Abstract
Among the bilateral animals, a centralized nervous system is found in both the deuterostome and protostome. To address the question of whether the CNS was derived from a common ancestor of deuterostomes and protostomes, it is essential to know kinds of genes existed in the CNS of the putative common ancestor and to trace the evolutionary divergence of genes expressed in the CNS. To answer these questions, we took a comparative approach using different species, particularly focusing on one of the lower bilateral animals, the planarian (Platyhelminthes, Tricladida), which is known to possess a CNS. We determined the nucleotide sequence of ESTs from the head portion of planarians, obtaining 3,101 nonredundant EST clones. As a result of homology searches, we found that 116 clones had significant similarity to known genes related to the nervous system. Here, we compared these 116 planarian EST clones with all ORFs of the complete genome sequences of the human, fruit fly, and nematode, and showed that >95% of these 116 nervous system-related genes, including genes involved in brain or neural morphogenesis, were commonly shared among these organisms, thus providing evidence at the molecular level for the existence of a common ancestral CNS. Interestingly, we found that ≈30% of planarian nervous system-related genes had homologous sequences in Arabidopsis and yeast, which do not possess a nervous system. This implies that the origin of nervous system-related genes greatly predated the emergence of the nervous system, and that these genes might have been recruited toward the nervous system.
For understanding the evolutionary divergence of metazoans, the CNS is a key organ because of its highly organized developmental patterning system. Comparative studies of the CNS should yield valuable insights into its evolutionary history. Among bilateral animals, although both of deuterostomes and protostomes possess a centralized nervous system (1), comparative embryology has suggested that their CNSs should have arisen from different origins, because the deuterostome CNS is formed from the dorsal neural tube, whereas that of protostomes develops from the ventral ectoderm (2). Recently discovered molecular evidence, however, has provided support for the opposite conclusion in this long-standing problem of evolutionary morphology, namely the conserved expression patterns of many regulatory genes (3). To answer the question of whether both of these types of CNS were derived from a common ancestor, it is essential to gain more extensive knowledge of genes in the CNS of more basal groups of bilaterians and to compare these genes with those of other organisms.
Planarians (Platyhelminthes, Tricladida) possess a CNS with a simple, primitive morphology (4, 5). The planarian CNS is composed of the cephalic ganglion and a pair of ventral nerve cords. The cephalic ganglion forms an inverted U-shaped structure with nine branches connecting to the sensory organs on each side (6, 7). The planarian cephalic ganglion exhibits many morphological features similar to the vertebrate CNS, such as multipolarized neurons (8). Moreover, planarian homologs of otx (or orthodenticle) are expressed specifically in the region of the cephalic ganglion (9). Therefore, the planarian cephalic ganglion can be considered to be a primitive brain.
Planarians belong to the phylum Platyhelminthes. Although the phylogenetic position of Platyhelminthes has not yet been completely established, Platyhelminthes is positioned at or near the root of bilateral animals, as determined based on its morphology and developmental biology (1, 10). Recently, Aguinaldo et al. (11) proposed a new taxonomy based on molecular analyses of 18S rRNA sequences; in this taxonomy, bilateral animals are classified into three groups: Deuterostomia, Ecdysozoa, and Lophotrochozoa. The traditional protozoan group is separated into Ecdysozoa and Lophotrochozoa. In their phylogenetic tree, Platyhelminthes is positioned near the root of the Lophotrochozoa. Although there are some contradictions between the molecular and morphological evidence (12), there is no doubt that planarians are among the descendants of early bilateral animals. For these reasons, planarians are suitable animals for studying the evolutionary history of the CNS, in particular, that of the brain.
Thus, in the present study, we identified nervous system-related (NS-related) genes of planarians by sequencing ESTs and took the approach of comparing planarian ESTs to genomic sequences of other organisms.
Materials and Methods
Animals. We used a clonal strain of planarian, Dugesia japonica, which was established previously by K. Watanabe (Himeji Technical Institute, Hyogo, Japan). Intact worms were cultured in autoclaved tap water at 22°C.
Construction of a cDNA Library. Planarians of length 5–7 mm that had starved for 7–10 days were used. More than 500 planarians were cut at the prepharyngeal region under a phase-contrast microscope, and their head portions were collected as the source of RNA that was used to produce a cDNA library that was constructed in lambda ZAP II (Stratagene) using oligo(dT) primers. Then clones containing inserts were selected for nucleotide sequencing. We did not amplify the cDNA by the PCR method, so this library was expected to reflect the mRNA frequency in vivo.
