Abstract
TASK3 gene (Kcnk9) is amplified and overexpressed in several types of human carcinomas. In this report, we demonstrate that a point mutation (G95E) within the consensus K+ filter of TASK3 not only abolished TASK3 potassium channel activity but also abrogated its oncogenic functions, including proliferation in low serum, resistance to apoptosis, and promotion of tumor growth. Furthermore, we provide evidence that TASK3G95E is a dominant-negative mutation, because coexpression of the wild-type and the mutant TASK3 resulted in inhibition of K+ current of wild-type TASK3 and its tumorigenicity in nude mice. These results establish a direct link between the potassium channel activity of TASK3 and its oncogenic functions and imply that blockers for this potassium channel may have therapeutic potential for the treatment of cancers.
TASK (TWIK-related acid-sensitive K+ channels) channels are members of the two-pore domain family of potassium channels, whose structure consists of two-pore forming regions flanked by four transmembrane domains (1, 2). Like other two-pore domain members, these channels show little time or voltage dependence; thus, they have characteristics of leaky K+ channels, generating background currents that contribute to membrane potential and the regulation of cell excitability. The activity of TASK channels is modulated by volatile anesthetics (3, 4), neurotransmitters (5, 6), as well as extracellular pH in the physiological range (7–11). TASK3 is expressed at very low levels among the normal tissues except in the brain, where high levels expression of TASK3 were detected (7–9). The physiological functions of TASK channels are largely unknown, though their roles in the regulation of breathing (12, 13), aldosterone secretion (5) and anesthetic-mediated neuronal activity (14) have been proposed. Recently, we showed that TASK3 is amplified in 10% of breast cancers and is overexpressed at a higher frequency of breast, lung, colon, and metastatic prostate cancers (15), suggesting that TASK3 may play a role in pathogenesis of some human carcinomas.
Is the dysregulated expression of TASK3 in tumor cells a consequence of their abnormal growth or is this K+ channel involved in promoting tumor growth? To begin to answer this question, we created an inactivating mutation of TASK3. We report here that TASK3G95E is a dominant-negative mutation that abolishes not only TASK3 K+ channel activity but also its oncogenic functions. These results provide molecular basis for developing specific blockers for this K+ channel in the treatment of cancer.
Materials and Methods
Plasmids and Mutagenesis. The coding region of TASK3 was cloned at BamHI and EcoRI sites of the pLPC retroviral expression vector (15) to generate pLPC-TASK3. Site-directed mutagenesis was performed to change Gly-95 to Glu to create pLPC-TASK3G95E, by using QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. pTracer-TASK 3G95E was generated by excising TASK3G95E from pLPC vector and cloned at KpnI and XbaI sites of pTracer (Invitrogen). To generate TASK3 expression vector for expression in Xenopus oocytes, the wild-type and mutant forms of TASK3 were excised from pLPC vector and cloned at BamHI and EcoRI sites of the pGEM3ZHEM vector.
Electrophysiological Recordings from Xenopus Oocytes. Capped cRNAs were synthesized in vitro from linearized plasmids by using T7 RNA polymerase (Epicenter, Madison, WI). Stage V and VI oocytes were defolliculated manually and kept in standard ND 96 solution (96 mM NaCl/2 mM KCl/1.8 mM CaCl2/2 mM MgCl2/5 mM Hepes, pH 7.4) supplemented with penicillin (100 units/ml), and streptomycin (100 μg/ml), at 16°C. Oocytes were injected with 5 ng per oocyte of the appropriate RNA solution, and electrophysiology studies were carried out 24–48 h after injection. Membrane currents were recorded by two-electrode voltage clamp. Currents were filtered at 1 kHz, digitally sampled at 5 kHz with a Digidata Interface (Axon Instruments, Foster City, CA). Recording and data analysis were performed by using pCLAMP software (Axon Instruments). Experiments were carried out at room temperature, and solutions were applied by a gravity-driven perfusion system. High [K+] solution contained 40 mM KCl (38 mM NaCl of ND96 solution was replaced by KCl).
