Abstract
Genome rearrangements are important in evolution, cancer, and other diseases. Precise mapping of the rearrangements is essential for identification of the involved genes, and many techniques have been developed for this purpose. We show here that end-sequence profiling (ESP) is particularly well suited to this purpose. ESP is accomplished by constructing a bacterial artificial chromosome (BAC) library from a test genome, measuring BAC end sequences, and mapping end-sequence pairs onto the normal genome sequence. Plots of BAC end-sequences density identify copy number abnormalities at high resolution. BACs spanning structural aberrations have end pairs that map abnormally far apart on the normal genome sequence. These pairs can then be sequenced to determine the involved genes and breakpoint sequences. ESP analysis of the breast cancer cell line MCF-7 demonstrated its utility for analysis of complex genomes. End sequencing of ≈8,000 clones (0.37-fold haploid genome clonal coverage) produced a comprehensive genome copy number map of the MCF-7 genome at better than 300-kb resolution and identified 381 genome breakpoints, a subset of which was verified by fluorescence in situ hybridization mapping and sequencing.
An increasing number of human disorders are linked to genomic rearrangements often involving unstable regions of the genome (see ref. 1 for review). Both structural and numeral aberrations are important in these diseases. Techniques like array comparative genomic hybridization (CGH; ref. 2), restriction landmark genome scanning (3), and high-throughput analysis of loss of heterozygosity (4) are well suited to detection of genome copy number changes but reveal little about structural changes. On the other hand, cytogenetic techniques such as spectral karyotyping (5) and banding analysis reveal both numerical and structural aberrations but are limited in genomic resolution to a few megabases. End-sequence profiling (ESP) as described here complements these techniques by providing high-resolution copy number and structural aberration maps on selected disease tissues. ESP is based on the concept of sequence-tagged connectors developed to facilitate de novo genome sequencing (6). We chose the MCF-7 breast cancer cell line as a demonstration system for ESP because the line was assessed by using both CGH and spectral karyotyping and is remarkable in its complexity (7).
Methods
Bacterial Artificial Chromosome (BAC) Library Construction. Full protocol is available as Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. Briefly, high molecular weight genomic DNA, isolated from MCF-7 cells, was partially digested with HindIII, size-fractionated, and cloned into pECBAC1 by Amplicon Express. Sizing of the inserts of 27 randomly selected clones showed that the average insert size for this library is 141 kb with SD of 36 kb.
BAC Sequencing. All sequencing was performed by S.Z. Detailed protocols for BAC end sequencing are available at http://iprotocol.mit.edu/protocol/231.htm.
ESP Analysis and Visualization. Public domain and custom software were used to assemble and visualize whole-genome ESP data. The public domain package wu-blast2 (http://sapiens.wustl.edu/blast) was used to map each BAC end sequence (BES) onto the normal genome sequence. A location was assigned if at least 50 bp of the BES align to the reference genome sequence with at least 97% identity. If the end sequence hit multiple locations in the genome, the position of the largest hit with highest identity was assigned and the BES was designated as being “ambiguously mapped.” Because the overall number of BES per given genomic interval is proportional to copy number, plotting these data generated a copy-number profile for the entire tumor genome. In addition, the software compiled a list of clones that detect aberrations. These include clones whose sizes deviated >3 SD from the mean insert size for MCF7_1 or had BES in the wrong polarity. Results were visualized with custom software. A map of MCF-7 BES onto the June, 28 2002, human genome sequence assembly can be viewed in Fig. 1B. The copy number profile generated by mapping tumor BES (dark-green line) is plotted on the x axis along the top of Fig. 1B. A red dashed line superimposed on the BES density plot shows the number of BES mapped per genomic interval, averaged across the whole genome, or averaged along each chromosome in a single-chromosome view. Clones that detect potential genome rearrangements are colorcoded according to the type of aberration described above. Clicking on a chromosome name brings up a pop-up of the chromosome-specific version of the ESP display showing the same elements at greater resolution (100 kb per pixel instead of 1 Mb per pixel for the whole-genome view). A mouse click on an amplicon peak, defined as BES number per genomic interval at least four times higher than the average number of BES per genomic interval, or on the end of an aberration spanning BAC clone opens the corresponding region of the University of California, Santa Cruz assembly.