Sequencing and Analyses of the cDNA Clones. Purified plasmid DNA was sequenced by using a 377, 310, or 3700 ABI sequencer and a Big-Dye terminator sequencing kit following the manufacturer's instructions (Applied Biosystems). The sequences were analyzed as follows, using our in-house EST-analysis system, the FinEST (K.M. K.I., and T.G., unpublished data). The sequence was used 500 bp long at the maximum, and clones <100 bp in length or with >5% of the nucleotides in their sequence not determined were omitted from the following analysis. The redundancy of sequences was checked by using the blastn program (13) with the criteria of ≥90% identity between ≥100-bp overlapping regions, and we thereby obtained a nonredundant data set. These nonredundant sequences were then searched against the available protein databases obtained by translation from the DDBJ/EMBL/GenBank nucleotide database using blastx programs. Here, we used the criterion of E value <10-4 for identifying homologous sequences based on these alignments.
Identification of NS-Related Genes. Genes homologous to NS-related genes were identified. For this process, we defined the NS-related genes as the homologous genes in the following categories: (A) neurotransmission, (B) neural network, (C) brain morphogenesis/neural differentiation, (D) sensory system, and (E) others. In particular, category E included clones that showed similarity to genes that are functionally unidentified but are known to be specifically or abundantly expressed in the brain or the nervous system, such as “adult brain protein 239.”
Whole-Mount in Situ Hybridization. Digoxigenin-labeled RNA probes were prepared according to the manufacturer's instructions (Boehringer Mannheim), using clones ID 04307_HH, 00517_HH, 01242_HH, 01791_HH, and 02467_HH cDNA as templates. We chose these clones as representative clones from the five categories noted above. Whole-mount in situ hybridization was performed as described by Umesono et al. (8)
Comparison of Planarian ESTs with Genes in Other Species. For the planarian clones similar to NS-related proteins, comparisons were performed against all translated ORFs in the complete genome sequences of Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, and Saccharomyces cerevisiae (obtained from EMBL proteome database, www.ebi.ac.uk/proteome/) by using blastx. In addition, we conducted an extensive search against H. sapiens, D. melanogaster, and C. elegans. This search was designated a “boomerang search” and done using the following two procedures: (i) a planarian EST was used as a query, and was searched against the public database with E value ≤10-4. (ii) Then, the highest match sequence was used as a query, and was searched against each genomic database with E value ≤10-4. We used this procedure, with some manual modification, to confirm the existence of similar sequences in the indicated bilateral animals.
Phylogenetic Analysis. Putative sequences homologous of planarian clone 00382_HN (homologous to the calcium/calmodulin-dependent kinase type II δ chain (CaMKII δ) of Rattus norvegius) in other species were obtained by a homology search using the blastx program. The sequences homologous to the CaMKII family in bilateral animals were subjected to the phylogenetic analysis, and all of the clones analyzed from A. thaliana belonged to the calcium dependent protein kinase family, according to the sequence annotation. After the identification of these homologous sequences, multiple alignment of the deduced amino acid sequences was performed by using the clustalx program (14) with the default parameters. Gapped regions were excluded for the distance calculation. We used a neighbor-joining method (15) for the tree construction.
Results
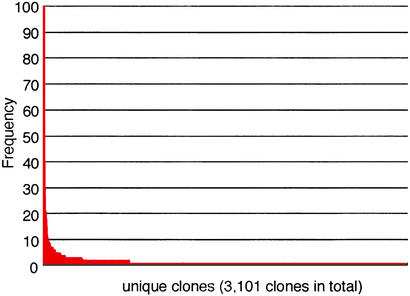
An Overview of ESTs Derived from the Planarian Head. We obtained 5,433 5′ ESTs, after removal of clones with no inserts, from a planarian head cDNA library. Taking into account the frequency of occurrence of different clones in these 5′ ESTs, we obtained 3,101 nonredundant clones (Table 1). The frequency distribution of nonredundant clones is shown in Fig. 1, based on the number of nonredundant clones and the frequency of each clone obtained. A majority (77%) of the clones were expressed as singletons. Among all of the nonredundant clones, 1,385 clones were homologous to functionally known genes in other organisms (groups a and b in Table 1).
Table 1. Summary of ESTs from the planarian head.
| Group | Category | No. of clones (%) | No. of nonredundant clones (%) |
|---|---|---|---|
| a | Recognized protein coding sequences | 2,779 (51.2) | 1,190 (38.4) |
| b | Putative but recognized coding sequences | 392 (7.2) | 195 (6.3) |
| c | Unidentified sequence (EST, clone ID, unknown or hypothetical protein) | 661 (12.2) | 416 (13.4) |
| d | No significant similarity to the protein database | 1,601 (29.5) | 1,300 (41.9) |
| Total | 5,433 | 3,101 |
Fig. 1.