Cell Culture and Transfection. Partially transformed mouse embryonic fibroblast cell line C8 was grown in DMEM/F12 (50:50) supplemented with 10% FBS. Retroviral packaging cell line Phoenix cells and lung carcinoma cell line Ben were cultured in DMEM with 10% FBS. Transfections were performed by using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's instructions. Retroviral infection was carried out by using described protocols (16). Transfectants were selected in puromycin (2 μg/ml) or Zeocin (500 μg/ml) for cells transfected with pLPC or pTracer expression vector, respectively. After 2–3 weeks in selective media, clones were pooled and analyzed for expression.
Immunofluorescence. Cells grown in chamber slides were fix in methonal/aceton (1:1) at -20°C for 10 min, and permeablized in 0.1% saponin and 0.1% BSA at room temperature for 10 min. Cells were then incubated with affinity purified anti-peptide antibody to TASK3 (REEEKLKAEEIRIKGKYNISSEDYRQ) (10 μg/ml) at room temperature for 1 h, washed three times with PBS, and then incubated with anti-rabbit FITC (Pierce) diluted 1:100 for 1 h at room temperature. After three washes in PBS, slides were dried and viewed by using a fluorescence microscope.
Electrophysiological Recordings from the Mouse Embryonic Fibroblast Cells. Cells were placed on the stage of an inverted microscope and continuously superfused with a solution containing 130 mM NaCl, 5 mM KCl, 10 mM Hepes, 10 mM glucose, 2 mM CaCl2, and 1 mM MgCl2 (pH 7.4) with NaOH. Patch electrodes were pulled from borosilicate glass to resistances of 2–6 megaohms. Internal solution contained 140 mM KCl, 4 mM NaCl, 1 mM MgCl2. 0.5 mM CaCl2, 10 mM Hepes, 10 mM EGTA, 3 mM ATP, and 0.3 mM GTP (pH 7.4) with NaOH. Recordings were obtained by using an Axopatch 200A amplifier and digitized with a Digidata 1200 A/D converter (Axon Instruments). Cells were held at a membrane potential of -60 mV and current evoked by voltage steps from -80 mV to +60 mV in 100-ms duration. Current responses filtered at 2 kHz and digitized at 10 kHz were stored for later analysis using PCLAMP software.
Membrane Potential Assay. The functional expression of TASK3 as a potassium channel in mammalian cells was determined by using a membrane potential sensitive dye on fluorescence imaging plate reader (FLIPR) (Molecular Devices). Cells were plated at 2 × 104 per well on black 384-well clear-bottom plates and incubated at 37°C overnight. Medium was then removed and the cells incubated with 50 μl of the membrane potential dye in assay buffer (Hanks' balanced salt solution plus 20 mM Hepes, pH 7.4) at 37°C for 30 min. Assays were carried out in the FLIPR at an excitation wavelength of 488 nm and by using an emission filter provided by Molecular Devices specifically for the fluorescence membrane potential (FMP) dye. After baseline readout for 10 s, 25 μl of a 3× concentration of KCl (270 mM) in assay buffer was added, and the change in fluorescence were measured for 90 s. Assays were carried out at room temperature.
Apoptosis and Cell Proliferation Assays. To induce apoptosis, cells were treated with 1 ng/ml tumor necrosis factor (TNF) for 16 h. Apoptosis was measured by DNA fragmentation assay using Cell Death Detection ELISA (Roche Diagnostics, Mannheim, Germany) following the manufacturer's instructions. For cell proliferation assay, cells were grown in media containing 1% FBS for 48 or 72 h, and the rate of cell proliferation was measured by using CellTiter Aqueous One Solution Cell Proliferation Assay kit (Promega).
Tumorigenicity Assays in Nude Mice. Cells from various stable cell lines were harvested and resuspended in PBS. The cell suspension (106 cells per injection) was injected s.c. into athymic nude mice. Mice were observed weekly for the visual appearance of tumors at injection sites, and tumor sizes were measured each week. At the end of the third week, mice were killed, and the tumors were excised and cultured.
