomers (18 mer) are added to the solution (10 < t < 20), they progressively bind to the bead. The bead moves away from the slide surface as the surface charge builds up. (b) Effect of ionic strength. The average position < h > of a bead tethered to the slide surface (by several ss 60 mer) depends on the salt concentration. Increasing salt screens the electrostatic repulsion so the bead moves closer to the slide. The horizontal scale is logarithmic, and the line is a linear fit to the data.
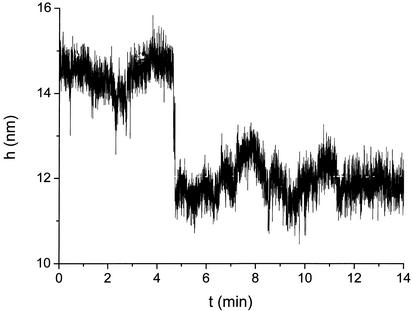
Fig. 2.
Relative bead–slide separation h, in nanometers, measured in the course of time by evanescent wave scattering. The bead is tethered by a 40 mer; a single hybridization event at t ≈ 4.7 with a complementary 30 mer pulls the bead ≈3 nm closer to the surface. Target concentration was 500 nM. The absolute h is not measured directly; it corresponds to an average value of the contact intensity Ic, determined separately.
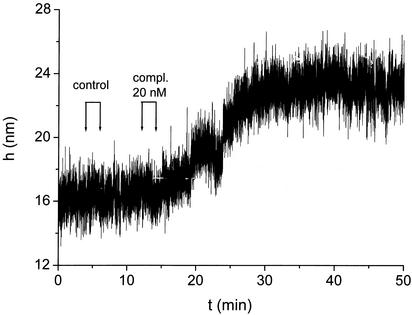
Fig. 4.
The stiffening of initially coiled tethers on hybridization displaces the bead away from the slide. The bead is held by multiple tethers (see Fig. 1a Lower); thus vertical fluctuations are smaller than in Fig. 3a. With the complementary present, the tethers hybridize and stretch out, pushing the bead away from the surface. The process is gradual in time, because several tethers are involved.
Single Tether, Stretched. In this case, we are able to detect a single hybridization event. If the ss tether is initially stretched to an end-to-end distance larger than the contour length of the corresponding ds, then on hybridization, the bead will be pulled closer to the slide. Here transient hairpins cannot form, due to sequence design and short length; then the required tension is ≥3 pN (21). Experiments were conducted as follows. Monitoring the behavior of a tethered bead (fluctuations, response to a flow) we can select with high probability beads that are correctly attached and do not show significant nonspecific sticking (1). First, a control is introduced in the flow cell, consisting of oligomers noncomplementary to the probe. After some time, the solution is exchanged for the same control with added target oligomers (complementary to the probe). We never observed any effects caused by the control oligomers.
Fig. 2 shows the signal obtained from the hybridization of a 30-mer target to a probe tether 40 bases long (C40). The figure shows the vertical position of a single bead in the course of time. At t ≈ 4.8 min, the hybridization event occurs, which pulls the bead toward the surface by Δh ≈ 2.7 nm. Thereafter, the bead remains in this state.
Fig. 3 shows the detection of single hybridization events where the tether is a 90 mer (C90) and the target a 60 mer complementary to the ss portion of C90. For 16 < t < 18 and 28 < t < 30, we make a flow in the cell to introduce first a control (a 60 mer unrelated to C90) and then the target (complementary to C90). For the duration of the flows, the bead is pushed closer to the slide by the drag force (1). The presence of the control has no effect; while a variable time after the complementary is introduced, a single event pulls the bead closer to the slide (t ≈ 36 in Fig. 3a). In this state, excursions away from the surface are more constrained by the ds tether. Thus in this configuration, the signature of hybridization consists of one abrupt jump, occurring some time after the target is introduced; this means that the jump is caused by a single hybridization event. This behavior was observed in nine different experiments with exactly these conditions; values of the beads' displacement ranged from 3 to 5.5 nm. In total, we obtained data showing the same effect for more than 20 experiments, performed under various conditions.