The gene expression profile in the planarian head. For each nonredundant clone, the frequency of occurrence was plotted. Highly expressed clones included clones homologous to β actin (100 clones), alpha actin (77 clones), alpha tubulin (70 clones), β tubulin (57 clones), and elongation factor-1 α (46 clones). A majority (77%) of the clones were expressed as singletons.
Search for NS-Related Genes in Planarians. To identify the gene set related to the CNS in planarians, we searched for NS-related genes. Among all of the clones corresponding to functionally known genes, we successfully identified 116 clones that exhibited significant similarity to NS-related genes previously characterized in other organisms (Table 2 and Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). Classifying these 116 clones, we found that these NS-related clones could be separated into the following five functional groups: (A) 42 clones were related to neurotransmission, including genes related to neurotransmitters, receptor/channels, synaptic vesicles and their transport, (B) 33 clones were related to the neural network, including the Ig cellular adhesion molecule family, the cadherin family and axon guidance, (C) 21 clones showed homology with genes for brain morphogenesis/neural differentiation such as the bone morphogenic protein (BMP) cascade, the Wnt cascade, the fibroblast growth factor (FGF) cascade, and the Notch cascade, (D) 11 clones were related to sensory systems, including the photosensory, chemosensory and mechanosensory systems, and (E) 9 clones were related to others proteins, such as brain protein AB239.
Table 2. NS-related genes in the planarian ESTs: representative clones.
| Clone ID | Frequency | GenBank accession no. | Protein with the best E value | E value |
|---|---|---|---|---|
| A. Neurotransmission (42) | ||||
| 00382_HN | 1 | NP_036651.1 | Calcium/calmodulin-dependent protein kinase type II δ chain (Rattus norvegicus) | 1.00E-73 |
| 01517_HH | 1 | BAA90484.1 | High-affinity choline transporter CHT1 (R. norvegicus) | 3.00E-40 |
| 04307_HH | 1 | AAD32697.1 | Putative nicotinic acetylcholine receptor α 7-1 subunit (Heliothis virescens) | 9.00E-19 |
| 06065_HH | 1 | NP_004701.1 | Synaptogyrin 2 (H. sapiens) | 1.00E-31 |
| 06173_HH | 1 | Q18179 | Putative neuropeptide Y receptor (C. elegans) | 2.00E-19 |
| B. Neural network (33) | ||||
| 00944_HH | 1 | P16170 | Neural cell adhesion molecule precursor (Xenopus laevis) | 4.00E-05 |
| 05189_HH | 1 | AAC83376.1 | Netrin precursor (Hirudo medicinalis) | 2.00E-48 |
| 06563_HH | 1 | NP_061241.1 | SemaF cytoplasmic domain associated protein 1 (Mus musculus) | 7.00E-23 |
| 06717_HH | 1 | AAF71926.1 | Dscam (D. melanogaster) | 8.00E-05 |
| 06802_HH | 1 | A46194 | Neurofilament protein NF-220, high-molecular-weight spliced form (Loligo pealei) | 8.00E-19 |
| C. Brain morphogenesis-neural differentiation (21) | ||||
| 00868_HN | 1 | AAB39211.1 | Mutant cysteine-rich FGF receptor (Gallus gallus) | 1.00E-15 |
| 01362_HH | 1 | AAF21645.1 | Frizzled homolog (Danio rerio) | 7.00E-06 |
| 01443_HH | 1 | BAA92185.1 | β-catenin (Ciona intestinalis) | 7.00E-08 |
| 03592_HH | 1 | AAD43133.1 | Noggin 2 (Danio rerio) | 3.00E-14 |
| 05855_HH | 1 | BAA22437.1 | BMP receptor (X. laevis) | 7.00E-32 |
| D. Sensory system (11) | ||||
| 00251_HH | 1 | NP_004032.1 | β-arrestin 1A (H. sapiens) | 2.00E-32 |
| 00287_HN | 2 | Q01062 | cGMP-dependent 3′,5′-cyclic phosphodiesterase (R. norvegicus) | 4.00E-47 |
| 01791_HH | 1 | AAC67569.1 | Cone transducin α subunit (Ambystoma tigrinum) | 6.00E-34 |
| 01997_HH | 1 | NP_038559.1 | Transducin beta chain 4 (M. musculus) | 8.00E-31 |
| 06472_HH | 1 | P24603 | Rhodopsin (Loligo forbesi) | 6.00E-08 |
| E. Others (9) | ||||
| 01279_HH | 1 | JE0209 | Brain-specific angiogenesis inhibitor-associated protein 1 (H. sapiens) | 1.00E-21 |
| 01704_HH | 7 | JC5759 | Brain-specific serine proteinase (M. musculus) | 7.00E-19 |
| 02467_HH | 1 | 015442 | Adult brain protein 239 (H. sapiens) | 4.00E-39 |
| 05177_HH | 1 | AAF14284.1 | Neural polypyrimidine tract binding protein (H. sapiens) | 9.00E-17 |
| 05723_HH | 1 | BAA35092.1 | Neural specific sr protein NSSR 1 (M. musculus) | 2.00E-15 |
We also conducted whole-mount in situ hybridization analysis of representative clones in all of the categories. The expression patterns of some representative clones are shown in Fig. 2. The results showed that these representative clones with significant similarity to NS-related proteins were expressed specifically in the CNS, though some other clones were not expressed specifically in the CNS or nervous system (data not shown).
