Abstract
Recognition of the 5′ splice site is an important step in mRNA splicing. To examine whether U1 approaches the 5′ splice site as a solitary snRNP or as part of a multi-snRNP complex, we used a simplified in vitro system in which a short RNA containing the 5′ splice site sequence served as a substrate in a binding reaction. This system allowed us to study the interactions of the snRNPs with the 5′ splice site without the effect of other cis-regulatory elements of precursor mRNA. We found that in HeLa cell nuclear extracts, five spliceosomal snRNPs form a complex that specifically binds the 5′ splice site through base pairing with the 5′ end of U1. This system can accommodate RNA-RNA rearrangements in which U5 replaces U1 binding to the 5′ splice site, a process that occurs naturally during the splicing reaction. The complex in which U1 and the 5′ splice site are base paired sediments in the 200S fraction of a glycerol gradient together with all five spliceosomal snRNPs. This fraction is functional in mRNA spliceosome assembly when supplemented with soluble nuclear proteins. The results argue that U1 can bind the 5′ splice site in a mammalian preassembled penta-snRNP complex.
Splicing of precursor mRNA (pre-mRNA) is a critical regulatory stage in which accurate recognition and removal of introns by the splicing machinery compose the correct code for protein production. The splicing reaction is carried out by the spliceosome, a dynamic complex of five small nuclear ribonucleoproteins (snRNPs)—U1, U2, U4, U5, and U6—and many auxiliary proteins. The spliceosome assembles de novo on each intron and, through a myriad of RNA-RNA, RNA-protein, and protein-protein interactions, acts to excise each intron and ligate the exons (reviewed in references 11, 20, 24, and 45).
Spliceosome assembly requires specific recognition of splice site signals: the 5′ splice site (5′ss) consensus sequence, which includes a conserved GU dinucleotide at the 5′ end of the intron; the 3′ splice site (3′ss) region, which consists of the polypyrimidine tract and a conserved AG dinucleotide at the 3′ end of the intron; and the branch site located ∼20 nucleotides upstream of the 3′ss, bearing a conserved adenosine (8).
According to the current model of spliceosome assembly on pre-mRNA, the snRNPs join the spliceosome in an ordered pathway. U1 snRNP initially recognizes the 5′ss by base pairing between U1 snRNA and the 5′ss exon-intron junction (at positions −3 to +6); these are highly complementary sequences (17, 35, 48, 50, 64). Non-snRNP splicing factors interact with the 3′ss, resulting in the 5′ss being brought to the proximity of the 3′ss. The U1/5′ss base pairing is then weakened in an ATP-dependent step (9, 21, 23, 29, 30, 53), allowing U2 snRNP to base pair with the branch site. Next, the U4/U5/U6 tri-snRNP complex is added, resulting in an apparent destabilization of U1 snRNP from the spliceosome (reviewed in reference 24), followed by several rearrangements in which U1 is replaced by U5 and U6 at the 5′ss (1, 4, 5, 22, 27, 32). The U4/U6 base pairing within the U4/U5/U6 complex is disrupted, U4 is released from the spliceosome, and U6 snRNA base pairs with U2 snRNA (3, 10, 26, 36, 37, 60). These rearrangements finally allow the two constitutive catalytic steps to generate mature mRNA and liberate the intron.
While the canonical view of ordered assembly has been supported by numerous studies, a growing number of reports have identified a variety of interactions that contradict the proposed chronology of events. Two examples are the findings that the E complex associates with U2 in the absence of ATP and does not require the branch site sequence (15) and that an early functional interaction between the U4/U5/U6 tri-snRNP complex and the 5′ss occurs independently of prior binding of U2 to the branch site (38). The latter finding also suggests that U1 and U5 snRNPs collaborate functionally to recognize and define the 5′ss, and this suggestion is supported by reports of both cross-linking between U1 and U5 snRNAs (4) and interactions between protein elements of U1 and U5 snRNPs (1, 6). In another contradiction to the canonical model, the U4/U5/U6 tri-snRNP complex was found to associate with the spliceosome when U1/5′ss base pairing was still stable (29, 53), suggesting that the assembly of U4/U5/U6 with the spliceosome is accompanied by the dissociation of U1 from the spliceosome. Along this line, specific protein elements of U1 and U5 were found within the prespliceosome (complex A), suggesting that U1 remains associated when U2 base pairs with the branch site and that U5 may enter prior to spliceosome assembly (complex B) (19). Most importantly, a 45S complex containing all five spliceosomal snRNPs and over 60 pre-mRNA splicing factors, named the penta-snRNP complex, was observed in yeast extracts (54). This complex was seen only when the purification procedure was performed with a maximum salt concentration of 50 mM, which is compatible with splicing in yeast extracts. Stevens et al. proposed that the five spliceosomal snRNPs associate prior to binding to a pre-mRNA substrate rather than binding in a stepwise manner as previously thought (54).
A U2/U4/U5/U6 complex was identified in mammalian cell nuclear extracts without the addition of external pre-mRNA during incubation at a high salt concentration (250 mM NH4Cl) (25). Also, a U1/U2/U4/U5/U6 complex was purified by the addition of an antisense oligonucleotide complementary to U5, and elevating the salt concentration from 50 to 250 mM NaCl led to the disassembly of that complex to U1/U4/U5 and U2/U6 complexes (5). These observations are difficult to reconcile with the canonical assembly pathway. They may represent temporal stable interactions within a penta-snRNP complex rather than stepwise spliceosome assembly. The one-step spliceosome assembly model is an intriguing model, especially in light of the different techniques used to identify each interaction (7, 43, 54).
To examine whether U1 approaches the 5′ splice site as a solitary snRNP or as part of a multi-snRNP complex, we used a simplified in vitro system in which a short RNA containing the 5′ss sequence served as a substrate in a binding reaction. This system allowed us to study the interactions of the snRNPs with the 5′ss in the absence of the effect of other cis-regulatory elements of pre-mRNA. Our results suggest that U1, U2, U4, U5, and U6 snRNPs preexist as a complex in mammalian cell nuclear extracts and that, when the 5′ss RNA is added, the complex specifically binds it through base pairing between the 5′ end of U1 and the 5′ss RNA. This system accommodates the RNA-RNA rearrangement in which U5 replaces U1 binding to the 5′ splice site. The complex in which U1 and the 5′ss are base paired sediments in the 200S fraction of a glycerol gradient together with all five spliceosomal snRNPs, can accommodate the replacement of U1 by U5 at the 5′ss, and can support pre-mRNA spliceosome assembly when supplemented with soluble nuclear proteins. These results indicate that U1 can bind the 5′ss within a penta-snRNP complex.
MATERIALS AND METHODS
Oligonucleotides
The RNA oligonucleotide OligoA (5′-AAAAAAUGGUAAGUAT) and the mutated 5′ss RNA oligonucleotides were synthesized by CyberSyn. The 5′ss RNA oligonucleotide Adeno (5′-CUGUUCAGGUAAGUAT) was synthesized by Dharmacon. All RNA oligonucleotides were deprotected, desalted, and purified by denaturing polyacrylamide gel electrophoresis (PAGE). The 3′-biotinylated RNA oligonucleotide OligoA was synthesized by Oligos-R-Us at Yale University Medical School. The DNA oligonucleotides were synthesized by Genset.
In vitro binding reaction.
A standard binding reaction mixture (25 μl) containing 60% HeLa cell nuclear extract (4C Biotech), 2.4 mM MgCl2, 0.5 mM ATP, 20 mM creatine phosphate, and 0.034 pmol of 32P-end-labeled 5′ss RNA/μl (≥100,000 cpm for cross-linking experiments and 5,000 to 10,000 cpm for complex analyses) was incubated for 15 min at 4°C (4, 5). To sequester the 5′ end of U1, the nuclear extract was preincubated with cofactors and with different concentrations of an antisense DNA oligonucleotide complementary to the first 14 nucleotides of U1 (U11-14) for 15 min at 4°C. Then, 32P-end-labeled 5′ss RNA was added, and incubation was continued for another 15 to 20 min at 4°C (28).
Native gel analysis, psoralen cross-linking, Northern blot analysis, RNase H digestion, primer extension, immunoprecipitation, affinity selection, splicing reactions, spliceosome analysis by native 4% PAGE, and micrococcal nuclease treatment.
Native agarose gel analysis was carried out as described by Das et al. (15), with the following modifications. The reaction mixtures were separated in a 0.75% agarose gel at 80 V for 8 h at 4°C. Cross-linking with psoralen and primer extension blockage were carried out as described by Ast and Weiner (4). Northern blot analysis, splicing reactions, and spliceosome analysis by native 4% PAGE were carried out as described by Shomron et al. (49). RNase H digestion, immunoprecipitation, and affinity selection were carried out as described by Ast and Weiner (5). Micrococcal nuclease treatment was performed as described by Liu et al. (33). Briefly, 1.5 U of micrococcal nuclease and 1 mM CaCl2 were added per 10 μl of nuclear extract. The reaction mixture was incubated at 30°C for 10 min, the reaction was stopped by the addition of 4 mM EDTA, and the reaction mixture was dialyzed against buffer D (see below).
Glycerol gradient fractionation.