We can analyze such time traces in more detail, as follows. Using part of the time series before the hybridization event (t < 36 in Fig. 3a), we calculate the interaction potential ϕss: because the bead moves in a field of force, the probability of finding it at position h is P(h) ∝ exp(–ϕ(h)/kT); P(h) is obtained from a histogram of the time series (1, 25). ϕss reflects the interaction potential when the bead is held by the ss tether; using part of the time series after the hybridization event, we can also obtain ϕds, the potential for the bead tethered by the ds tether. These potentials are shown in Fig. 3b; the arbitrary additive constant has been chosen so that the repulsive part of the potentials coincide (the repulsion is due to the polymer layers on the surface and is the same in the two cases). The work necessary to pull the bead to its new equilibrium position, in this case, is only ≈2 kT, which is easily supplied by the free energy difference of the hybridization reaction. The attractive part of ϕss represents a region of the force-extension curve of these; Fig. 3 shows that thermal fluctuations occasionally stretch the ss beyond the ds contour length (i.e., E > 1). The tether is a 90 mer, but 15 bases at either end are hybridized to the adaptor oligos on the surfaces (Fig. 1b Right); thus the ss segment of the tether is 60 bases long. After hybridization with the 60-mer target, the contour length of this segment is 20 nm (the contour length of a 60-mer duplex). Thus we compute the relative extension for a point [h, ϕss(h)] in Fig. 3b as: E = 1 + (h – h0)/20, where h0 is the maximum excursion in the ds state (h0 = 28 in Fig. 3b; the assumption is that h0 corresponds to the maximum extension of the ds, because the ds length is << persistence length). In this way, we extract E referring not to the absolute distance h but only to the differences Δh, which we measure with high resolution. The force corresponding to the point [h, ϕss(h)] is computed from the derivative of ϕss at that point; to the right of the minimum of ϕss, this force is increasingly dominated by the tether elasticity. The force-extension curve thus obtained is shown in Fig. 3c, where the open circles are obtained from three different experiments (including the data shown in Fig. 3 a and b), and the filled squares are data from figure 4 of Dessinges et al. (21). Those data were obtained by using 11 kb of DNA and magnetic beads after chemical treatment to suppress hairpin formation; then the force-extension curve is largely independent of salt concentration (21). We compare with these conditions because our synthetic sequence is designed to minimize hairpin formation. It is noteworthy that our data agree with measurements on DNA molecules 200×longer. Moreover, figure 4 of ref. 21 demonstrates that the beads were indeed held by single tethers.
All hybridization assays presently in use employ a relatively large number of probe molecules, e.g., typically 1012 in the reaction volume of an assay based on beacons. A sufficient number of probes must be hybridized to be detectable; for example, for the beacons, this is of order 1%. In the present experiment, the entire signal comes from the hybridization of a single probe and is therefore independent of the total amount or concentration of target. There is still a limitation in the minimum concentration that is practical in terms of the on rate of hybridization. However, in a microfluidic environment, where relevant volumes are of order ≈1 nanoliter, the ≈1 nM target concentration used here corresponds to a total amount of 10–18 mol of target DNA, which should be detectable without labeling.
Multiple Tethers, Coiled. In this configuration, several tethers are pulling on the bead (estimated number >10), and we do not have the 70 mer on the surface, which was providing a relatively strong repulsion in the previous case. Each individual tether is under small tension and closer to a random coil state (relative extension E < 1); therefore on hybridization, the end-to-end distance increases, and the bead is pushed away from the surface (Fig. 4). The process is gradual in time, because it involves several tethers. However, we often can still distinguish discrete steps in the bead's vertical position (at least two are visible in Fig. 4). Thermal fluctuations are smaller than in the single tether case, reflecting a more constrained bead.
The magnitude of the bead's displacement (Δh ≈ 7 nm in Fig. 4) is different for different experiments, because it depends on the initial extension of the tethers, which in turn depends on the number of tethers and the bead-slide repulsive force. The upper limit of the expected Δh is given by comparing the rms size of a ss 60 mer in the random coil state, ≈9 nm (using the Flory theory with a persistence length lp ≈ 1.6 nm) to the contour length of the ds, ≈20 nm; thus Δhmax ≈ 11 nm. In a series of eight different experiments, we observed displacements Δh between 3 and 10 nm.