Fig. 1.
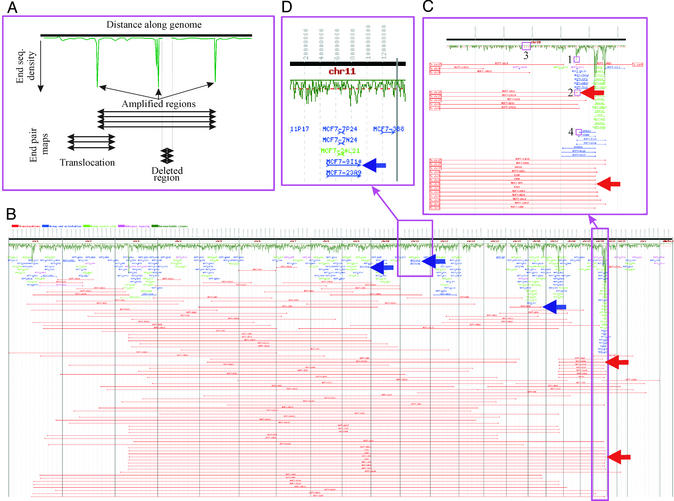
Genome analysis of MCF-7 using ESP. (A) Schematic representation of the generated types of data. End-sequence density plotted as a function of distance along the genome shows regions of copy number increase and decrease. Horizontal lines connecting BES pairs show regions of the genome that are within ≈141 kb in the test genome. Abnormally long lines indicate regions of the test genome joined by structural changes. Abnormalities that can be detected with this approach include translocations, inversions, deletions, and complex structures that develop during gene amplification. (B) End-sequence density and end-sequence pair plots for the MCF-7 genome. A total of 8,320 end-sequence pairs were mapped onto the normal human genome sequence (represented as a horizontal line along the top). The dark green plot represents the number of end sequences per 1-Mb interval. BAC end pairs with ends mapping to different chromosomes are shown as horizontal red lines. BAC clones with ends in the wrong orientation (not pointing toward each other) are shown in blue. BAC clones with ends mapping >3 SDs farther apart than the average BAC insert are shown in green. Ambiguously mapped end sequences are shown in purple. Blue arrows indicate BAC clones linking inversions and translocations validated by FISH, and red arrows indicate BAC clones detecting complex structural rearrangements associated with gene amplification that were confirmed by FISH and sequencing (see Figs. 3 and 4). (C) Expanded view of chromosome 20. Plot symbols and annotation are as described for B. Copy number data are presented in 100-kb windows. (D) Enlarged view of chromosome 11. Plot symbols and annotation are as described for B.
Fluorescence in Situ Hybridization (FISH). FISH analyses were performed as described (8).
Results
A BAC library was constructed from MCF-7 comprised of 64,896 BAC clones (≈3-fold redundancy) arrayed in 384-well microtiter plates. The average insert size of the library as determined by NotI digestion and pulsed-field electrophoresis of a subset of the clones was 141 kb. A total of 8,320 random BAC clones were end-sequenced, resulting in 15,156 end sequences. Sequences were obtained from both ends of 4,196 clones and from one end of 3,267 clones, resulting in ≈11 Mb of total sequence. Ten percent of the clones did not have useable end sequences, a rate of failure well within accepted parameters for BAC end sequencing. Fig. 1A illustrates schematically how BES information is displayed to reveal structural and numerical abnormalities. Additional information is available in Supporting Text.