Results
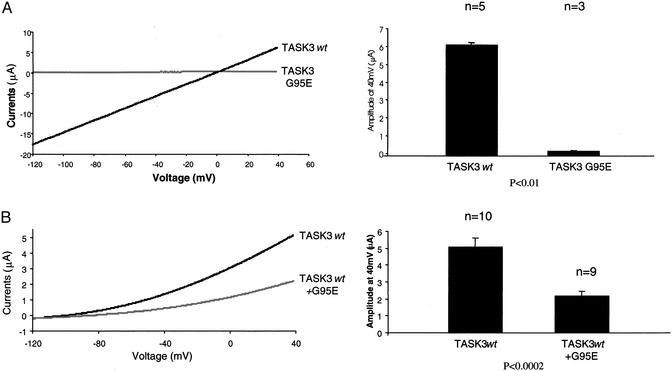
Point Mutation Within the GYG K+ Selective Filter Results in an Inactive TASK3 Channel. We used site-directed mutagenesis to change glycine at amino acid 95 within the conserved GYG K+ filter (17) to glutamate and tested the potassium channel activity of the mutant TASK3 in Xenopus oocyte and in mammalian cells. When wild-type TASK3 cRNA was injected in oocyte, it induced a K+ current of 17 μA at -120 mV (in 40 mM KCl, n = 5), whereas injection of TASK3G95E did not induce any measurable K+ current (n = 3) (Fig. 1A). When wild-type TASK3 was coinjected with mutant TASK3G95E, macroscopic current amplitudes were reduced from 5.2 μA (wild type alone, at + 30 mV, in 2 mM KCl, n = 10) to 2.05 μA (Fig. 1B). On the other hand, coexpression of TASK3G95E with the closely related channel TASK-1 did not affect TASK-1 current (data not shown). These results suggest that TASK3G95E is an inactive K+ channel when expressed in Xenopus oocytes and that the expression of the mutant TASK3 inhibits the channel activity of the wild-type TASK3.
Fig. 1.
Effect of the point mutation within the consensus K+ filter sequence on K+ current of TASK3 expressed in Xenopus oocyte. (A) TASK3G95E does not exhibit any measurable K+ current. (B) Expression of TASK3G95E inhibits K+ current of the wild-type TASK3. Shown are whole cell currents in response to voltage ramp between -120 and +40 mV recorded from Xenopus oocytes 48 h after injection with individual wild-type or mutant TASK3 cRNA or in combination as indicated. (Left) Representative traces of whole cell currents recorded in response to 175-ms voltage ramp between -120 and +40 mV. (Right) Average currents (±SE) of all of the oocytes tested in each group. P value was determined by a Student's t test. The extracellular [K+] concentration was 40 mM (A) and 2 mM (B).
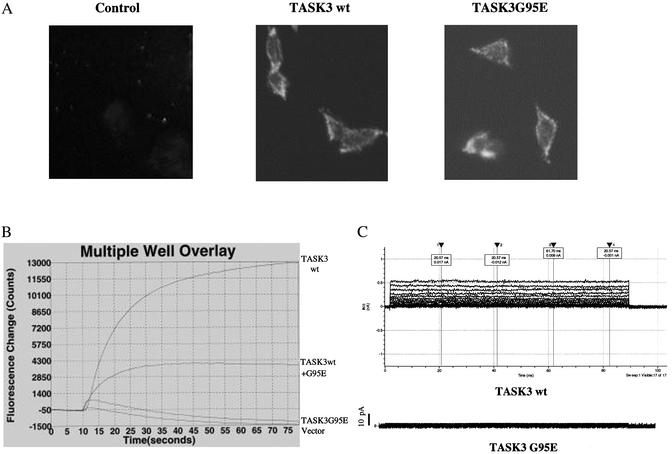
To determine the effect of this point mutation on TASK3 channel activity in mammalian cells, we generated stable cell lines in a partially transformed mouse embryonic fibroblast C8 cells that overexpress either the wild-type or mutant form of TASK3. We verified cell surface expression of the channel protein by indirect immunofluorescence assay (Fig. 2A). To test whether these cell lines express a functional channel, we used a membrane potential sensitive dye (Figs. 4 and 5, which are published as supporting information on the PNAS web site, www.pnas.org) to measure the change in fluorescence signals on depolarization of cells by KCl. Expression of wild-type TASK3 resulted in increase in fluorescence signal (≈13,000 counts) on addition of 90 mM KCl (Fig. 2B). In contrast, cells overexpressing TASK3G95E showed no change in fluorescence signal comparing to cells expressing the empty vector alone (Fig. 2B). When the wild-type and the mutant TASK3 were cotransfected into cells, the fluorescence signals were reduced 75% compare with wild-type TASK3 alone (Fig. 2B). We also determined the functional channel expression in stable cell lines by whole cell current recording. Cell membrane potential was held at -60 mV and stepped to various potentials for 100-ms duration. In cells stably transfected with the mutant form TASK3, only very small currents of <50 pA were recorded (Fig. 2C). In cells stably expressing wild-type TASK3, the same voltage step produced large instantaneous and noninactivating currents (Fig. 2C). These results agree with the results in Xenopus oocytes, suggesting that TASK3G95E is a dominant negative mutant.