Glycerol gradients were formed in SW41 ultracentrifuge tubes (Beckman). A 10% glycerol solution (10% glycerol, 50 mM KCl, 1 mM MgCl2, 20 mM HEPES [pH 7.9]) was layered onto an equal volume of a 50% glycerol solution. The tubes were covered with Parafilm, placed in a horizontal position for 2 h at room temperature, and returned to the original vertical position for 5 to 8 h at 4°C (58). Ten- or 16-fold-psoralen-cross-linked standard binding reaction mixtures (250 or 400 μl) were loaded by replacing the top 400 μl. Gradients were centrifuged at 15,000 rpm for 12 h at 4°C, and 450-μl fractions were removed sequentially from top to bottom.
Fractions 15 to 20 (200S fraction) were combined and placed in a dialysis bag (Spectra/Por MW-CO; 12,000 to 14,000 Da). The bag was placed in buffer D (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol) (16) and dialyzed for 8 h at 4°C. The dialyzed fractions were concentrated to 200 μl (protein concentration, ∼0.5 mg/ml) by using a Centricon apparatus (Amicon MW-CO; 10,000 Da) and stored at −70°C.
RESULTS
U1 binds the 5′ss within an ∼45S complex.
The canonical model of spliceosome assembly proposes that the snRNPs join the spliceosome in an ordered pathway in which U1 base pairs with the 5′ss as an individual snRNP prior to the binding of the remaining snRNPs. In view of evidence suggesting spliceosome assembly in one step, it was intriguing to investigate the spliceosomal complex associated with the 5′ss when U1 base pairs with the 5′ss.
In order to detect potential interactions of spliceosomal snRNPs with the 5′ss, we used a simplified in vitro system developed by Konarska et al. (28), in which a short 5′ss RNA is used as a substrate in an in vitro binding reaction. This system enables the interactions of the snRNPs with the 5′ss in the absence of the effect of other cis-splicing regulatory elements of the pre-mRNA, such as the 3′ss, polypyrimidine tract, branch site, or exon-intron enhancer elements (27, 28). The 5′ss RNA oligonucleotide (5′ss RNA) is 16 nucleotides long, spans eight exon and eight intron nucleotides, and contains the optimal 5′ss conserved sequence from positions −1 to +8 (for clarity, positions “−” and “+” represent nucleotides upstream and downstream from the 5′ss, respectively). We used two different 5′ss RNAs: Adeno, the authentic 5′ss sequence of the adenovirus type 2 major late transcript, and OligoA, which was designed to enhance base pairing between the uridines of the invariant loop of U5 and the adenosines of the 5′ss exon sequence. Both are 16-nucleotide RNAs that contain the 5′ss conserved sequence from positions −1 to +8 and differ in the nonconserved sequence from positions −2 to −8 (Fig. 1A).
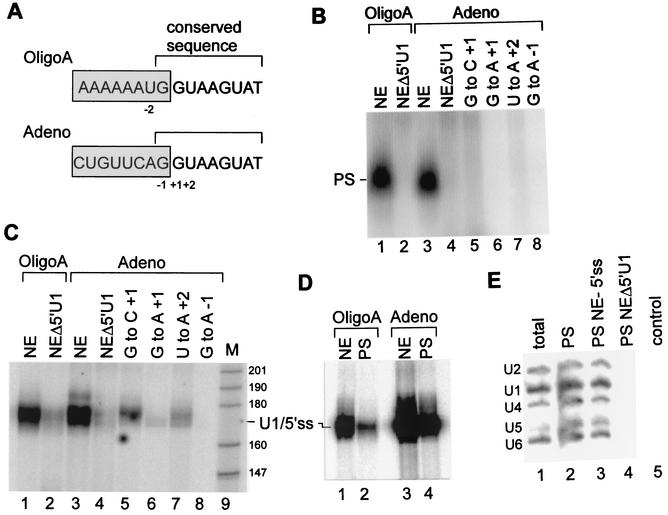
FIG. 1.
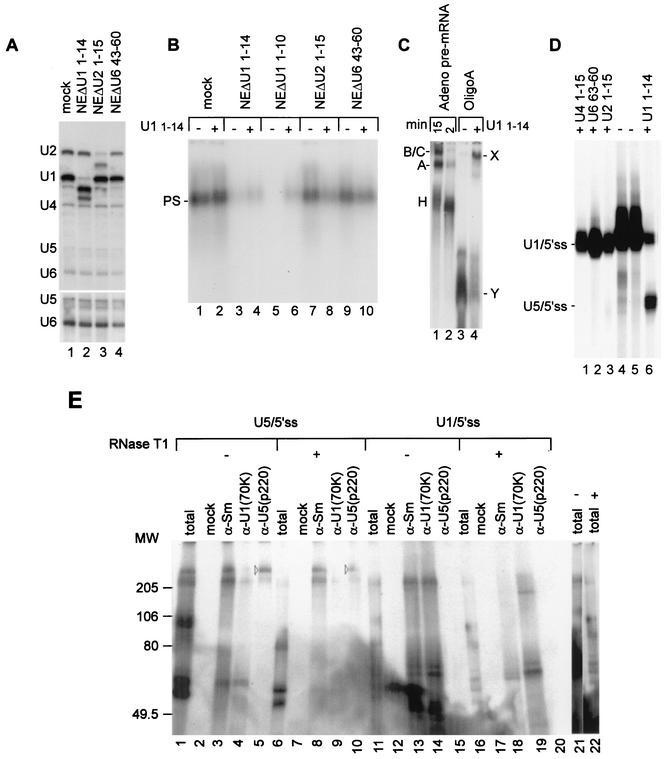
U1 base pairs the 5′ss RNA in a large complex. (A) Two 5′ss RNA oligonucleotides used for experiments. Boxed and nonboxed sequences represent exon and intron regions, respectively. The sequences of the two oligonucleotides are identical from positions −1 to +8 and differ in the exon sequence such that OligoA contains a single uridine in the exon sequence (position −2). Positions −2 to +2 are marked. (B) U1 is assembled with the 5′ss RNA in a complex that requires the 5′ end of U1 snRNA and the conserved nucleotides of the 5′ss. 32P-labeled OligoA or Adeno was added to HeLa cell nuclear extracts (NE) and incubated for 15 min at 4°C under in vitro binding conditions, and the reaction mixtures were separated in a native 0.75% agarose gel. The complex assembled on the 5′ss RNA is indicated by PS. In lanes 2 and 4, the 5′ end of U1 snRNA was removed by antisense DNA oligonucleotide-directed RNase H cleavage. In lanes 5 to 8, Adeno was mutated at the three most conserved nucleotides of the 5′ss, as indicated in panel A. (C) U1 binds the 5′ss RNA. 32P-labeled OligoA or Adeno was added to HeLa cell nuclear extracts as described for panel B. Following incubation, the reaction mixtures were cross-linked with psoralen, and the RNA was purified and separated by denaturing 6% PAGE. The numbers on the right represent sizes in nucleotides of a pBR MspI marker (M). (D) U1/5′ss cross-linking occurs within the PS complex. Nuclear extracts were incubated with 32P-labeled OligoA or Adeno, cross-linked with psoralen, and separated in a native 0.75% agarose gel. The RNA from the PS complex was eluted, purified, and separated by denaturing 6% PAGE (lanes 2 and 4). Lanes 1 and 3, control standard reactions. (E) RNA from the PS complex that assembled on OligoA was eluted from the native agarose gel, separated by denaturing 10% PAGE, and Northern blotted with radiolabeled antisense riboprobes complementary to U1, U2, U4, U5, and U6 snRNAs (lane 2). Lane 1, total nuclear extract RNA. Lane 3, nuclear extracts incubated under splicing conditions in the absence of the 5′ss RNA; the RNA was eluted from the same position on the gel as the PS complex. Lane 4, similar to lane 3, except that the 5′ end of U1 snRNA was digested by RNase H treatment of nuclear extracts. Lane 5, analysis of a control region located above the PS complex from the reaction mixture shown in lane 2.
To explore the interactions of spliceosomal snRNPs with the 5′ss, 32P-labeled 5′ss RNAs (OligoA and Adeno) were incubated in HeLa cell nuclear extracts under in vitro binding conditions, and the reaction mixtures were separated in a native 0.75% agarose gel (15). A complex with a similar mobility (∼45S; data not shown) was formed on the 5′ss RNAs (possible penta-snRNP [PS] complex) (Fig. 1B, lanes 1 and 3). The assembly of these two complexes with similar mobilities on the two 5′ss RNAs indicates that the complex formation is specific for the 5′ss conserved sequence and that the exon sequence, which differs in the two 5′ss RNAs, is not essential for complex formation. We verified the specificity of the complex for the 5′ss conserved sequence by using Adeno mutated at the three most conserved positions of the 5′ss: −1, +1, and +2 (Fig. 1A); these mutations eliminated the formation of the complex (Fig. 1B, lanes 5 to 8). To examine the role of U1, we cleaved the 5′ end of U1 snRNA in the nuclear extracts by treating the nuclear extracts with RNase H directed by an antisense DNA oligonucleotide complementary to the 5′ end of U1 snRNA (positions 1 to 14). When the two 32P-labeled 5′ss RNAs were incubated in treated nuclear extracts, no complex was formed on the labeled 5′ss RNAs (Fig. 1B, lanes 2 and 4), indicating that the formation of the complex or its binding to labeled 5′ss RNAs requires the 5′ end of U1 snRNA.