The end-sequence density plots in Fig. 1 B and D show copy number increases in MCF-7 genome at 1p21, 3p14, 17q23–24, 20q12, and 20q13.2 and resolve the 20q13.2 amplification into at least five independent peaks (Fig. 1C). These results are remarkably concordant with data obtained with array CGH with contigs of overlapping clones as illustrated in Fig. 2. More importantly, ESP provides information on structural changes that are not apparent in the CGH data. A total of 381 genome breakpoints were identified with 108 being interchromosomal junctions. Of these, 18 join amplicons on 1p13, 3p14, 17q23–24, and 20q. A total of 273 BAC clones span intrachromosomal breakpoints including 211 potential inversions, 24 of which occur in amplicon peaks. Fig. 1B shows numerous MCF-7 BAC clones with ends mapping on the reference genome at distances, deviating >3 SD from the mean size of the MCF-7 BAC clone. This finding indicates that they contain regions of the MCF-7 genome that span rearrangement junctions. Because sufficiently detailed cytogenetic information generally is not available, we designate these regions with the letter j to note the junction plus the chromosome locations of the ends from http://genome.ucsc.edu. Cloned junctions identified in MCF-7 by using ESP that join genome segments on different chromosomes include j(1p13;20q13), j(8q21;11q21), j(2p13;3p23), and j(15q11;16q22). Junctions between these chromosomes were previously detected with spectral karyotyping (7). However, ESP analysis has localized each breakpoint with much higher resolution than is possible with spectral karyotyping (typically within ≈140 kb) and identified BAC clones containing the junction fragment so that the exact breakpoints and associated genes could be determined by sequencing a single BAC. Four of five tested interchromosomal rearrangements predicted by ESP were confirmed by FISH, suggesting that the current genome assembly is sufficiently accurate. Fig. 3A, for example, shows FISH confirmation of the j(15q11;16q22) translocation.
Fig. 2.
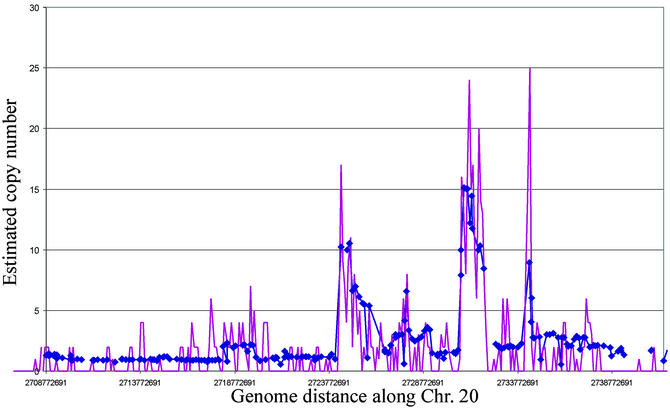
Comparison of measurements of genome copy number along chromosome 20q in MCF-7 cells made by using CGH array and end-sequence density analysis. CGH array was comprised of minimal tiling path BAC clones along 20q. End-sequence density plots were generated by enumerating the numbers of end sequences in 1-Mb-wide bins. Blue diamonds linked by solid blue lines indicate CGH measurements. Pink lines show end-sequence densities measured by using ESP. The concordance between the two assay techniques is remarkable.
Fig. 3.
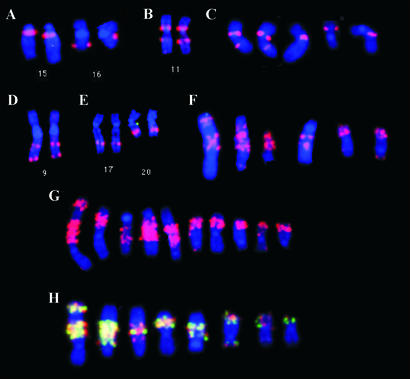
FISH-based validation of genome rearrangements identified by ESP. Complete metaphase images can be viewed as Figs. 6-16, which are published as supporting information on the PNAS web site. (A) FISH analysis of the hybridization of BAC clone MCF7_1–5H15 to normal human metaphase chromosomes confirms the ESP analysis indicating that this clone connected chromosomes 15q11.2 and 16q22.2. This clone also hybridized to chromosome 1, suggesting a more complex rearrangement (data not shown). (B) FISH analysis of the hybridization of BAC clone MCF7_1–9I10 to normal human metaphase chromosomes. The FISH analysis confirms the ESP determination that this clone joins 11p11.2 to 11q14.3. (C) Hybridization of BAC clone MCF7_1–9I10 to MCF-7 metaphase chromosomes. This clone hybridizes