Fig. 2.
Effect of the point mutation within the consensus K+ filter sequence on TASK3 currents expressed in mouse embryonic fibroblast cells. (A) Cell surface expression of the wild-type and mutant TASK3 by indirect immunofluorescence. (B) K+ channel activity of the stable cell lines expressing wild-type or mutant TASK3 individually or together measured by FLIPR assay. Twenty thousand cells were plated in a 384-well plate and grown for 24 h. Cells were loaded with the voltage-sensitive fluorescence dye for 30 min and assayed on FLIPR in assay buffer containing 90 mM [K+]. (C) TASK3 whole cell currents were recorded from the stable cell line expressing either wild-type or mutant TASK3. Pipette and bath solutions contained 140 and 5 mM K+, respectively. Membrane potential was held at -60 mV and stepped from -80 to +60 mV in 10-mV increments.
TASK3G95E Loses Oncogenic Potential and Inhibits Oncogenic Function of the Wild-Type TASK3. To determine whether the oncogenic activity of TASK3 is correlated with its function as a potassium channel, we tested the effects of overexpression of wild-type TASK3 and TASK3G95E mutant on cell proliferation in low serum, resistance to TNF-induced apoptosis as well as tumorigenicity in nude mice.
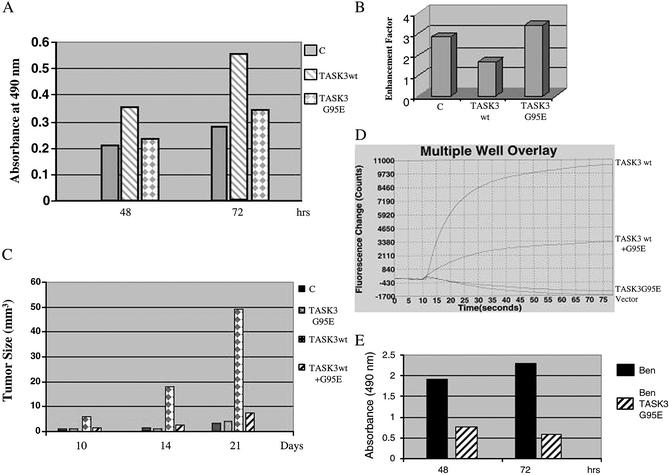
To elucidate the role of TASK3 in cell proliferation, mouse embryonic fibroblast cells expressing either the wild-type or mutant TASK3 were cultured in 1% serum for 72 h, and the growth rate was quantified by using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. Cells expressing either vector alone or the TASK3G95E mutant proliferate in a much slower rate comparing to cells expressing wild-type TASK3, resulting in a 2-fold increase in cell number in TASK3 overexpressing cells after 72 h in culture (Fig. 3A). These results suggest that overexpression of TASK3 confers growth advantage in low serum and that this function requires a functional potassium channel.
Fig. 3.
The inactivating mutation of TASK3 abolished its oncogenic functions. (A) Cell proliferation in low serum. Cell growth rate is expressed as absorbance at 490 nm. Data are representative of two independent experiments (n = 8 for each experiment). (B) Apoptosis assay (DNA fragmentation ELISA). Apoptosis was induced by treatment of cells with 1 ng/ml TNF for 16 h. Cell apoptosis is expressed as enhancement factor (fold over untreated cells in each group). Data are representative of three independent experiments (n = 3 for each experiment). (C) Tumorigenicity in nude mice. Mouse embryonic fibroblast cells stably transfected with vector alone, wild-type, or mutant TASK3 individually or together were injected (106 cells per injection) into nude mice (two experiments, n = 10 for each cell line). Tumor formation was monitored. The tumors were measured each week at their longest points by using a caliper. The bar in the graph represents the average tumor size for each group. (D)K+ channel activity in excised tumors. Tumors were excised and cultured for a week, and the TASK3 channel activity was measured by FLIPR assay as describe in the legend for Fig. 2. (E) Ben cell proliferation. Five thousand Ben or BenTASK3G95E cells were plated in a 96-well plate, and the cell proliferation rated was determined as in A.