RNA-RNA base-pairing interactions of snRNAs with the 5′ss RNA within the PS complex were examined by using cross-linking with psoralen (4, 57). 32P-labeled Adeno and OligoA were incubated in nuclear extract as in the experiment shown in Fig. 1B, psoralen was added, and the reaction mixtures were irradiated at 365 nm. The RNA was purified and separated in a denaturing gel. Figure 1C shows the region of the gel in the size range of 140 to 200 nucleotides. Inasmuch as the labeled 5′ss RNAs are 16 nucleotides long, the ∼180-nucleotide band observed (Fig. 1C, lanes 1 and 3) must be a labeled 5′ss RNA cross-linked to a longer molecule. As shown in Fig. 2, this cross-link was mapped to between the uridine at position 10 of U1 snRNA and the uridine at position −2 of the 5′ss RNA and thus was designated U1/5′ss. When OligoA and Adeno were incubated in nuclear extracts in which the 5′ end of U1 snRNA was removed, U1/5′ss cross-linking was 84 and 92% reduced, respectively (Fig. 1C, lanes 2 and 4) (quantification was done by phosphorimager analysis). To confirm the specificity of U1/5′ss cross-linking to the 5′ss conserved sequence, we again used the Adeno mutant. All of the mutations in the conserved sequence of Adeno diminished or totally abolished U1/5′ss cross-link formation (Fig. 1C, lanes 5 to 8). Interestingly, the mutation of G to C at position +1 decreased U1/5′ss cross-linking by 82%, while the same mutation eliminated the PS complex (compare lanes 5 in Fig. 1C and B). The mutation presumably destabilized the binding of U1 to the 5′ss within the PS complex, although a certain level of U1/5′ss base pairing was detected. The overall effect of these mutations indicates that the U1/5′ss cross-links are specific for the 5′ss conserved sequence. Furthermore, the U1/5′ss cross-link reached a maximum level at 0.3 pmol of Adeno/μl, which is also the concentration of U1 in the splicing reaction mixture (reference 18 and data not shown).
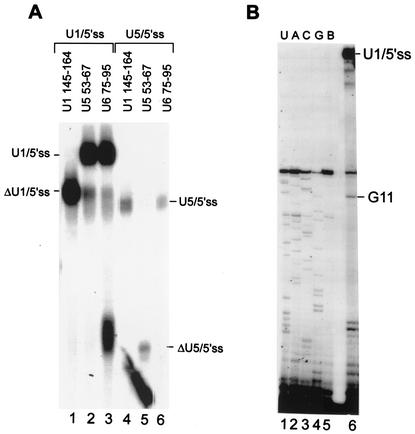
FIG. 2.
The U1/5′ss cross-link occurs at position 10 of U1 snRNA. (A) Cross-links of U1/5′ss and U5/5′ss RNAs were eluted from the gel, treated with the indicated antisense DNA oligonucleotides, and then RNase H digested. The RNA was separated by denaturing 6% PAGE. The partial digestion of the U1/5′ss cross-link by U6 75- 95 is probably nonspecific. (B) The RNA of the U1/5′ss cross-link was eluted from the gel and used as a template for reverse transcription primed by 5′-end-labeled DNA oligonucleotide U164-75 (lane 6). A block to the extension of the primer was observed at position 11 of U1 snRNA. The sequencing ladder shown on the left was generated with the same primer in the presence of the appropriate dideoxynucleoside triphosphates (ddNTPs) (lanes 1 to 4) or in the absence of ddNTPs (lane 5; B, blank).
To confirm that the U1/5′ss cross-link occurs within the PS complex, we performed the same reaction as that shown in Fig. 1B, except that the complex was psoralen cross-linked prior to being resolved in the native agarose gel. The RNA from the cross-linked PS complex was eluted and separated in a denaturing gel. The U1/5′ss cross-link was detected within the PS complex (Fig. 1D, lanes 2 and 4), indicating that U1/5′ss base pairing occurs within the PS complex. These results show that when the 5′ss RNA was added to nuclear extracts, it was assembled within the PS complex through base pairing between the 5′ end of U1 and the 5′ss RNA in an interaction specific for the 5′ss conserved sequence.
Next we examined which snRNPs are present in the PS complex that forms on the 5′ss RNA. We eluted the RNA from the gel region containing the PS complex and performed a Northern blot analysis with 32P-labeled antisense riboprobes complementary to each of the five snRNAs: U1, U2, U4, U5, and U6. The PS complex gel region contained all five snRNAs in almost stoicheometric amounts (Fig. 1E, lane 2). However, when nuclear extracts were incubated under splicing conditions in the absence of 5′ss RNA and the RNA was eluted from the same region of the agarose gel as the PS complex, all five snRNAs were also observed (Fig. 1E, lane 3). Interestingly, when we used nuclear extracts in which the 5′ end of U1 snRNA had been removed by RNase H cleavage, there were no snRNAs in the same region of the agarose gel as the PS complex (Fig. 1E, lane 4). A control region located above the PS complex of the agarose gel was also devoid of snRNAs (Fig. 1E, lane 5). These results suggest that all five snRNPs comigrate in the PS complex position of the gel in the absence of added 5′ss RNA; that the PS complex can interact with the 5′ss through base pairing with the 5′ end of U1 snRNA; and that the integrity of this complex requires the 5′ end of U1 snRNA.
The U1/5′ss cross-link occurs between position 10 of U1 and position −2 of the OligoA exon.
To identify the cross-link species observed in Fig. 1C (U1/5′ss) and the U5/5′ss cross-link (to be discussed later), the RNAs of the U1/5′ss (OligoA) and U5/5′ss (OligoA) cross-links were eluted from the denaturing gel and treated with RNase H directed by antisense DNA oligonucleotides complementary to specific regions on U1, U5, or U6 snRNAs. RNase H cleavage with the U1 145-164 antisense oligonucleotide (U1 positions 145 to 164) but not the U5 and U6 antisense oligonucleotides affected the U1/5′ss cross-link by changing its mobility in the gel (Fig. 2A, compare lane 1 to lanes 2 and 3), indicating that this cross-link is between U1 snRNA and OligoA. RNase H cleavage with the U5 53-67 antisense oligonucleotide but not the U1 and U6 antisense oligonucleotides affected the U5/5′ss cross-link by changing its mobility in the gel (Fig. 2A, compare lane 5 to lanes 4 and 6), indicating that this cross-link is between U5 snRNA and OligoA.
We then undertook localization of the precise position on U1 that cross-links to OligoA. The cross-link reaction mixture was separated by denaturing 12% PAGE, the cross-link was eluted, and the RNA was used as a template for reverse transcription primed by a 32P-labeled DNA oligonucleotide complementary to U1 positions 64 to 75. A block to the extension of the primer was observed at position 11 of U1 snRNA (Fig. 2B, lane 6). Inasmuch as the extension stops 1 nucleotide before the cross-link site, the block at position 11 of U1 pinpoints the cross-link site to the uridine at position 10 of U1.
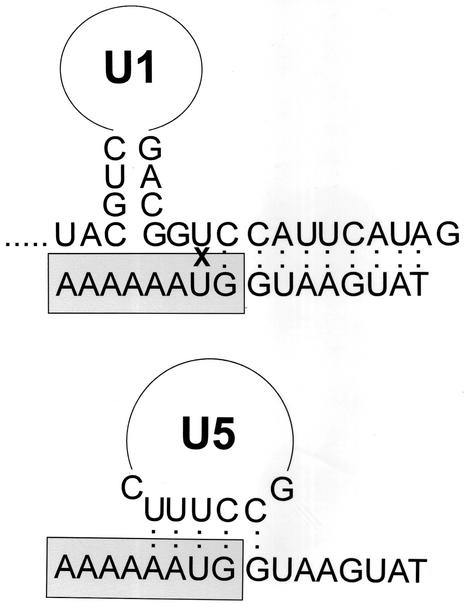
The precise nucleotide on OligoA that cross-links to U1 was studied by RNase T1 mapping and reversal cross-linking at 254 nm of the U1/5′ss and U5/5′ss cross-links. The results indicated that both U1 and U5 cross-linked to the exon portion of OligoA (data not shown). Inasmuch as psoralen usually cross-links between uridines (4, 57), these results suggest that the precise nucleotide on OligoA that cross-links to U1 and to U5 is the only uridine in the exon sequence of OligoA located at position −2 (Fig. 3). RNase T1 mapping of the U5/5′ss cross-link demonstrated that an unspecified nucleotide between positions 25 and 50 of U5 cross-links to OligoA (data not shown). This region contains the invariant loop of U5 which was shown to base pair with positions −1 and −2 of the 5′ss (42, 51, 59).
FIG. 3.
Proposed schematic representation of the U1/5′ss and U5/5′ss base-pairing interactions. The U1/5′ss cross-link site is indicated by X. Boxed and nonboxed sequences represent exon and intron regions, respectively.
The PS complex accommodates RNA-RNA rearrangements.
During the splicing reaction, U5 and U6 replace U1 at the 5′ss. Thus, we examined whether progress from the stage of early binding of U1 to the 5′ss to the stage at which U5 replaces U1 and base pairs with positions −1 and −2 of the 5′ss also occurs in the PS complex.