to five chromosomes. (D) Hybridization of BAC clone MCF7_1–5K16 to normal human metaphase chromosomes. The FISH analysis confirms the ESP determination that this clone joins 9q22.3 and 9q34.1. The distal breakpoint is located within the first intron of the ABL oncogene. (E) FISH analysis of the hybridization of BAC MCF7_1–1A11 to normal human metaphase chromosomes. This BAC was determined by ESP to connect amplicons on 20q13.2 and 17q23. Hybridization to 17q23 and 20q13.2 confirms the ESP analysis. (F) Hybridization of BAC MCF7_1–1A11 to MCF-7 metaphase chromosomes reveals multiple loci of amplification. (G) Hybridization of BAC clone MCF7_1–3F5 to MCF-7 metaphase chromosomes detects high-level amplification. ESP mapping shows that this clone has one end sequence at the ZNF217 locus at 20q13.2 and another at 3p14. FISH with BAC MCF7_1–3F5 on normal metaphase chromosomes confirmed the ESP analysis (data not shown). (H) Dual-color FISH using normal BACs spanning BMP7 (red) and ZNF217 (green) to MCF-7 metaphase chromosomes. Yellow FISH signals show coamplification and colocalization of these loci in MCF-7 genome.
ESP also identified junctions that join widely separated regions of the same chromosome, for example j(11p11;11q14) and j(9q22;9q34) (Fig. 3 B–D). The former may have been caused by pericentric inversion. FISH analysis confirmed j(11p11;11q14) as illustrated in Fig. 3 B and C. Interestingly, analysis of the sequences at the j(11p11;11q14) junction showed that the breakpoints were within 99% conserved duplicon sequences at 11p11 and 11q14. This finding suggests that recombination between paralogous elements mediated this inversion. The j(9q22;9q34) junction occurred within the first intron of the ABL oncogene (Fig. 3D). Translocations involving ABL are seen in virtually all cases of chronic myelogenous leukemia (9). The majority of these also occur in the first intron. This is an indication of the possible involvement of ABL in human breast cancer.
Another remarkable finding from ESP was the existence of numerous BACs with ends mapping to different regions of amplification. For example, BAC MCF7_1–12O5 has ends that map to regions of amplification at 1p21 and 20q13.2, whereas BAC MCF7_1–1A11 has ends that map to regions of amplification at 20q13.2 and 17q23. This finding indicates that genome sequences from these regions of amplification are located within ≈140 kb in the MCF-7 genome. Other BACs link these regions to three other regions of amplification. Colocalization of coamplified genes is an established phenomenon (10, 11). However, it is remarkable that five normally separate regions of the genome, amplified in MCF-7, appear to be located together in one or more superstructures. FISH analyses in Fig. 3 confirm the colocalization of the amplified sequences from 20q13.2 and 17q23 (Fig. 3E) and show that both sequences are amplified at high level in the MCF-7 genome (Fig. 3F). Availability of a BAC contig spanning the amplicon superstructure(s) will make possible studies aimed at determining whether they arose through extrachomosomal replication and recombination followed by integration, or alternatively, were created by independent episomes integrating into common receptive sites. Sequencing the structure(s) will allow identification of novel oncogenes and perhaps chimeric oncogenes driving tumor evolution and associated regulatory elements and origins of replication.