To test whether overexpression of TASK3 would result in resistance to apoptosis, we treated embryonic fibroblast cells with TNF and measured TNF-induced apoptosis by using a DNA fragmentation ELISA assay. Overexpression of wild-type TASK3 led to a 50% reduction of TNF-induced apoptosis, whereas expression of the mutant TASK3 had no effects (Fig. 3B). This result suggests that TASK3 confers partial resistance to TNF-induced apoptosis and that the K+ channel activity of TASK3 is required for this function.
To determine the effect of TASK3G95E mutation on TASK3 tumorigenicity, we injected athymic nude mice with C8 cells expressing vector alone, wild-type or mutant form of TASK3, as well as C8 cells expressing both the wild-type and the mutant TASK3 together. All animals injected with C8 cells expressing wild-type TASK3 developed large tumors within 2 weeks. Although mice injected with cells expressing vector alone or cells expressing mutant TASK3 also developed tumor after 3 weeks, the size of the tumors are much smaller in comparison to tumors expressing wild-type TASK3 (Fig. 3C). Furthermore, cell coexpressing both the wild-type and mutant form of TASK3 only developed small tumors as seen in the vector control (Fig. 3C). To determine whether the tumor cells continue to express a function K+ channel, the tumors were excised, cultured for 1 week and then assayed for K+ channel activity by FLIPR. The tumor cells overexpressing the wild-type TASK3 maintain a functional K+ channel, whereas no channel activity was detected for tumor cells overexpressing the mutant TASK3 (Fig. 3D). These results suggest that K+ channel activity is required for TASK3 to promote tumor formation in nude mice and that TASK3G95E acts as a dominant-negative mutant for expression of the mutant protein inhibited tumorigenicity of the wild-type TASK3.
To test whether expression of TASK3G95E would affect functions of endogenous TASK3, we stably transfected TASK3G95E into human lung carcinoma cell line Ben that overexpresses TASK3 (Fig. 6, which is published as supporting information on the PNAS web site). As shown in Fig. 3E, expression of TASK3G95E significantly reduced Ben-cell proliferation within 48 h, suggesting that TASK3G95E interferes with endogenous TASK3 function to inhibit proliferation of these lung carcinomas cells.
Discussion
K+ channel activity plays important roles in the signaling pathways that regulate cell proliferation and apoptosis. Inhibition of K+ channels by pharmacological agents has been found to inhibit cell proliferation in normal human lymphocytes (18–21), human melanoma cells (22, 23), small cell lung cancer (24), breast (25), and prostate (26) cancer cells. The role of K+ channels in cellular proliferation has been thought to be indirect, by either the possible influence of K+ channels on the intracellular Ca2+ concentration (27) or, alternatively, the control of cell volume via K+ channels (28). Recent studies have shown that enhancement of K+ current is directly involved in apoptosis (29, 30) and oncogenesis (31). In mouse neocortical neurons, treatment of cell with the K+ ionophore valinomycin or KATP channel opener cromakalin induced apoptosis (29), whereas inhibition of outward K+ currents with tetraethylammonium, but not with blockers of Ca2+, or Cl- reduced apoptosis (30). The human ether-a-go-go related (HERG) K+ channel is overexpressed in a variety of tumor cell lines (32), and HERG conductance markedly promotes H2O2-induced apoptosis of various tumor cells (33). The expression of rEAG favors tumor progression when the transfected cells are injected in nude mice, and overexpression of rEAG K+ in NIH 3T3 cells induces significant features characteristic of malignant transformation (31). Taken together, these studies suggest that K+ channels play a crucial role in oncogenesis.