Electrostatic Effects. The repulsive force that stretches the ssDNA in the single tether configuration comes from electrostatics. To display this effect, we show in Fig. 5a a streptavidin functionalized bead tethered through several ss biotinylated 60 mer; the other end of the tethers is covalently linked to the slide surface, which is covered with DNA 18 mer. When an unrelated biotinylated 18 mer is added to the solution (such as the 18 mer adaptors of Fig. 1b), the DNA oligomers gradually bind to the bead, increasing the surface charge. Consequently, the bead moves away from the surface. A control experiment with immobilized beads shows that this is not an optical effect due to the change in dielectric properties of the bead.
Fig. 5b shows the effect of varying salt concentration in a configuration where charged polymers on the surface provide a repulsive force; the bead is tethered by several ssDNA strands (60 mer) covalently linked to the surfaces. We plot the average position of the bead < h > vs. salt concentration. Increasing salt screens the electrostatic repulsion, thus the bead moves closer to the slide surface; < h > varies approximately logarithmically with [NaCl].
Discussion
We have shown that the method presented here can detect nanometer-scale conformational changes of a single 10- to 30-nm-long DNA oligonucleotide. We have applied the technique to the detection of single hybridization events, through the contour length shortening of the probe oligomer. Mechanical manipulations of single DNA molecules have been performed previously, but at larger scales (λ-DNA, ≈15 μm long) (19, 20, 29–34). Nanometer-scale conformational changes of single molecules have been observed by FRET (2) and atomic-force microscopy (AFM) (35). However, the method described here can detect conformational motion between parts of a molecule, which are beyond the useful range for FRET (>10 nm); this is the case for the end-to-end distance of our ≈30-nm-long oligomers. Compared with the AFM, the advantage of the method is that it is technically simpler and suitable for high-throughput applications.
The size of our probe (40–90 bases) is adapted to hybridization studies; because single-hybridization events are detected, the method holds the promise of a very low detection limit in terms of total amount of target. We therefore envision applications as a platform for sensitive DNA arrays. A further advantageous aspect of the method is that it delivers a quantitative measurement of probe shortening, rather than detecting only the presence of the target. We expect that hybridization of partial homologues can be discriminated on this basis.
Our further steps will include moving from detection alone to measuring the amount of target, which will involve collecting the signal from many smaller beads. Other aspects to be improved include: more stable (covalent) attachment of the probe oligomers to the surfaces, better surface chemistry to minimize nonspecific sticking of the beads, perhaps the possibility of controlling bead–slide interactions, and hybridization rates through an electric field (12).
Finally, we believe this method can address a wider range of applications, in particular the detection of conformational changes in DNA oligomers induced by protein binding.
Acknowledgments
We thank David Bensimon for illuminating discussions and the referees for constructive comments. This work was supported by National Science Foundation Grant DMR-0105903.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FRET, fluorescence resonance energy transfer; ss, single strand; ds, double strand; DIG, digoxigenin.
References
- 1.Zocchi, G. (2001) Biophys. J. 81, 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhuang, X., Bartley, L. E., Babcock, H. P., Russell, R., Ha, T., Herschlag, D. & Chu, S. (2000) Science 288, 2048–2051. [DOI] [PubMed] [Google Scholar]