Fig. 2.
Whole-mount in situ hybridization of the NS-related clones in intact planarians. (A) Clone ID: 04307_HH (homologous to the nicotinic acetylcholine receptor α 7-1 subunit). (B) Clone ID: 00517_HH (homologous to the protein tyrosine phosphatase X receptor). (C) Clone ID: 01242_HH (homologous to the lethal giant larvae). (D) Clone ID: 01791_HH (homologous to the cone transducin α subunit). (E) Clone ID: 02467_HH (homologous to the adult brain protein AB239). The letters correspond to the functional categories in Table 2.
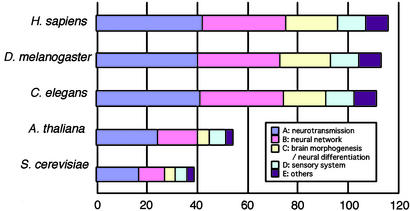
Extreme Conservation of the NS-Related Gene Set Among Bilateral Animals. As the number of species whose complete genome sequences are available has increased, it has become possible to see whether genes of interest exist or not in a given species. To study the evolutionary emergence of the gene set and the features of the CNS in various species, we took the approach of comparative genomics. Sequences homologous to the 116 NS-related planarian clones were extensively searched against all ORFs in the complete genome sequences of human, D. melanogaster and C. elegans, all of which possess a CNS (Fig. 3).
Fig. 3.
Comparisons between NS-related genes of the planarian and all ORFs in the complete genomes of S. cerevisiae, A. thaliana, C. elegans, D. melanogaster, and H. sapiens. The horizontal axis represents the number of sequences homologous to the planarian NS-related clones (116 clones in total) in the genome examined. The functional categories are indicated by different colors. C. elegans, D. melanogaster, and H. sapiens were examined by a boomerang search.
We found that 110 of the total of 116 planarian head genes (i.e., >95%) were shared by all of the bilateral animals examined (C. elegans, D. melanogaster, and humans) (Fig. 3). These clones included genes from all of the five functional categories noted above. Interestingly, all of the 116 genes examined were completely shared between humans and planarians (Fig. 3), whereas six of these clones were absent in C. elegans and/or D. melanogaster. The clones not shared by all of the species examined were shown in Table 3. Sequences homologous to sortin (planarian ID 00546_HN) and noggin (03592_HH) could not be found in either C. elegans or D. melanogaster, whereas sequence homologous to synaptophysin (05093_HH) could not be found in D. melanogaster, and sequences homologous to aFGF intracellular binding protein (02975_HH), glia maturation factor β (07031_HH), and brain platelet-activating factor acetyl hydrolase (03408_HH) could not be found in C. elegans.
Table 3. Planarian nervous system-related genes that did not have similar sequences in certain organisms.