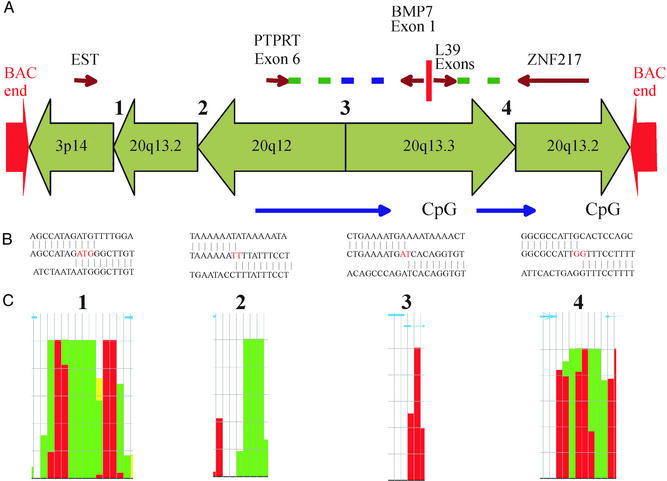
Because the five major regions of amplification appeared to be “packaged” together, we isolated and end-sequenced BAC clones from the region of amplification at 20q13.2. Clones were picked by using filter-based hybridization of arrayed MCF-7 BAC library with a probe for the ZNF217 gene, selected because it mapped to the region of very high end-sequence density (Figs. 1C and 2). Sequence-tagged site content mapping demonstrated that 16 of 38 independent clones isolated contained both ZNF217 and BMP7, genes normally separated by ≈5 Mb. Subsequent FISH analyses confirmed these results (Fig. 3 G and H). One of these clones, BAC MCF7_1–3F5, was sequenced and fully assembled, revealing a remarkably complex and enigmatic structure. Genomic sequence from four widely separated loci on 20q were packaged together and fused to a region of chromosome 3p14. The organization is represented diagrammatically in Fig. 4. The 3p14 sequence encoded an anonymous cDNA. The adjacent 20q13.2 sequence was ≈3 kb long and comprised of highly AT-rich intergenic DNA originating just distal to CYP24/PFDN4. The 20q12 sequence encoded only exon 6 of PTPRT. The 20q13.3 sequence encoded truncated BMP7 and L39 genes with only their shared promoter being intact. Finally, ZNF217 was intact, consistent with its selection as a gene important in tumor progression (12, 13). Genomic sequence was generated across each of the four junctions (Fig. 4B). Junctions 1, 2, and 4 occurred within repetitive elements, whereas junction 3 occurred within nonrepetitive DNA (Fig. 4C). These data support the role of recombination between repetitive sequences in gene amplification.
Fig. 4.
A graphical representation of the structural organization of BAC clone MCF7_1–3F5 selected to contain ZNF217 at 20q13.2. Red arrows show end sequences 3p14 and 20q13.2. Sequence-tagged site content mapping localized the 5′ region of the BMP7 gene within the BAC. This is 1 of 26 independent clones in the library juxtaposing ZNF217 and BMP7, and 1 of 4 that also contain end sequences in a region of amplification at 3p14. Full sequence analysis of MCF7_1–3F5 identified five widely separated chromosomal regions fused together in the orientations shown. Only ZNF217 is structurally intact. PTPRT, BMP7, and L39 all are truncated. PTPRT intron 6 is fused to BMP7 intron 1 in opposing polarity. L39 is fused to nontranscribed DNA 3′of ZNF217. A large CpG island shared by BMP7 and L39 is structurally intact. genscan and fgenes predict at least two novel genes created by these genome rearrangements (blue arrows). (B) Sequences spanning each genome breakpoint are presented with the fusion site in red. (C) Genome cryptographer (19) plot of the density and classification of repetitive elements (Alu, elements red; L1, green; LTR, blue, etc., details in ref. 19) at each breakpoint shown for each 150-bp window on a scale of 0–100%. Blue arrows across the top show position and orientation of the normal genome reference sequence. Thus, breakpoints 1, 2, and 4 occur in regions of very high repetitive element density, whereas breakpoint 3 occurs in single-copy DNA.
Discussion
We have demonstrated the utility of ESP by analysis of structural and numerical aberrations in the MCF7 genome. ESP takes advantage of efficient techniques for BAC library construction (14, 15) and BAC end sequencing (6, 16) developed in support of the human genome project, and it fully uses the nearly complete human genome sequence (17, 18). Still, ESP might seem too expensive and labor intensive to be practical. However, it has several advantages that are compelling. First, creation of the BAC library effectively immortalizes the target genome. This may be particularly important for rare genomes, small samples for which extensive analyses are planned, or genomes that appear from other analyses to contain important structural or numerical aberrations that warrant detailed study. Second, arrayed BAC libraries can be distributed readily. Several public and private organizations are already in place for this purpose. Third, arrayed BAC libraries can be screened for BACs from specific regions of interest. Thus, it is not necessary to perform large-scale end sequencing to gain important information. This was done in the present study to identify BAC clones from regions of amplification on chromosome 20q13. This may be particularly appealing to individuals interested in studying sequences and genes involved in specific genomic rearrangements. Fourth, individual BACs can be fully sequenced to identify genes involved in rearrangements, as was done for BAC MCF7_1–3F5 in the present study. Finally, although not pursued in this study, the BACs carrying rearranged genomic segments are biological reagents that can be transfected in vitro or in vivo to assess functional consequences of the genomic aberrations captured in the BAC.