The complex assembled on the 5′ss RNA was tested by using the technique developed by Konarska et al. (28). The 5′ end of U1 snRNA was sequestered by preincubating nuclear extracts with U11-14 prior to the addition of the 5′ss RNA. Konforti et al. demonstrated that sequestering the 5′ end of U1 decreases U1/5′ss binding, leading to the binding of U6 at positions +4 to +6 (the intron portion) of the 5′ss RNA (28) and U5(p220) protein to the GU dinucleotide at positions +1 and +2 (46, 47). These authors used an 11-mer 5′ss RNA oligonucleotide spanning three exon and eight intron nucleotides. Thus, we examined whether sequestering the 5′ end of U1 snRNA by DNA oligonucleotide U11-14 can induce the replacement of U1 by U5 at the 5′ss RNA. Inasmuch as the base-pairing interaction between U5 snRNA and the 5′ss occurs between uridines at the invariant loop of U5 and position −2 of the 5′ss (Fig. 3, lower panel), we used OligoA, which contains a single uridine in the exon sequence located at position −2. This strategy allowed us to capture the U5/5′ss base pairing by cross-linking with psoralen.
Nuclear extracts were preincubated with increasing concentrations of U11-14, 32P-labeled OligoA was added, the reaction mixture was cross-linked with psoralen, and the purified RNA was separated in a denaturing gel. There was a reduction in U1/5′ss cross-linking as the concentration of U11-14 was increased, and a new cross-link identified as U5/5′ss appeared (Fig. 4A). The U5/5′ss cross-link appeared at 0.32 pmol of U11-14/μl, peaked at 0.64 pmol/μl, and declined when higher U11-14 concentrations were added (Fig. 4A and graph in Fig. 4C). The addition of Adeno above the concentration of U1 in the binding reaction mixture (∼0.3 pmol/μl) (18) did not lead to the formation of the U5/5′ss cross-link (data not shown). However, this result does not rule out the possibility that the decrease in U5/5′ss cross-linking with higher U11-14 concentrations is an indirect consequence of the overall decrease in recognition by U1 to start with or a nonspecific effect of too much DNA in the reaction mixture (see Discussion).
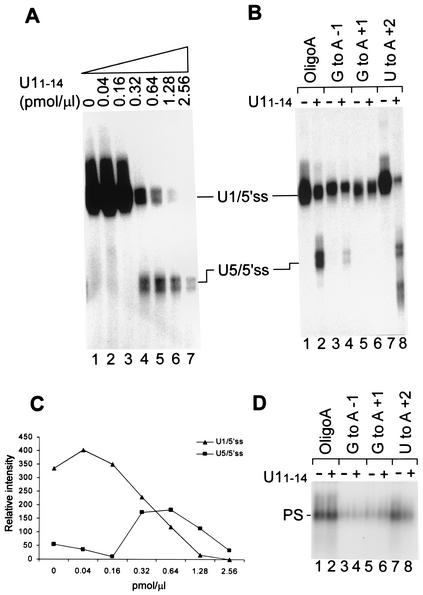
FIG. 4.
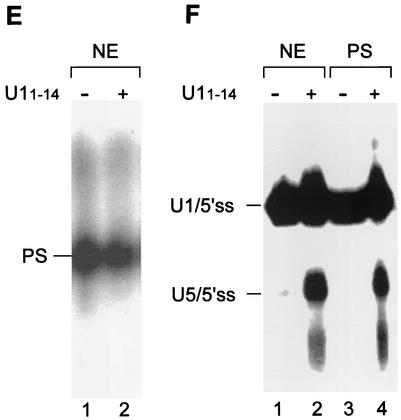
U5 replaces U1 at the 5′ss RNA. (A) Nuclear extracts were preincubated with increasing concentrations of U11-14, followed by the addition of 32P-labeled OligoA. The reaction mixtures were further incubated for 15 min at 4°C and psoralen cross-linked, and the RNA was purified and separated by denaturing 6% PAGE. (B) 32P-labeled OligoA mutated at the three most conserved nucleotides of the 5′ss was incubated with nuclear extracts that had been preincubated in the absence or presence of 0.64 pmol of U11-14/μl. The reaction mixtures were cross-linked with psoralen, and the RNA was purified and separated by denaturing 6% PAGE. (C) Quantification of the U1/5′ss and U5/5′ss cross-links in increasing concentrations of U11-14 as shown in panel A. Quantification was done by phosphorimager analysis with TINA software. (D) 32P-labeled OligoA mutated at the same nucleotides as in panel B was added to nuclear extracts that had been preincubated in the presence or absence of U11-14, and the reaction mixtures were separated in a native 0.75% agarose gel. (E) The PS complex accommodates the replacement of U5 by U1 at the 5′ss RNA. Nuclear extracts (NE) were preincubated in the presence or absence of 0.64 pmol of U11-14/μl, 32P-labeled OligoA was added, and the reaction mixtures were further incubated for 15 min at 4°C and separated in a 0.75% agarose gel. (F) Similar to panel E, except that the reaction mixtures were psoralen cross-linked prior to separation in a 0.75% agarose gel. The RNA from the PS complex was eluted, purified, and separated by denaturing 6% PAGE (lanes 3 and 4). Lanes 1 and 2, total reactions in which OligoA was incubated in nuclear extracts in the absence or presence of 0.64 pmol of U11-14/μl.
The specificity of the U5/5′ss interaction with the 5′ss conserved sequence was verified by mutating OligoA at the three most conserved positions: −1, +1, and +2. When the labeled mutated 5′ss RNAs were incubated in nuclear extracts that had been preincubated with U11-14, the U5/5′ss cross-link was diminished or abolished completely: the G-to-A mutation at position −1 diminished the U5/5′ss cross-link by 90% (Fig. 4B, compare lane 4 to lane 2), the G-to-A mutation at position +1 completely abolished the U5/5′ss cross-link (Fig. 4B, compare lane 6 to lane 2), and the U-to-A mutation at position +2 diminished the U1/5′ss cross-link by 65% (Fig. 4B, compare lane 8 to lane 2). The results show that U11-14 induces the replacement of U1 by U5 at the 5′ss RNA and that this replacement is specific for the 5′ss conserved sequence.
To explore the effects of mutated OligoA on PS complex assembly, HeLa cell nuclear extracts were incubated in the absence or presence of U11-14, 32P-labeled OligoA was added, and the reaction mixtures were separated in a native 0.75% agarose gel (15). All of the mutations decreased the formation of the PS complex by an average of 85% (Fig. 4D). One exception was the U-to-A mutation at position +2 which, when incubated in the absence of U11-14, reduced the formation of the PS complex by only 40% (Fig. 4D, lane 7). These results indicate that the formation of this complex requires the 5′ss conserved sequence.
Interestingly, we observed different effects of the mutations at positions −1 to +2 of OligoA and Adeno on U1/5′ss cross-linking and on PS complex formation. Mutations at positions −1 to +2 of Adeno reduced U1/5′ss cross-linking by more than 85%. The same mutations in OligoA resulted in only 60 and 75% reductions for G-to-A mutations at positions −1 and +1, respectively (compare Fig. 1C to 4B), and a 15% reduction for the U-to-A mutation at position +2 of OligoA versus a 99% reduction for the same mutation in Adeno (lanes 7 in Fig. 4B and 1C, respectively). Similarly, a mutation at position +2 of Adeno eliminated the formation of the PS complex, compared to the 40% reduction obtained with the same mutation in OligoA (lanes 7 in Fig. 1B and 4D, respectively). Inasmuch as the only difference between Adeno and OligoA is the exon sequence, these findings may mean that different levels of the U1/5′ss cross-link are affected by the exon sequence. That difference may be related to the binding of U1 snRNP proteins, such as U1(70K), SmD1, and SmD3, upstream of the 5′ss and their effect on the stabilization of 5′ss (62, 63). However, it may also be due to different efficiencies of U1 and U5 cross-linking to the different mutants.
To examine whether the replacement of U1 by U5 occurs within the PS complex, we separated the reaction mixture containing the U1/5′ss interaction in a native agarose gel. 32P-labeled OligoA was incubated in nuclear extracts that had been preincubated in the presence or absence of 0.64 pmol of U11-14/μl. The reaction mixtures were psoralen cross-linked and separated in a native 0.75% agarose gel. The PS complex assembled on the 5′ss RNA in the presence or absence of U11-14 reached to the same position in the gel (Fig. 4E). To detect the cross-linked RNA within the PS complex, we eluted and purified the RNA from the complex and separated it in a denaturing gel. The PS complex assembled on OligoA contained the U1/5′ss cross-link in the absence of sequestering of the U1 5′ end (Fig. 4F, lane 3), and the U1/5′ss and U5/5′ss cross-links were detected after sequestering of the U1 5′ end (Fig. 4F, lane 4). Hence, the replacement of U1 by U5 at the 5′ss RNA occurs within the PS complex, suggesting that U1 binding to the 5′ss is replaced by U5 within the PS complex.