| Clone ID | Homologous protein | E value | Category | C. elegans | D. melanogaster | H. sapiens |
|---|---|---|---|---|---|---|
| 00546_HN | Sortilin 1 (Mus musculus) | 5.00E-35 | A | - | - | * |
| 05093_HH | Synaptophysin (Bos taurus) | 9.00E-11 | A | * | - | * |
| 03592_HH | Noggin 2 (Danio rerio) | 3.00E-14 | C | - | - | * |
| 02975_HH | aFGF intracellular binding protein (M. musculus) | 2.00E-18 | C | - | * | * |
| 07031_HH | Glia maturation factor beta (Cyprinus carpio) | 6.00E-33 | C | - | * | * |
| 03408_HH | Brain platelet-activating factor acetyl hydrolase (B. taurus) | 5.00E-41 | C | - | * | * |
Asterisks indicate the existence of a similar sequence with a significant E value, and a bar indicates that no similar sequence was identified using the same criteria. Clone 00805_HH (homologous to gap junction protein pannexin (Clione limacine), E value: 4.0E-18) did not show significant similarity with any human protein (sequence with the highest E value was 1.0E-3) in our search, however, there was a report about human pannexin (GenBank accession number: BC016931 and others), so we concluded that 00805_HH existed in all of the species examined. Therefore, we excluded clone of 00805_HH from Table 3. This misidentification may have been caused by the long evolutionary distance between Clione and humans (see Materials and Methods). The categories correspond to those in Table 2
NS-Related Genes Shared by Organisms Not Possessing a CNS. To examine the evolutionary origin of the CNS-related gene set, we also compared these NS-related genes of planarians with all ORFs of the complete genomes of S. cerevisiae and A. thaliana, which do not possess a nervous system. As a result, we discovered that 30% of the 116 genes were shared between S. cerevisiae and planarians, and 37% were shared between A. thaliana and planarians (Fig. 3). We found that the functional category with the largest number of shared clones was category A, neurotransmission, which included 17 clones homologous to ORFs of S. cerevisiae and 24 clones homologous to ORFs of A. thaliana, though some genes were also found in the other categories. We then conducted a phylogenetic analysis based on these NS-related genes shared with S. cerevisiae and A. thaliana. As an example of the clones that were shared with S. cerevisiae and A. thaliana, we chose a planarian clone homologous to the CaMKII δ chain (planarian ID 00382_HN). The phylogenetic tree constructed thereby is shown in Fig. 4. CaMKII homologs in S. cerevisiae and A. thaliana were clustered in each lineage, showing that sequence divergence had occurred after the speciation.
Fig. 4.
A phylogenetic tree based on homology to clone 00382_HN (homologous to the calcium/CaMKII δ chain). Hsa, H. sapiens; Rno, R. norvegicus; Dme, D. melanogaster; Cel, C. elegans; Ath, A. thaliana; and Sce, S. cerevisiae. The genes with the highest blast score in each species are shown by bold, underlined letters. Rno (NP036651) was the gene that showed the highest blast score with clone 00382_HN at the original homology search, and is shown only by underlining. The accession numbers of the DDBJ/EMBL/GenBank database are stated in parentheses after the species name. The tree was constructed by the neighbor-joining method. Bootstrap confidence levels are shown at the node of each branch.
Discussion
Complexity of the Planarian CNS Revealed by the NS-Related Genes. The planarian NS-related genes identified here fell into various functional groups known to be important for the CNS (Tables 2 and 4). In particular, the clones in category C of brain morphogenesis/neural differentiation are known to be essential for the CNS development in higher organisms. These NS-related clones in planarians may not all be orthologous genes of functionally known NS-related genes in other organisms, some of them may instead be paralogous genes. In either case, the planarian sequences were significantly similar to those of functionally known genes in other organisms. The expression patterns of these clones as revealed by in situ hybridization (Fig. 2) showed that the CNS of planarians consists of a wide variety of subtypes of neuronal cells (16–18), suggesting that the planarian CNS has complex cytoarchitecture and that these clones will be useful as important molecular markers for further neuroanatomical research. In addition, the planarian has a greater variety of NS-related genes than we expected (Tables 2 and 4), indicating the complex structure and molecular composition of the planarian CNS.
The Evolutionary Origin of the CNS. We found that 110 of a total of 116 genes (95%) were shared among all of the bilateral animals examined (Fig. 3 and Table 3). The species examined belong to the three different groups of bilateral animals (Deuterostomia, Ecdysozoa, and Lophotrochozoa) (11) and all possess a CNS (1). The NS-related genes identified appeared to include (i) genes related to the nervous system, and (ii) genes related specifically to the centralized nervous system. Among these NS-related genes shared by the bilateral animals, we found genes essential for the CNS. These included many common genes involved in brain morphogenesis and neural network formation (functional categories B and C in Tables 2 and 4) such as genes for FGF signaling molecules, noggin, frizzled, and genes of Ig cellular adhesion molecule/cadherin family (Fig. 3, Tables 2 and 4). These genes specifically related to the CNS provide a good means of examining the evolution of the CNS. Recently, we have shown that FGF signaling may have an important role in the formation of the planarian brain, as shown by the observations that FGF receptors are strongly expressed in the brain (19) and that loss of function of a FGF receptor-related gene, nou-darake (Djndk), caused ectopic brain formation in the trunk region (20). In addition, BMP signaling is also known to have a crucial role in the CNS. Noggin is known to be an important gene in the BMP signaling pathway for the CNS development of chordate (21). Interestingly, a planarian homolog of noggin, Djnlg, was found in our ESTs and expressed in both the ventral side of the regenerating planarian and the stump region of the lateral brain (22), thus showing functional similarity between chordates and planarians. We also found by analyzing our ESTs that planarian homologs of the Wnt receptor frizzled and the Ig cellular adhesion molecule and cadherin family genes, also known to be genes with important functions in CNS formation in other organisms, were specifically expressed in the CNS (unpublished observations by C. Kobayashi and K. Takeuchi, respectively). These findings of the functional conservation of the genes involved in brain morphogenesis and neural network formation strongly support the idea that the CNSs in all bilateral animals may have been derived from a common origin.