In principle, chimeric BAC clones could introduce an unacceptably high rate of false positives in this and other ESP studies. DNA from different loci can become conjoined by three mechanisms. Composite BAC clones arise because of genome instability in the tumor and their subsequent cloning during library construction. Such clones may cluster if they occur in regions of increased copy number. Chimeric BAC clones represent either cloning or sequencing artifacts. In either case, there should be no clustering because this process is random. It is possible that a chimeric BAC clone could be highly overrepresented in the library. However, all such clones should be identical and we have never observed this. Published estimates of the frequency of chimeric clones in BAC libraries range from 4% to 11% and we believe our library is typical. To assess the quality of Amplicon Express libraries we end-sequenced 200 BAC clones from a normal human BAC library, detecting no chimerism. Thus, chimerism is not likely to represent a significant factor in this study.
The information obtained from ESP is remarkable. End sequences from only ≈8,000 BACs demonstrated that regions of high-level amplification are contiguous in the MCF-7 genome and that the amplified genomic regions are far more complex than anticipated. Complete sequence analysis of an involved BAC suggested that recombination between repetitive elements and duplicons may play a role in the formation of these aberrations. These observations raise the intriguing possibility that amplification-mediated genome rearrangements may alter gene function and/or regulation of genes important for tumor progression independent of gene dosage. However, the results presented in this article represent a fraction of what can be learned from a more detailed analysis of the available data. Possibilities include the use of BESs to assess pangenomic mutation rates, as well as those in different genomic subregions (i.e., amplified and nonamplified loci), further junction sequencing to identify specific DNA sequence elements that are involved in breakpoint formation, and the use of BAC clones as biological reagents for functional assessment of complex genome rearrangements. Accordingly, all ESP data (which are published as Supporting Text and Fig. 8, which can be found at http://shark.ucsf.edu/seq_search/MCF7/15KJune.100k.4x.html) and BAC clones (www.genomex.com/AEX_zone/index.html) are available for additional studies.
Conclusion
Full functional interpretation of ESP data will require analysis of additional tumor cell lines and primary tumors to identify recurrent structural aberrations among the multitude of random and functionally irrelevant aberrations. This will become increasingly tractable as technologies for BAC library and BES analysis increase in efficiency and decrease in cost. However, even with current technology, ESP analysis of 100–200 tumors at >150-kb resolution can be accomplished for approximately the cost of sequencing a single mammalian genome. Being sequence-based, it can be integrated with expression profile and proteomic data to colocalize aberrantly expressed genes with genome rearrangements. Such studies will make possible high throughput and comprehensive identification of currently elusive genome rearrangements and associated genes that play important roles in tumor progression. This will enable a systems view of tumor biology and lead to identification of numerous novel prognostic markers and therapeutic targets with associated biomarkers. Of course, ESP need not be limited to tumors. It appears equally useful for assessment of genomic rearrangements and/or aberrations in any closely related or derivative genomes. In this regard, it will be interesting to learn whether DNA sequence elements, associated with structural rearrangements, in cancer are involved in evolution and nonmalignant disease.
Supplementary Material
Acknowledgments
This work was performed with support from Department of Defense Grant DAMD100110500, Breast Cancer Research Program Grant 8WB0054, Bay Area Breast Cancer Special Program of Research Excellence Grant CA58207, and the Avon Foundation.