To study the roles of U2 and U6 in the formation of the PS complex, we cleaved the 5′ end of U2 snRNA or positions 43 to 60 of U6 snRNA in the nuclear extracts by treating the nuclear extracts with RNase H directed by an antisense DNA oligonucleotide. Ninety-eight percent removal of the U1 snRNA 5′ end resulted in no PS complex formation in the absence of U11-14, as well as 92% inhibition of the PS complex when the digested nuclear extracts were preincubated with U11-14 (compare Fig. 5A, lan e 2, to Fig. 5B, lanes 3 to 6). However, 89% removal of the U2 snRNA 5′ end (Fig. 5A, lane 3) resulted in 35 and 72% reductions in PS complex formation in the absence and presence of preincubation of the digested nuclear extracts with U11-14, respectively (Fig. 5B, lanes 7 and 8). Furthermore, digestion of the nuclear extracts with the U6 43-60 antisense DNA oligonucleotide resulted in 60% digestion of U6 snRNA but also in 25 to 47% reductions in the levels of U1, U2, U4, and U5 snRNAs (Fig. 5A, lane 4). These digested nuclear extracts supported 82 and 48% assembly of the PS complex in the absence and presence of preincubation of those nuclear extracts with U11-14, respectively (Fig. 5B, lanes 9 and 10). These results suggest that digestion of specific regions of U2 and U6 snRNAs has a modest effect on the initial binding of U1 to the 5′ end of U1 within the PS complex but that, following a presumably conformational change from U1/5′ss to U5/5′ss binding, both of these regions of U2 and U6 snRNAs are essential for the stability of the PS complex. It is unclear whether U2 and U6 remain as intact particles after site-specific digestion.
FIG. 5.
Complexity of U1 and U5 binding to OligoA. (A) Site-specific digestion of U1, U2, and U6 snRNAs in nuclear extracts. U1, U2, and U6 snRNAs were digested by antisense DNA oligonucleotide-directed RNase H cleavage. Mock, same treatment, but in the absence of the antisense DNA oligonucleotide. Total treated nuclear extract RNA (5 μl) was separated by denaturing 10% PAGE and tested by Northern blotting with radiolabeled antisense riboprobes complementary to U1, U2, U4, U5, and U6 snRNAs. A longer exposure of the U5 and U6 regions of the gel is shown at the bottom. (B) Effect of selective digestion of U1, U2, and U6 snRNAs on PS complex assembly. Treated nuclear extracts from panel A were preincubated in the absence (−) or presence (+) of U11-14. Then, end-labeled OligoA was added and incubation was continued for another 20 min at 4°C, followed by separation in a 0.75% agarose gel. (C) Analysis of binding reaction mixtures by native 4% PAGE. Splicing reaction mixtures containing labeled Adeno pre-mRNA were incubated at 30°C for 15 and 2 min (lanes 1 and 2, respectively). Also, binding reaction mixtures were incubated at 4°C for 20 min in the absence and in the presence of U11-14 (lanes 3 and 4, respectively). The reaction mixtures were separated by native 4% PAGE. The positions of spliceosomal H, A, and B/C complexes are shown on the left. The positions of OligoA X and Y complexes are shown on the right. (D) Specificity of the U5/5′ss cross-link for preincubation with U11-14. Nuclear extracts were incubated with the indicated cDNA oligonucleotides. Following incubation with end-labeled OligoA, the reaction mixtures were cross-linked with psoralen, and the RNA was purified and separated by denaturing 6% PAGE. (E) U5(p220) cross-links downstream of the 5′ss. Nuclear extracts were incubated in the absence and in the presence of U11-14 (U1/5′ss and U5/5′ss, respectively). Following incubation with 5′-end-labeled OligoA, the reaction mixtures were cross-linked at 254 nm and then immunoprecipitated either with anti-Sm, anti-U1(70K), or anti-U5(p220) antibodies or no antibodies (mock). Half of each precipitated reaction mixture was digested with RNase T1 (indicated by a plus sign; 25 U, 1 h, 30°C), and then the reaction mixtures were separated by sodium dodecyl sulfate-8% PAGE. Lanes 21 and 22 are longer exposures of lanes 11 and 16, respectively. The sizes of the molecular weight (MW) markers are shown on the left (in thousands).
We also examined the types of complexes assembled on OligoA when the reaction mixtures were separated by native 4% PAGE analysis instead of native agarose gel analysis. A rapidly migrating complex was replaced by a slowly migrating complex when the binding reaction mixtures were incubated in the absence or presence of U11-14 (marked as Y and X, respectively) (Fig. 5C, lanes 3 and 4). The X complex migrates slightly faster than spliceosomal complexes B and C (Fig. 5C, lanes 1 and 3). This result indicates both that OligoA is assembled within a multi-snRNP complex(s) after incubation in the presence of U11-14 and that the formation of this complex can take place at 4°C. The X complex is presumably the U2/U4/U5/U6/5′ss RNA complex (reference 28 and data not shown). The Y complex migrates in the region of U1 in the gel. This mobility is only slightly slower than the mobility of U1 snRNA alone (data not shown). This result suggests that the conditions used for the separation of U1 snRNP in the native 4% acrylamide gel destabilize U1 snRNP assembly as well as U1 binding to other snRNPs. This destabilization effect is presumably due to the use of heparin in the sample buffer and the nonphysiological pH used in the running buffer (Tris-glycine) (26).
We have also shown that the formation of the U5/5′ss cross-link complex is specific for the sequestering of the 5′ end of U1 snRNA by antisense DNA oligonucleotides but not for the sequestering of specific regions of U2, U4, or U6 snRNA (Fig. 5D). Interestingly, sequestering of the 5′ end of U1 snRNA by DNA oligonucleotides that are complementary to either positions 1 to 8 or positions 1 to 10 induced U5/5′ss cross-linking in a fashion similar to that of U11-14 (data not shown). In addition, preincubation of nuclear extracts with U11-14 has no effect on the integrity of the 5′ end of U1 snRNA when incubation is done at 4°C. However, sequestering of the 5′ end of U1 at 4°C and then incubation of the binding reaction mixture at 30°C have an outcome similar to that of incubation at 4°C in terms of U1/5′ss and U5/5′ss cross-link formation. Incubation at 30°C leads to the degradation of OligoA after 15 min of incubation, presumably by endogenous nucleases which are activated at 30°C but which are inactive at 4°C (data not shown). Therefore, we used incubation at 4°C, which prevents OligoA degradation and allows the assembly of complexes identified on native gels and the formation of U1/5′ss and U5/5′ss base-pairing interactions.
To examine whether U1/5′ss and U5/5′ss cross-links reflect either simple antisense-snRNA base-pairing interactions of complementary sequences or elaborate interactions in which specific proteins of U1 and U5 also bind the 5′ss RNA, we examined the latter by using cross-linking at 254 nm, which allows the identification and characterization of protein-RNA interactions. Therefore, nuclear extracts were incubated in the absence or presence of U11-14 (Fig. 5E; U1/5′ss indicates incubation in the absence of U11-14, while U5/5′ss indicates incubation in the presence of U11-14). We used conditions for U5/5′ss similar to those in Fig. 4A, lane 6, to ensure that U5/5′ss base pairing would constitute the major complex within the reaction. Following the addition of 5′-end-labeled OligoA and incubation for 20 min at 4°C, the reaction mixtures were immunoprecipitated with various antibodies. In OligoA, the first site which is digested by RNase T1 (cuts after a guanosine), downstream of the 5′-end label, is at the 5′ss. To differentiate among proteins that cross-link to the exon or intron portion of OligoA, half of each precipitated reaction mixture was digested with RNase T1. The precipitated proteins were then separated by sodium dodecyl sulfate-PAGE, and the cross-linked proteins were detected by autoradiography. We showed that two different sets of proteins bind the 5′ss RNA oligonucleotide when either U1 or U5 snRNA base pairs with the 5′ss RNA. Three U1-associated proteins cross-link to the 5′ss RNA (170, 62, and 68 kDa) (Fig. 5E, lane 14). They are replaced by a set of Sm-associated proteins that cross-link to the 5′ss RNA when U5 base pairs with the 5′ss exon (220, 200, and 55 kDa) (Fig. 5E, lane 3). One of these proteins is U5(p220), which cross-links downstream from the 5′ss RNA (Fig. 5E, compare lanes 5 and 10), presumably to the GU dinucleotide (47). However, it is unclear whether the p200 precipitated by the anti-Sm antibodies is U5(p200) (Fig. 5E, lanes 3 and 8) (30, 31). The complex precipitated by the anti-Sm antibodies probably prevents RNase T1 from digesting OligoA. These results can explain why, within the total reaction, the p220 and p200 cross-links which were labeled before RNase T1 digestion became unlabeled after that digestion but remained labeled after RNase T1 treatment of the Sm-precipitated reaction mixture (Fig. 5E, compare lanes 1, 6, and 8). These results indicate that U5 snRNP binds simultaneously across the 5′ss; the invariant loop of U5 snRNA base pairs to the last nucleotides of the exonic portion of the 5′ss, and the U5 snRNP protein, p220 and perhaps also p200, bind to the intronic portion of the 5′ss.
The U1/5′ss and U5/5′ss cross-links sediment as a 200S complex in glycerol gradients together with all five snRNPs.