- 3.Tyagi, S. & Kramer, F. R. (1996) Nat. Biotechnol. 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi, S., Bratu, D. P. & Kramer, F. R. (1997. Nat. Biotechnol. 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet, G., Tyagi, S., Libchaber, A. & Kramer, F. R. (1999) Proc. Natl. Acad. Sci. USA 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marras, S. A. E., Kramer, F. R. & Tyagi, S. (1999) Genet. Anal. Biomol. E 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 7.Elghanian, R., Storhoff, J. J., Mucic, R. C., Letsinger, R. L. & Mirkin, C. A. (1997) Science 277, 1078–1081. [DOI] [PubMed] [Google Scholar]
- 8.Andrew, T. A., Mirkin, C. A. & Letsinger, R. L. (2000) Science 289, 1757–1760. [DOI] [PubMed] [Google Scholar]
- 9.Schultz, S., Smith, D. R., Mock, J. J. & Schultz, D. A. (2000) Proc. Natl. Acad. Sci. USA 97, 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubertret, B., Calame, M. & Libchaber, A. (2001. Nat. Biotechnol. 19, 365–370. [DOI] [PubMed] [Google Scholar]
- 11.Peterlinz, K. A., Georgiadis, R. M., Herne, T. M. & Tarlov, M. J. (1997) J. Am. Chem. Soc. 119, 3401–3402. [Google Scholar]
- 12.Heaton, R. J., Peterson, A. W. & Georgiadis, R. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3701–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stimpson, D. I., Hoijer, J. V., Hsieh, W., Jou, C., Gordon, J., Theriault, T., Gamble, R. & Baldeschwieler, J. D. (1995) Proc. Natl. Acad. Sci. USA 92, 6379–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, J. A., Boles, T. C., Adams, C. P. & Walt, D. R. A. (1996) Nat. Biotechnol. 14, 1681–1684. [DOI] [PubMed] [Google Scholar]
- 15.Steemers, F. J., Ferguson, J. A. & Walt, D. R. (2000) Nat. Biotechnol. 18, 91–94. [DOI] [PubMed] [Google Scholar]
- 16.Yeakley, J. M., Fan, J. B., Doucet, D., Luo, L., Wickham, E., Ye, Z., Chee, M. S. & Fu, X. D. (2002) Nat. Biotechnol. 20, 353–358. [DOI] [PubMed] [Google Scholar]
- 17.Patolsky, F., Lichtenstein, A. & Willner, I. (2001) Nat. Biotechnol. 19, 253–257. [DOI] [PubMed] [Google Scholar]
- 18.Park, S. J., Taton, T. A. & Mirkin, C. A. (2002) Science 295, 1503–1506. [DOI] [PubMed] [Google Scholar]
- 19.Cluzel, P., Lebrun, A., Heller, C., Lavery, R., Viovy, J. L., Chatenay, D. & Caron, F. (1996) Science 271, 792–794. [DOI] [PubMed] [Google Scholar]
- 20.Smith, S. B. & Cui, C. (1996) Science 271, 795–799. [DOI] [PubMed] [Google Scholar]
- 21.Dessinges, M. N., Maier, B., Zhang, Y., Peliti, M., Bensimon, D. & Croquette, V. (2002) Phys. Rev. Lett. 89, 248102. [DOI] [PubMed] [Google Scholar]
- 22.Strick, T., Allemand, J., Croquette, V. & Bensimon, D. (2000) Prog. Biophys. Mol. Biol. 74, 115–140. [DOI] [PubMed] [Google Scholar]
- 23.Prieve, D. C. & Frej, N. A. (1990) Langmuir 6, 396–403. [Google Scholar]
- 24.Prieve, D. C. & Walz, J. Y. (1993) Appl. Opt. 32, 1629–1641. [DOI] [PubMed] [Google Scholar]
- 25.Singh-Zocchi, M., Andreasen, A. & Zocchi, G. (1999) Proc. Natl. Acad. Sci. USA 96, 6711–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebert, R. B. & Prieve, D. C. (1995) Biophys. J. 69, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh-Zocchi, M., Hanne, J. & Zocchi, G. (2002) Biophys. J. 83, 2211–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Israelachvili, J. (1991) Intermolecular and Surface Forces (Academic, London).
- 29.Smith, S. B., Finzi, L. & Bustamante, C. (1992) Science 258, 1122–1126. [DOI] [PubMed] [Google Scholar]
- 30.Essevaz-Roulet, B., Bockelmann, U. & Heslot, F. (1997) Proc. Natl. Acad. Sci. USA 94, 11935–11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rief, M., Clausen-Schaumann, H. & Gaub, H. (1999) Nat. Struct. Biol. 6, 346–349. [DOI] [PubMed] [Google Scholar]
- 32.Wuite, G. J., Smith, S. B., Young, M., Keller, D. & Bustamante, C. (2000) Nature 404, 103–106. [DOI] [PubMed] [Google Scholar]
- 33.Maier, B., Bensimon, D. & Croquette, V. (2000) Proc. Natl. Acad. Sci. USA 97, 12002–12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strick, T. R., Croquette, V. & Bensimon, D. (2000) Nature 404, 901–904. [DOI] [PubMed] [Google Scholar]
- 35.Radmacher, M., Fritz, M., Hansma, H. G. & Hansma, P. K. (1994) Science 265, 1577–1579. [DOI] [PubMed] [Google Scholar]