Abbreviations: ESP, end-sequence profiling; BAC, bacterial artificial chromosome; BES, BAC end sequence; CGH, comparative genomic hybridization; FISH, fluorescence in situ hybridization.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. BZ597614–BZ612944 and AC116668).
References
- 1.Stankiewicz, P. & Lupski, J. R. (2002) Trends Genet. 18, 74-82. [DOI] [PubMed] [Google Scholar]
- 2.Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W. L., Chen, C., Zhai, Y., et al. (1998) Nat. Genet. 20, 207-211. [DOI] [PubMed] [Google Scholar]
- 3.Imoto, H., Hirotsune, S., Muramatsu, M., Okuda, K., Sugimoto, O., Chapman, V. M. & Hayashizaki, Y. (1994) DNA Res. 1, 239-243. [DOI] [PubMed] [Google Scholar]
- 4.Hampton, G. M., Larson, A. A., Baergen, R. N., Sommers, R. L., Kern, S. & Cavenee, W. K. (1996) Proc. Natl. Acad. Sci. USA 93, 6704-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrock, E., du Manoir, S., Veldman, T., Schoell, B., Wienberg, J., Ferguson-Smith, M. A., Ning, Y., Ledbetter, D. H., Bar-Am, I., Soenksen, D., et al. (1996) Science 273, 494-497. [DOI] [PubMed] [Google Scholar]
- 6.Mahairas, G. G., Wallace, J. C., Smith, K., Swartzell, S., Holzman, T., Keller, A., Shaker, R., Furlong, J., Young, J., Zhao, S., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 9739-9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kytola, S., Rummukainen, J., Nordgren, A., Karhu, R., Farnebo, F., Isola, J. & Larsson, C. (2000) Genes Chromosomes Cancer 28, 308-317. [DOI] [PubMed] [Google Scholar]
- 8.Pinkel, D., Landegent, J., Collins, C., Fuscoe, J., Segraves, R., Lucas, J. & Gray, J. (1988) Proc. Natl. Acad. Sci. USA 85, 9138-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sattler, M. & Salgia, R. (1997) Cytokine Growth Factor Rev. 8, 63-79. [DOI] [PubMed] [Google Scholar]
- 10.Tanner, M. M., Tirkkonen, M., Kallioniemi, A., Isola, J., Kuukasjarvi, T., Collins, C., Kowbel, D., Guan, X. Y., Trent, J., Gray, J. W., et al. (1996) Cancer Res. 56, 3441-3445. [PubMed] [Google Scholar]
- 11.Courjal, F., Cuny, M., Simony-Lafontaine, J., Louason, G., Speiser, P., Zeillinger, R., Rodriguez, C. & Theillet, C. (1997) Cancer Res. 57, 4360-4367. [PubMed] [Google Scholar]
- 12.Nonet, G. H., Stampfer, M. R., Chin, K., Gray, J. W., Collins, C. C. & Yaswen, P. (2001) Cancer Res. 61, 1250-1254. [PubMed] [Google Scholar]
- 13.Collins, C., Rommens, J. M., Kowbel, D., Godfrey, T., Tanner, M., Hwang, S. I., Polikoff, D., Nonet, G., Cochran, J., Myambo, K., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 8703-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, U. J., Birren, B. W., Slepak, T., Mancino, V., Boysen, C., Kang, H. L., Simon, M. I. & Shizuya, H. (1996) Genomics 34, 213-218. [DOI] [PubMed] [Google Scholar]
- 15.Osoegawa, K., Woon, P. Y., Zhao, B., Frengen, E., Tateno, M., Catanese, J. J. & de Jong, P. J. (1998) Genomics 52, 1-8. [DOI] [PubMed] [Google Scholar]
- 16.Kelley, J. M., Field, C. E., Craven, M. B., Bocskai, D., Kim, U. J., Rounsley, S. D. & Adams, M. D. (1999) Nucleic Acids Res. 27, 1539-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 18.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 19.Collins, C., Volik, S., Kowbel, D., Ginzinger, D., Ylstra, B., Cloutier, T., Hawkins, T., Predki, P., Martin, C., Wernick, M., et al. (2001) Genome Res. 11, 1034-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.