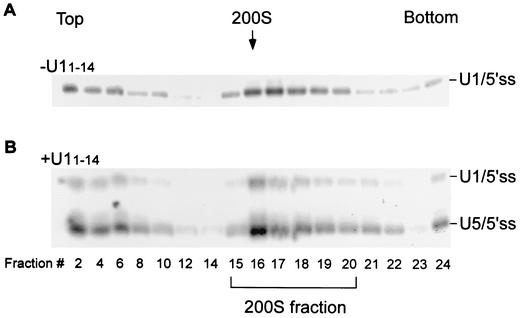
To examine the homogeneity of the complex formed on the 5′ss, we undertook a two-step separation approach by using sedimentation in glycerol gradients followed by affinity selection. Nuclear extracts were incubated in the absence or presence of U11-14 (Fig. 6A and B, respectively), 32P-labeled OligoA was added, and the reaction mixtures were cross-linked with psoralen and loaded on top of a 10 to 50% glycerol gradient (58). Following centrifugation, each gradient was divided into 24 fractions, and the RNA from each fraction was purified and separated in a denaturing gel. About 65% of the U1/5′ss and U5/5′ss cross-links remained at the top of the gradient (Fig. 6A and B, fractions 2 and 4), but about 35% of the U1/5′ss (Fig. 6A) and both U1/5′ss and U5/5′ss (Fig. 6B) cross-links sedimented as a peak around the 200S region (fractions 15 to 20). The cosedimentation of U1/5′ss and U5/5′ss cross-links within the 200S region further supports the observed similarities between complexes in which U5 and U1 bind the 5′ss.
FIG. 6.
The U1/5′ss cross-link sediments as a 200S complex in glycerol gradients together with all five spliceosomal snRNPs. Nuclear extracts were incubated in the absence (A) or presence (B) of U11-14. 32P-labeled OligoA was added, and the reaction mixtures were further incubated for 15 min at 4°C under binding conditions. Following cross-linking with psoralen, the reaction mixtures were loaded on a 10 to 50% glycerol gradient and centrifuged for 12 h at 4°C. The gradient was divided into 24 fractions; the RNA was purified and separated by denaturing 6% PAGE. One-fifth of the RNA of fractions 2 and 4, with respect to the other fractions, was loaded on the gel. The 200S position was determined by sedimentation of TMV in a parallel gradient. (C) Gradients similar to those in panels A and B, except that biotinylated OligoA was used and the reaction mixtures were not cross-linked prior to sedimentation. Fractions 15 to 20 (200S) were combined, dialyzed against nuclear extract buffer D, and affinity selected on Streptavidin-agarose beads. The selected RNA was purified, separated by denaturing 10% PAGE, and Northern blotted with radiolabeled antisense riboprobes complementary to U1, U2, U4, U5, and U6 snRNAs. (D) The 200S fractions containing U1/5′ss cross-links (lane 2) or U1/5′ss and U5/5′ss cross-links (lanes 1 and 3) were immunoprecipitated with either anti-U1(70K) antibodies or no antibodies (lane 1). The immunoprecipitated RNA was separated by denaturing 5% PAGE. One-fifth of the RNA in lane 2, with respect to the other lanes, was loaded on the gel.
The 200S complex was previously identified by Sperling et al. as a large RNP complex released from mammalian cell nuclei containing all five snRNPs and many proteins required for RNA processing (2, 39, 40, 52, 61). Wassarman and Steitz (58) demonstrated that RNP complexes of HeLa cell nuclear extracts incubated under splicing conditions in the absence of a substrate sedimented at >150S in a glycerol gradient (peaks at fraction 15). Using the same glycerol gradient conditions as Wassarman and Steitz (58), we identified the peak of the U1/5′ss cross-link in the same fraction region as the >150S complexes (peaks at fraction 16) and demonstrated that the peak of tomato mosaic virus (TMV), which was used as a 200S marker, sedimented at fraction 16. These results indicate that the >150S RNP complexes (58) and the U1/5′ss complex are probably identical RNP complexes. They also suggest that the native 200S large nuclear ribonucleoprotein (lnRNP) complex (52) and the nuclear extract >150S complex (58) are identical.
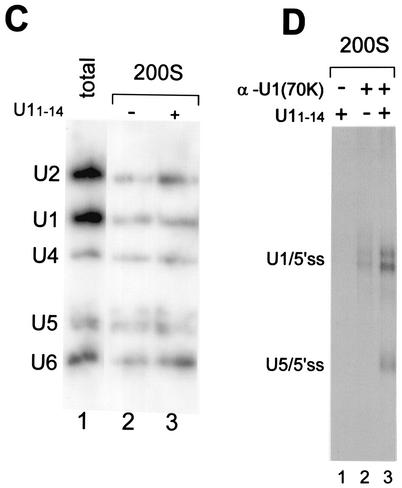
The next step was to identify the snRNPs within the 200S fraction containing either U1/5′ss or both U1/5′ss and U5/5′ss complexes. A two-step separation was used to purify the complexes. Nuclear extracts were incubated in the absence or presence of U11-14 under binding conditions, biotinylated OligoA was added, the reaction mixtures were separated in a glycerol gradient, and the 200S fraction was collected and dialyzed against buffer D (16). In the second step, the 200S fraction was affinity selected by using Streptavidin-agarose beads, and the selected RNA was purified and examined by Northern blot analysis with 32P-labeled antisense riboprobes against the five snRNAs: U1, U2, U4, U5, and U6. The U1/5′ss cross-link complex of the 200S fraction was shown to contain all five spliceosomal snRNPs in stoichiometric amounts (Fig. 6C, lane 2). In addition, affinity selection of the 200S fraction containing both U1/5′ss and U5/5′ss cross-links revealed the five spliceosomal snRNPs in stoichiometric amounts (Fig. 6C, lane 3). These results show that U1 base paired with the 5′ss within a penta-snRNP complex that was part of the 200S particles. The 200S fraction that contained both U1/5′ss and U5/5′ss cross-links also showed the same profile of snRNA as the 200S fraction containing the U1/5′ss cross-link alone. Both labeled U1/5′ss and U5/5′ss cross-links were affinity selected from the 200S fraction (data not shown), suggesting that the U5/5′ss and U1/5′ss complexes have similar amounts of the snRNP attached. Also, nonbiotinylated OligoA did not precipitate any snRNAs, indicating the specificity of that selection (data not shown).
The possibility that U1 remains attached to the U5/5′ss complex was examined by an immunoprecipitation assay. The 200S fraction (from Fig. 6A and B) was immunoprecipitated with antibodies directed against the U1-specific protein 70K [anti-U1(70K)]. The RNA was purified and separated by denaturing PAGE. Both U1/5′ss and U5/5′ss cross-links were immunoprecipitated when the reaction mixture was incubated with anti-U1(70K) (Fig. 6D, lane 3), indicating that U1, or at least U1(70K), remains associated with the complexes containing the U5/5′ss cross-link.
The 200S fraction is functional in spliceosome assembly and in U1-to-U5 replacement.
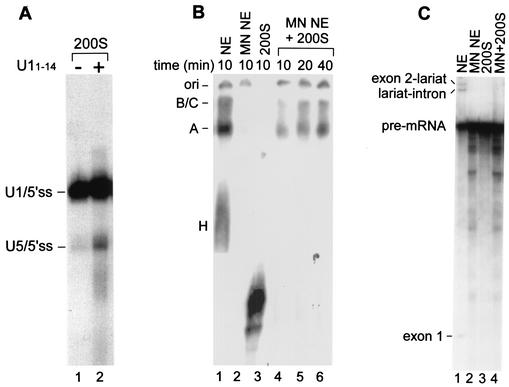
To examine whether the U1/5′ss complex can advance to the U5/5′ss complex, we performed a chase experiment. Nuclear extracts were preincubated under binding conditions with labeled OligoA, followed by separation in a 10 to 50% glycerol gradient. The 200S fraction was collected and dialyzed against buffer D (16). One-half of the purified 200S fraction was cross-linked with psoralen, and the other half was first incubated with U11-14 for 45 min at 4°C and then cross-linked with psoralen. The RNA was purified and separated in a denaturing gel (Fig. 7A). The U1/5′ss complex was able to advance to the U5/5′ss complex following incubation with U11-14, indicating that the 5′ss RNA can advance from U1 to U5 binding within the 200S fraction and that a factor(s) essential for the replacement process is present in this fraction.
FIG. 7.
U1/5′ss and U5/5′ss cross-links in the 200S fraction support spliceosome assembly. (A) Nuclear extracts were incubated under binding conditions in the presence of labeled OligoA, followed by separation in a 10 to 50% glycerol gradient as described in the legend to Fig. 6. The 200S region was dialyzed against nuclear extract buffer and concentrated. The dialyzed 200S fraction was incubated for 45 min at 4°C in the absence and in the presence of U11-14 (lanes 1 and 2, respectively) and cross-linked with psoralen, and the RNA was purified and separated by denaturing 6% PAGE. (B) Full-length and labeled Adeno pre-mRNAs were incubated under in vitro splicing conditions at 30°C for the indicated times. Nuclear extracts (NE) were replaced with MN-treated NE (lane 2), the 200S fraction (lane 3), or the 200S fraction supplemented with MN-treated NE (lanes 4 to 6). The reaction mixtures were separated by native 4% PAGE. The positions of H, A, and B/C complexes are shown on the left. The origin of the gel (ori) is indicated. (C) Adeno pre-mRNA was incubated under in vitro splicing conditions at 30°C for 60 min. Nuclear extracts (NE) were replaced by MN-treated NE (lane 2), the 200S fraction (lane 3), or the 200S fraction supplemented with MN-treated NE (lane 4). The RNA was extracted and separated in a denaturing 8% gel. The positions of splicing intermediates and pre-mRNA are shown on the left.