On the other hand, the strong conservation of NS-related genes observed here implies that these genes have remained unchanged in the evolutionary lineage from the ancestral CNS in the common ancestor of bilaterians to the highly ordered human brain. It has been reported that the evolutionary rates of NS-related genes are, in general, very slow, resulting in a high degree of conservation at the sequence level (23). Thus, our finding of strong conservation (95%) of the gene composition of the NS-related genes identified here suggests that the genes related to the nervous system might have functional constraints not only on the nature of their sequences but also on the gene composition during evolution.
Our next question about the CNS evolution was how the nervous system become centralized. As we mentioned above, Djndk functions to restrict brain tissues to the head region of planarians (20). Based on this result, we speculate that Djndk is one of the key genes for the centralization of the nervous system. The emergence of the function of Djndk or other key genes might have led from the diffuse nervous system such as in Cnidaria to the centralization of nerve cells into specific portions of the body and then to the construction of a centralized nervous system early in the process of evolution of the CNS.
The Early Origin of NS-Related Genes and Evidence Supporting Possible Gene Recruitment. CaMKII is known to be a gene essential for long-term memory in animals (24). A planarian clone homologous to the CaMKII δ gene (00382_HN) was also strongly expressed in the CNS (data not shown). However, sequences homologous to 00382_HN were also found in A. thaliana and yeast, which do not possess a nervous system (Fig. 4). Also, we found many synaptic vesicle transport-related genes in A. thaliana and yeast; these genes are possibly used in the Golgi transport system in these organisms. These results imply possible functional similarity between the Golgi transport system in eukaryotic cells and the synaptic vesicle transport system in the nervous system (25).
The divergence time among A. thaliana, S. cerevisiae, and metazoans has been estimated as ≈1,700 million years ago (26). The emergence of the CNS is generally assumed to have concomitant with the emergence of bilateral animals (before the Cambrian explosion; >543 million years ago; ref. 27). Because A. thaliana and S. cerevisiae do not possess the CNS or nervous system, our results imply that the evolutionary origin of NS-related genes greatly predates the emergence of the nervous system and CNS. Based on our observations, we speculate that during evolution many genes that are functional in the nervous system or CNS may have been recruited from genes used in unicellular systems.
Gene Loss During Evolution of the CNS. As shown in Table 3, we identified six NS-related genes that were shared between planarians and human, but that must have been lost or have greatly diverged in the evolutionary lineages that branched toward C. elegans or D. melanogaster, though there are probably strong functional constraints among the NS-related genes.
Among those six genes, we will first discuss the noggin gene as an example. Noggin is an important inducer of neural tissues from the dorsal ectoderm in chordates, and acts by blocking the BMP signaling pathway (21). We failed to identify any noggin homolog in D. melanogaster and C. elegans by extensive searches using blastn/x and psi-blast. However, we found that the planarian has a sequence similar to noggin (22). On the other hand, bilateral animals possess chordin (in deuterostomes) and sog (in protostomes) (28) as antagonists of the BMP signaling pathway. These genes function in alternative pathways to that of the noggin gene. Because the planarian also has BMP and its receptor homologs (29) (Table 2), the noggin cascade may exist in planarians. These findings imply that the system of neural induction in the planarian is more similar to that in chordates than that in protostomes. No homolog of the chordin/sog gene has been found in the planarian so far. Taken together, our findings imply that there was flexibility in the usage of various gene sets in organisms during the evolution of the CNS. The transgenic experiments have shown that Xenopus noggin can function in D. melanogaster (30), though noggin has not been found so far in D. melanogaster. This suggests that a function analogous to that of noggin may play a role in the CNS in D. melanogaster.
Another example of gene loss observed here was loss of the gene for glia maturation factor β. No gene for glia maturation factor β was found in C. elegans, though a homolog of glia maturation factor β was found in Brugia malayi (31), which belongs to the same Nematoda family. This suggests that glia maturation factor β might have been lost or have greatly diverged in the lineage of C. elegans after the divergence between C. elegans and B. malayi. This result further supports our idea that some genes might have greatly diverged or been lost during the evolution of the CNS.