Recently, we showed that TASK3 (Kcnk9) is amplified in 10% of breast cancers and is overexpressed at a higher frequency of breast, lung, colon, and metastatic prostate cancers (15). This was the first time that a genetic modification in K+ channels has been identified in primary human carcinomas. The data reported here expended the results in this previous study and established a direct link between K+ channel activity of TASK3 and its oncogenic function. We have characterized a dominant-negative mutation of TASK3 that not only abolishes its K+ channel activity but also abrogates its oncogenic functions. Our data showed that wild-type TASK3 confers growth advantage to cells overexpress this K+ channel, whereas the inactivating mutant had no effect on cell growth, suggesting that TASK3 K+ channel activity is directly involved in cell proliferation. We provided evidence that TASK3 overexpression resulted in partial resistance to TNF-induced apoptosis, a mechanism that causes cancer cells to escape host immune defenses. However, cells expressing the mutant TASK3 are sensitive to TNF-induced apoptosis, suggesting the requirement for a functional K+ channel. Most importantly, we demonstrated the TASK3 mutant not only was incapable of promoting tumor growth in nude mice, but also inhibited the tumor promotion function of the wild-type TASK3. We cannot at the present completely rule out the possibility that this dominant-negative mutant of TASK3 could interfere with the activity of other TASK channels through heterodimerization (34). A previous study showed the same TASK3 mutant did not have effect on TASK1 current when coexpressed in Xenopus oocytes (35), and we confirmed this observation in this study (data not shown). In addition, TASK1 gene amplification or overexpression was not detected in tumor samples where TASK3 was amplified (15).
In summary, the experimental results presented in this study provide the biological basis for the development of TASK3 antagonists that should potentially inhibit growth of certain human carcinomas and therefore would represent a new family of anticancer therapeutics.
Supplementary Material
Acknowledgments
We thank Chris Mathes (Axon Instruments) for help with whole cell recording in mammalian cells.
Abbreviations: TNF, tumor necrosis factor; FLIPR, fluorescence imaging plate reader.
References
- 1.Goldstein, S. A., Bockenhauer, D., O'Kelly, I. & Zilbergerg, N. (2001) Nat. Rev. Neurosci. 2, 175-184. [DOI] [PubMed] [Google Scholar]
- 2.Lesage, F. & Lazdunski, M. (2000) Am. J. Physiol. 279, F973-F801. [DOI] [PubMed] [Google Scholar]
- 3.Meadows, H. J. & Randall, A. D. (2001) Neuropharmacology 40, 551-559. [DOI] [PubMed] [Google Scholar]
- 4.Patel, A. J., Honore, E., Lesage F., Fink, M., Romey, G. & Lazdunski, M. (1999) Nat. Neurosci. 2, 422-426. [DOI] [PubMed] [Google Scholar]
- 5.Czirjak, G., Fischer, T., Spat, A. & Enyedi, P. (2000) Mol. Endocrinol. 14, 863-874. [DOI] [PubMed] [Google Scholar]
- 6.Talley, E. M. & Bayliss, D. A. (2002) J. Biol. Chem. 277, 17733-17742. [DOI] [PubMed] [Google Scholar]
- 7.Duprat F., Lesage, F., Fink, M., Reyes, R., Heurteaux, C. & Lazdunski, M. (1997) EMBO J. 16, 5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman, C. G., Meadows, H. J., Godden, R. J., Campbell, D. A., Duckworth, M., Kelsell, R. E., Murdock, P. R., Randall, A. D., Rennie, G. I. & Gloger, I. S. (2000) Mol. Brain Res. 82, 74-83. [DOI] [PubMed] [Google Scholar]
- 9.Kim, G., Bang, H. & Kim, D. (2000) J. Biol. Chem. 275, 9340-9347. [DOI] [PubMed] [Google Scholar]
- 10.Rajan, S., Wischmeyer, E., Liu, G. X., Preisig-Muller, R., Daut, J., Karschin, A. & Derst, C. (2000) J. Biol. Chem. 275, 16650-16657. [DOI] [PubMed] [Google Scholar]
- 11.Lopes, C. M., Zilbergerg, N. & Goldstein, S. A. (2002) J. Biol. Chem. 276, 24449-24452. [DOI] [PubMed] [Google Scholar]