Finally, the ability of the 200S fraction to associate with pre-mRNA and catalyze pre-mRNA splicing was assayed by using standard in vitro splicing conditions. Nuclear extracts were preincubated under binding conditions in the absence of a substrate followed by separation in a 10 to 50% glycerol gradient. The 200S fraction was collected and dialyzed against buffer D (16). The purified 200S fraction was incubated under in vitro splicing conditions with full-length and labeled Adeno pre-mRNAs. The reaction mixture was separated by native 4% PAGE (Fig. 7B), or the RNA was purified and separated in a denaturing gel (Fig. 7C). The 200S fraction alone did not support spliceosomal complex assembly or pre-mRNA splicing (lanes 3 in Fig. 7B and C). The addition of micrococcal nuclease (MN)-treated nuclear extracts, themselves unable to splice (Fig. 7C, lane 2), to the 200S fraction supported spliceosome A and spliceosome B/C complex assembly but not mRNA splicing (Fig. 7B, lanes 4 to 6, and Fig. 7C, lane 4, respectively). Although in a longer exposure of the gel a low level of mRNA splicing activity was detected (data not shown), the ability of the 200S fraction to support spliceosome assembly but not mRNA splicing might be attributed to the low concentrations of proteins in the 200S fraction (0.5 mg/ml versus 6 mg/ml in the nuclear extracts) or to the lack of an essential factor(s) in the 200S fraction and the MN-treated nuclear extracts; alternatively, the assembly of external pre-mRNA into the 200S particles is somehow impaired if not coupled with the transcription process. The ability of the 200S fraction to support spliceosome complex assembly when supplemented with soluble nuclear proteins indicates that the snRNPs within the 200S fraction are functional in spliceosome assembly.
DISCUSSION
We have shown that incubation of HeLa cell nuclear extracts in in vitro binding conditions results in association of the endogenous snRNP particles to a stable complex composed of U1, U2, U4, U5, and U6. This complex specifically binds the 5′ss RNA through base pairing with the 5′ end of U1 and advances to U5 binding following sequestering of the 5′ end of U1. These results suggest that U1 approaches the 5′ss as part of a penta-snRNP complex and, following replacement of U1 by U5 at the 5′ss, U1 presumably remains attached to the complex.
The 5′ss RNA is assembled into a large complex that contains all five snRNPs and that sediments at the 200S region in glycerol gradients. This complex is probably identical to the 200S lnRNP complex that can be released from mammalian nuclei, and its size was found to be independent of the length and number of introns in the RNA it packages (40, 52). It is also presumably identical to the >150S complex found in mammalian cell nuclear extracts incubated under splicing conditions in the absence of a substrate (58) and is consistent with the latest observations of the penta-snRNPs complex formed in yeast extracts under in vitro splicing conditions (54). The results presented here indicate that the 200S complex that forms in mammalian cell nuclear extract and that contains the five spliceosomal snRNPs can bind the 5′ss via sequence-specific base pairing with the 5′ end of U1 snRNA, is able to advance to U5/5′ss binding, and supports spliceosome assembly when supplemented with soluble nuclear proteins; thus, it may be a holospliceosome.
The 5′ss RNA was shown in previous studies to bind U1 snRNP and to induce a U2/U4/U5/U6 complex in HeLa cell nuclear extracts following sequestering of the 5′ end of U1 snRNA by U11-14 (18). However, the complexes were resolved by native PAGE, which causes destabilization of U1 from all of the complexes resolved by this gel (Fig. 5C) (26). We prevented the dissociation of U1 from the complexes by using the technique of Das and Reed (14) for native separation in which the splicing complexes are separated in a native agarose gel without the addition of heparin. We found that the complex that contained heparin migrated faster than the complex that was separated without heparin (data not shown).
When the 5′ end of U1 snRNA was removed, the U1/5′ss cross-link was significantly diminished. The small amount of the U1/5′ss cross-link that is still seen is probably a result of a small amount of full-length U1 snRNA molecules (1 to 2%) that survived the RNase H activity. In this treated extract, the PS complex was not observed and the five snRNAs were not present in the region of the PS complex in the native agarose gel. Moreover, digestion of the U1 5′ end led to the disassembly of the 200S complex and sedimentation of the snRNP around the top of the gradient (fractions 1 to 9) (data not shown). This suggests that the PS complex and the mammalian cell nuclear extract >150S complex (58) are present in the nuclear extract as a penta-snRNP complex in the absence of substrate addition. However, the integrity of the PS complex requires the 5′ end of U1. This finding is also consistent with evidence that a 5′ truncated form of U1 is not present in the >150S complex (58). We anticipate that the discrepancy between the dispensability of the 5′ end of U1 for the assembly of the U2/U4/U5/U6 on the 5′ss RNA oligonucleotide (28) and the requirement of that end for maintaining the integrity of the preassembled penta-snRNP (58; this study) is due to the different separation techniques, or related to the splicing of certain pre-mRNAs in U1 snRNP-inactivated or -depleted extracts (12, 13, 55, 56). Another possibility is that the base pairing between the 5′ end of U1 and the DNA oligonucleotide, even for a short period before that RNase H removes that end, serves as a signal for the advance to U5 5′ss binding.
We cannot exclude the possibility that the PS complex consists of several different complexes that migrate to the same position in the native gel, each composed of a different combination of snRNPs that bind the 5′ss RNA or migrate to that position in the gel. However, increasing the gel percentage from 0.75 to 1.5% and even to 2% failed to lead to separation of the PS complex into several subcomplexes (data not shown). Thus, the U1/5′ss complex had to be isolated by a different technique that would address the homogeneity of the U1/5′ss complex. The two-step separation method—glycerol gradient followed by affinity selection of the 5′ss RNA within the 200S fraction—indicates that all five spliceosomal snRNPs are present in the U1/5′ss complex in stoichiometric amounts, suggesting that U1 approaches the 5′ss within a penta-snRNP complex. Mass measurements of the 200S lnRNP complexes revealed a similarity between the lnRNP particles and the uniformity of their subunits that contain all five spliceosomal snRNPs (41).
The binding reaction with the HeLa cell nuclear extracts and the simple RNA oligonucleotide containing a consensus 5′ss revealed that the base-pairing interaction with U1 can take place at 4°C as well as 30°C (Fig. 1C and data not shown). Interestingly, when yeast extracts were incubated with a 72-nucleotide transcript containing the entire 5′ exon and the first 49 nucleotides of the intron, the U1/5′ss cross-link with psoralen was specific to incubation at 25°C but not at 0°C; the U1/5′ss cross-link does form at both temperatures when U1-specific protein U1(C)-depleted extract is used (17). This suggests that the U1/5′ss interaction is in some way different in yeasts and humans or that other elements located farther away from the 5′ss of the 72-nucleotide transcripts affect the base-pairing interaction of U1 with the 5′ss in a way which depends on U1(C) (34). Such an effect might be attributed to the binding of a yeast CAP binding protein, yCBP80, to both the 5′ exon and U1 (63).
According to our results, the 5′ss RNA was packed into the 200S complex that contained the five spliceosomal snRNPs, in which U1 base pairs with the 5′ss RNA in a fashion similar to the PS complex. However, the PS complex observed in the agarose gel was ∼40 to 65S in size (data not shown), consistent with the size of a single spliceosome (40) and with the 45S penta-snRNPs complex in yeasts (54). TMV, with a sedimentation coefficient of 200S, was unable to enter the agarose gel (data not shown). Thus, we suggest that the PS complex is a subcomplex of the 200S particle that contains four spliceosomal substructures (44) and that is unable to enter the agarose gel as an intact particle. Parts of that large complex are probably stuck at the top of the native 4% acrylamide gel and need to be “stripped” by heparin or a nonphysiological pH (such as Tris-glycine running buffer) to enter the gel (data not shown).
We have shown that the replacement of U1 by U5 at the 5′ss, which is known to occur at an early stage of the splicing reaction, also occurs on the 5′ss RNA in 200S complexes. This indicates that the same 5′ss RNA can advance from U1 to U5 binding and that a factor(s) essential for the replacement procedure is present in the 200S fraction. This implies that the purified 200S complex is functional for U5 binding to the 5′ss RNA and for pre-mRNA spliceosome assembly when supplemented with soluble nuclear proteins. This is consistent with the functionality of the penta-snRNP complex observed in yeast extracts, a complex shown to be capable of pre-mRNA splicing when also supplied with soluble factors (54). Based on our observations for mammalian cell extracts and the observations of Stevens et al. for yeast extracts (54), we suggest that a penta-snRNP complex assembles prior to binding the pre-mRNA, recognizes the 5′ss, and subsequently accommodates the RNA-RNA rearrangements required for the splicing activity and that all of these events can take place within the same penta-snRNP complex. The 200S complex might represent the holospliceosome that houses single spliceosomal complexes.