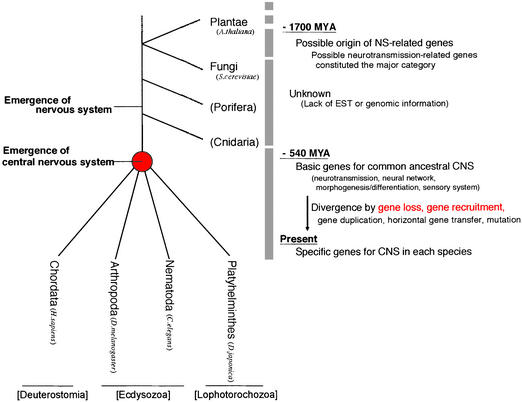
A Possible Scenario of the Evolution of the CNS from Planarian NS-Related Genes. Based on our results, we suggest a model for CNS evolution from NS-related genes (Fig. 5). We found here that NS-related genes emerged much earlier than the nervous system itself, and were subject to evolutionary forces such as gene loss or marked divergence (Table 3) and gene recruitment in addition to the traditional evolutionary forces (mutation or gene duplication) in the process of giving rise to the sophisticated CNS functions in ways specific to various species. Here we would like to point out that, as shown in Fig. 4, large-scale genetic data are lacking about the diffuse nervous systems such as in Cnidaria or much older species such as Porifera. The identification and characterization of the genes in those species should cast light on the question of how the centralization of the nervous system occurred during evolution.
Fig. 5.
A possible scenario of the process of evolution of the CNS. The phyla not analyzed are indicated in parenthesis. Based on our analysis, possible events during evolution of the NS-related genes are noted.
In planarians, powerful experimental methods, such as the single cell PCR method (K.A., unpublished data), RNA interference method (32) and DNA microarray method (18), have been developed. Moreover, we have now obtained >3,000 nonredundant EST clones of planarians (as shown in this study), and also ≈2,000 clones for genes from a closely related species (Schmidtea mediterranea) (33). By using a combination of these experimental approaches and large-scale genetic analyses, further comparative studies of planarians and other organisms should yield profound insights into the evolution of complex brain systems.
Supplementary Material
Acknowledgments
We thank Mr. Masao Agui, Ms. Tomomi Kudome-Takamatsu, Ms. Chie Iwamoto, Ms. Hiroko Ohizumi, Ms. Kyoko Terada, Ms. Akemi Mizuguchi, and Ms. Yumi Umehara-Takezawa for technical assistance in the sequence determination. We are grateful to Mr. Kiheita Konno and Mr. Toshinori Yagi (Hitachi Software Engineering Co., Tokyo) for kindly helping with our computer analysis. This research was supported by Japan Society for the Promotion of Science research fellowships (to M.N.), Special Coordination Funds for Promoting Science and Technology (to K.A.), Grants-in-Aid for Scientific Research on Priority Areas (C) (to T.G.) and (A) (to K.A.), and Grants-in-Aid for Creative Basic Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to K.I., K.A., and T.G.).
Abbreviations: FGF, fibroblast growth factor; BMP, bone morphogenic protein; CaMKII calmodulin-dependent kinase type II; NS-related, nervous system related.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. BP185095–BP191538).
References
- 1.Neilsen, C. (2001) Animal Evolution (Oxford Univ. Press, Oxford), 2nd Ed.
- 2.Bateson, W. (1885) Quant. J. Microsc. Sci. 26, 535-576. [Google Scholar]
- 3.Arendt, D. & Nubler-Jung, K. (1996) BioEssays 18, 255-259. [DOI] [PubMed] [Google Scholar]
- 4.Baguna, J. (1998) in Cellular and Molecular Basis of Regeneration: From Invertebrates to Humans, eds. Ferretti, P. & Geradieu, J. (Wiley, Chichester, U.K.).
- 5.Riger, R. M., Tyler, S., Smith, J. P. S., III, & Riger, G. E. (1991) in Microscopic Anatomy of Invertebrates, ed. Harrison, F. W. (Wiley-Liss, New York), Vol. 3.
- 6.Tazaki, A., Gaudieri, S., Ikeo, K., Gojobori, T., Watanabe, K. & Agata, K. (1999) Biochem. Biophys. Res. Commun. 260, 426-432. [DOI] [PubMed] [Google Scholar]
- 7.Agata, K., Soejima, K., Kato, K., Kobayashi, C., Umesono, Y. & Watanabe, K. (1998) Zool. Sci. 15, 433-440. [DOI] [PubMed] [Google Scholar]
- 8.Sarnat, H. B. & Netsky, M. G. (1985) Can. J. Neurol. Sci. 12, 296-302. [DOI] [PubMed] [Google Scholar]
- 9.Umesono, Y., Watanabe, K. & Agata, K. (1999) Dev. Genes Evol. 209, 18-30. [DOI] [PubMed] [Google Scholar]
- 10.Hyman, L. H. (1951) Platyhelminthes and Rhynchocoela: The Acoelomate Bilateria, The Invertebrates (McGraw–Hill, New York), Vol. II.