- 12.Hartness, M. E., Lewis, A., Searle, G. J., O'Kelley, I., Peers, C. & Kemp, P. J. (2002) J. Biol. Chem. 276, 26499-26508. [DOI] [PubMed] [Google Scholar]
- 13.Buckler K. J., Williams, B. A. & Honore, E. (2000) J. Physiol. 15, 135-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sirois, J. E., Lei, Q., Talley, E. M., Lynch, C., III, & Bayliss, D. A. (2000) J. Neurosci. 20, 6347-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu, D., Chen, L., Zhang, X., See, L. H., Koch, C. M., Yen, C., Tong, J. J., Spiegel, L., Nguyen, K. C., Servoss, A., et al. (2003) Cancer Cell 3, 297-302. [DOI] [PubMed] [Google Scholar]
- 16.Pei, L. Peng, Y., Yang, Y., Ling, X. B., Van Eyndhoven, W. G., Nguyen, K. C., Rubin, M., Hoey, T., Powers, S. & Li, J. (2002) Cancer Res. 62, 5420-5424. [PubMed] [Google Scholar]
- 17.Heginbotham, L., Lu, Z., Abramson, T. & MacKinnon, R. (1994) Biophys. J. 66, 1061-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amigorena, S., Choquet, D., Teillaud, J. L., Korn, H. & Friedman, W. H. (1990) Mol. Immunol. 27, 1259-1268. [DOI] [PubMed] [Google Scholar]
- 19.Lin, C. S., Boltz, R. C., Blake, J. T., Nguyen, M., Talento, A., Fischer, P. A., Springer, M. S., Sigal, N. H., Slaughter, R. S., Garcia, M. L., et al. (1993) J. Exp. Med. 177, 637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rader, R. K., Kahn, L. E., Anderson, G. D., Martin, C. L., Chinn, K. S. & Gregory, S. A. (1996) J. Immunol. 156, 1425-1430. [PubMed] [Google Scholar]
- 21.Jensen, B. S., Odum, N., Jorgensen, N. K., Christophersen, P. & Olesen, S. P. (1999) Proc. Natl. Acad. Sci. USA 96, 10917-10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilius, B. & Wohlrab, W. (1992) J. Physiol. 445, 537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lepple-Wienhues, A., Berweck, S., Bohmig, M., Leo, C. P., Meyling, B., Garbe, C. & Wiederholt, M. (1996) J. Membr. Biol. 151, 149-157. [DOI] [PubMed] [Google Scholar]
- 24.Pancrazio, J. J., Tabbara, I. A. & Kim, Y. I. (1993) Anticancer Res. 13, 1231-1234. [PubMed] [Google Scholar]
- 25.Woodfork K. A., Wonderlin, W. F., Peterson, V. A. & Strobl, J. S. (1995) J. Cell. Physiol. 162, 163-171. [DOI] [PubMed] [Google Scholar]
- 26.Skryma, R. N., Prevarskaya, N. B., Dufy-Barbe, L., Odessa, M. F., Audin, J. & Dufy, B. (1997) Prostate 33, 112-122. [DOI] [PubMed] [Google Scholar]
- 27.Santella, L. (1998) Biochem. Biophys. Res. Commun. 244, 317-324. [DOI] [PubMed] [Google Scholar]
- 28.Rouzaire-Dubois, B. & Bubois, J. M. (1998) J. Physiol. 510, 93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, S. P., Yeh, C. H., Sensi, S. L., Gwag, B. J., Canzoniero, L. M., Farhangrazi, Z. S., Ying, H. S., Tian, M., Dugan, L. L. & Choi, D. W. (1997) Science 278, 114-117. [DOI] [PubMed] [Google Scholar]
- 30.Yu, S. P., Yeh, C. H., Gottron, F., Wang, X., Grabb, M. C. & Choi, D. W. (1999) J. Neurochem. 73, 933-941. [DOI] [PubMed] [Google Scholar]
- 31.Pardo, L. A., del Camino, D., Sanchez, A., Alves, F., Bruggemann, A., Beckh, S. & Stuhmer, W. (1999) EMBO J. 18, 5540-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianchi, L., Wible, B., Arcangeli, A., Taglialatela, M., Morra, F., Castaldo, P., Crociani, O., Rosati, B., Faravelli, L., Olivotto, M. & Wanke, E. (1998) Cancer Res. 58, 815-822. [PubMed] [Google Scholar]
- 33.Wang, H., Zhang, Y., Cao, L., Han, H., Wang, J., Yang, B., Nattel, S. & Wang, Z. (2002) Cancer Res. 62, 4843-4848. [PubMed] [Google Scholar]
- 34.Czirjak, G. & Enyedi, P. (2002) J. Biol. Chem. 277, 5426-5432. [DOI] [PubMed] [Google Scholar]
- 35.Karschin, C., Wischmeyer, E., Preisig-Muller, R., Rajan, S., Derst, C., Grzeschik, K-H., Daut, J. & Karschin, A. (2001) Mol. Cell. Neurosci. 18, 632-648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.