- 11.Aguinaldo, A. M. A., Turbeville, J. M., Linford, L. S., Rivera, M. C., Garey. J. R., Raff, R. A. & Lake, J. A. (1997) Nature 387, 489-493. [DOI] [PubMed] [Google Scholar]
- 12.Adoutte, A., Balavoine, G., Lartillot, N., Lespinet, O., Prud'homme, B. & de Rosa, R. (2000) Proc. Natl. Acad. Sci. USA 97, 4453-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 24, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitou, N. & Nei, M. (1986) J. Mol. Evol. 24, 189-204. [DOI] [PubMed] [Google Scholar]
- 16.Cebrià, F., Nakazawa, M., Mineta, K., Ikeo, K., Gojobori, T. & Agata, K. (2002) Dev. Growth Differ. 44, 135-146 [DOI] [PubMed] [Google Scholar]
- 17.Cebrià, F., Kudome, T., Nakazawa, M., Mineta, K., Ikeo, K., Gojobori, T. & Agata, K. (2002) Mech. Dev. 116, 199-204. [DOI] [PubMed] [Google Scholar]
- 18.Nakazawa, M., Cebrià, F., Mineta, K., Ikeo, K., Agata, K. & Gojobori, T. (2003) Mol. Biol. Evol., in press. [DOI] [PubMed]
- 19.Ogawa, K., Kobayashi, C., Hayashi, T., Orii, H., Watanabe, K. & Agata, K. (2002) Dev. Growth Differ. 44, 191-204. [DOI] [PubMed] [Google Scholar]
- 20.Cebrià, F., Kobayashi, C., Umesono, Y., Nakazawa, M., Mineta, K., Ikeo, K., Gojobori, T., Itoh, M., Taira, M., Alvarado, A. S. & Agata, K. (2002) Nature 491, 620-624. [DOI] [PubMed] [Google Scholar]
- 21.Smith, W. C., Knecht, A. K., Wu, M. & Harland, R. M. (1993) Nature 361, 547-549. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa, K., Ishihara, K., Ishihara, S., Saito, Y., Mineta, K., Nakazawa, M., Ikeo, K., Gojobori, T., Watanabe, K. & Agata, K. (2002) Dev. Biol. 250, 59-70. [DOI] [PubMed] [Google Scholar]
- 23.Kuma. K., Iwabe. N. & Miyata, T. (1995) Mol. Biol. Evol. 12, 123-130. [DOI] [PubMed] [Google Scholar]
- 24.Fink, C. C. & Meyer, T. (2002) Curr. Opin. Neurobiol. 12, 293-299. [DOI] [PubMed] [Google Scholar]
- 25.Kandel, E. R. (2000) in Principles of Neural Science, eds. Kandel, E. R., Scheartz, J. H. & Jessell, T. M. (McGraw–Hill, New York), 4th Ed, pp. 253-279.
- 26.Nei, M., Xu, P. & Glazko, G. (2001) Proc. Natl. Acad. Sci. USA 98, 2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowring, S. A., Grotzinger, J. P., Isachsen, C. E., Knoll, A. H., Pelechaty, S. M. & Kolosov, P. (1993) Science 261, 1293-1298. [DOI] [PubMed] [Google Scholar]
- 28.Holley, S. A., Jackson, P. D., Sasai, Y., Lu, B., De Robertis, E. M., Hoffmann, F. M. & Ferguson, E. L. (1995) Nature 376, 249-253. [DOI] [PubMed] [Google Scholar]
- 29.Orii, H., Kato, K., Agata, K & Watanabe, K. (1998) Zool. Sci. 15, 861-869. [Google Scholar]
- 30.Holley, S. A., Neul, J. L., Attisano, L., Wrana, J. L., Sasai, Y., O'Connor, M. B., De Robbertis, E. M. & Ferguson, E. L. (1996) Cell 86, 607-617. [DOI] [PubMed] [Google Scholar]
- 31.Liu, L. X., Xu, H., Weller, P. F., Shi, A. & Debnath, I. (1997) Gene 186, 1-5. [DOI] [PubMed] [Google Scholar]
- 32.Alvarado, A. S. & Newmark, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 5049-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarado, A. S., Newmark, P. A., Robb, S. M. C. & Juste, R. (2002) Development (Cambridge, U.K.) 129, 5659-5665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.