Acknowledgments
We thank Joseph Sperling, Ruth Sperling, and Juan Valcarcel for comments on the manuscript; Reinhard Luhrmann for the U1(70K) antibody; Melissa Moore for the U5(p220) antibody; Joan Steitz for the anti-Sm antibody; and Hezy Antignus for TMV.
This work was supported by a grant from the Israel Academy of Science and in part by a grant from the Israel Cancer Association to G.A.
REFERENCES
- 1.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 2.Ast, G., D. Goldblatt, D. Offen, J. Sperling, and R. Sperling. 1991. A novel splicing factor is an integral component of 200S large nuclear ribonucleoprotein (lnRNP) particles. EMBO J. 10:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ast, G., T. Pavelitz, and A. M. Weiner. 2001. Sequences upstream of the branch site are required to form helix II between U2 and U6 snRNA in a trans-splicing reaction. Nucleic Acids Res. 29:1741-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ast, G., and A. M. Weiner. 1997. A novel U1/U5 interaction indicates proximity between U1 and U5 snRNAs during an early step of mRNA splicing. RNA 3:371-381. [PMC free article] [PubMed] [Google Scholar]
- 5.Ast, G., and A. M. Weiner. 1996. A U1/U4/U5 snRNP complex induced by a 2′-O-methyl-oligoribonucleotide complementary to U5 snRNA. Science 272:881-884. [DOI] [PubMed] [Google Scholar]
- 6.Awasthi, S., R. Palmer, M. Castro, C. D. Mobarak, and S. W. Ruby. 2001. New roles for the Snp1 and Exo84 proteins in yeast pre-mRNA splicing. J. Biol. Chem. 276:31004-31015. [DOI] [PubMed] [Google Scholar]
- 7.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 8.Burge, C. B., R. A. Padgett, and P. A. Sharp. 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2:773-785. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. Y., L. Stands, J. P. Staley, R. R. Jackups, Jr., L. J. Latus, and T. H. Chang. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7:227-232. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, S. C., and J. Abelson. 1987. Spliceosome assembly in yeast. Genes Dev. 1:1014-1027. [DOI] [PubMed] [Google Scholar]
- 11.Collins, C. A., and C. Guthrie. 2000. The question remains: is the spliceosome a ribozyme? Nat. Struct. Biol. 7:850-854. [DOI] [PubMed] [Google Scholar]
- 12.Crispino, J. D., J. E. Mermoud, A. I. Lamond, and P. A. Sharp. 1996. cis-Acting elements distinct from the 5′ splice site promote U1-independent pre-mRNA splicing. RNA 2:664-673. [PMC free article] [PubMed] [Google Scholar]
- 13.Crispino, J. D., and P. A. Sharp. 1995. A U6 snRNA:pre-mRNA interaction can be rate-limiting for U1-independent splicing. Genes Dev. 9:2314-2323. [DOI] [PubMed] [Google Scholar]
- 14.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, R., Z. Zhou, and R. Reed. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5:779-787. [DOI] [PubMed] [Google Scholar]
- 16.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419:86-90. [DOI] [PubMed] [Google Scholar]
- 18.Hall, K. B., and M. M. Konarska. 1992. The 5′ splice site consensus RNA oligonucleotide induces assembly of U2/U4/U5/U6 small nuclear ribonucleoprotein complexes. Proc. Natl. Acad. Sci. USA 89:10969-10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Luhrmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 21.Hong, W., M. Bennett, Y. Xiao, R. Feld Kramer, C. Wang, and R. Reed. 1997. Association of U2 snRNP with the spliceosomal complex E. Nucleic Acids Res. 25:354-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kandels-Lewis, S., and B. Seraphin. 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262:2035-2039. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D. H., and J. J. Rossi. 1999. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 5:959-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konarska, M. M. 1998. Recognition of the 5′ splice site by the spliceosome. Acta Biochim. Pol. 45:869-881. [PubMed] [Google Scholar]
- 25.Konarska, M. M., and P. A. Sharp. 1988. Association of U2, U4, U5, and U6 small nuclear ribonucleoproteins in a spliceosome-type complex in absence of precursor RNA. Proc. Natl. Acad. Sci. USA 85:5459-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konarska, M. M., and P. A. Sharp. 1987. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell 49:763-774. [DOI] [PubMed] [Google Scholar]
- 27.Konforti, B. B., and M. M. Konarska. 1994. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes Dev. 8:1962-1973. [DOI] [PubMed] [Google Scholar]
- 28.Konforti, B. B., M. J. Koziolkiewicz, and M. M. Konarska. 1993. Disruption of base pairing between the 5′ splice site and the 5′ end of U1 snRNA is required for spliceosome assembly. Cell 75:863-873. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn, A. N., Z. Li, and D. A. Brow. 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell 3:65-75. [DOI] [PubMed] [Google Scholar]
- 30.Laggerbauer, B., T. Achsel, and R. Luhrmann. 1998. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA 95:4188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauber, J., P. Fabrizio, S. Teigelkamp, W. S. Lane, E. Hartmann, and R. Luhrmann. 1996. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 15:4001-4015. [PMC free article] [PubMed] [Google Scholar]
- 32.Lesser, C. F., and C. Guthrie. 1993. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science 262:1982-1988. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H., W. Zhang, R. B. Reed, W. Liu, and P. J. Grabowski. 2002. Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA 8:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long, M., S. J. de Souza, and W. Gilbert. 1997. The yeast splice site revisited: new exon consensus from genomic analysis. Cell 91:739-740. [DOI] [PubMed] [Google Scholar]
- 35.Lund, M., and J. Kjems. 2002. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA 8:166-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madhani, H. D., and C. Guthrie. 1994. Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 28:1-26. [DOI] [PubMed] [Google Scholar]
- 37.Madhani, H. D., and C. Guthrie. 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71:803-817. [DOI] [PubMed] [Google Scholar]
- 38.Maroney, P. A., C. M. Romfo, and T. W. Nilsen. 2000. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 6:317-328. [DOI] [PubMed] [Google Scholar]
- 39.Medalia, O., A. J. Koster, A. Tocilij, M. Angenitzki, J. Sperling, Z. Berkovitch-Yellin, and R. Sperling. 1997. Automated electron tomography of large nuclear RNP (lnRNP) particles—the naturally assembled complexes of precursor messenger RNA and splicing factors. J. Struct. Biol. 120:228-236. [DOI] [PubMed] [Google Scholar]
- 40.Miriami, E., M. Angenitzki, R. Sperling, and J. Sperling. 1995. Magnesium cations are required for the association of U small nuclear ribonucleoproteins and SR proteins with pre-mRNA in 200 S large nuclear ribonucleoprotein particles. J. Mol. Biol. 246:254-263. [DOI] [PubMed] [Google Scholar]
- 41.Muller, S., B. Wolpensinger, M. Angenitzki, A. Engel, J. Sperling, and R. Sperling. 1998. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J. Mol. Biol. 283:383-394. [DOI] [PubMed] [Google Scholar]
- 42.Newman, A. J., and C. Norman. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68:743-754. [DOI] [PubMed] [Google Scholar]
- 43.Nilsen, T. W. 2002. The spliceosome: no assembly required? Mol. Cell 9:8-9. [DOI] [PubMed] [Google Scholar]
- 44.Raitskin, O., M. Angenitzki, J. Sperling, and R. Sperling. 2002. Large nuclear RNP particles—the nuclear pre-mRNA processing machine. J. Struct. Biol. 140:123-130. [DOI] [PubMed] [Google Scholar]
- 45.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 46.Reyes, J. L., E. H. Gustafson, H. R. Luo, M. J. Moore, and M. M. Konarska. 1999. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA 5:167-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reyes, J. L., P. Kois, B. B. Konforti, and M. M. Konarska. 1996. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA 2:213-225. [PMC free article] [PubMed] [Google Scholar]
- 48.Seraphin, B., L. Kretzner, and M. Rosbash. 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 7:2533-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shomron, N., H. Malca, I. Vig, and G. Ast. 2002. Reversible inhibition of the second step of splicing suggests a possible role of zinc in the second step of splicing. Nucleic Acids Res. 30:4127-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siliciano, P. G., and C. Guthrie. 1988. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 2:1258-1267. [DOI] [PubMed] [Google Scholar]
- 51.Sontheimer, E. J., and J. A. Steitz. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262:1989-1996. [DOI] [PubMed] [Google Scholar]
- 52.Spann, P., M. Feinerman, J. Sperling, and R. Sperling. 1989. Isolation and visualization of large compact ribonucleoprotein particles of specific nuclear RNAs. Proc. Natl. Acad. Sci. USA 86:466-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staley, J. P., and C. Guthrie. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3:55-64. [DOI] [PubMed] [Google Scholar]
- 54.Stevens, S. W., D. E. Ryan, H. Y. Ge, R. E. Moore, M. K. Young, T. D. Lee, and J. Abelson. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9:31-44. [DOI] [PubMed] [Google Scholar]
- 55.Tarn, W. Y., and J. A. Steitz. 1995. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc. Natl. Acad. Sci. USA 92:2504-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarn, W. Y., and J. A. Steitz. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 8:2704-2717. [DOI] [PubMed] [Google Scholar]
- 57.Wassarman, D. A. 1993. Psoralen crosslinking of small RNAs in vitro. Mol. Biol. Rep. 17:143-151. [DOI] [PubMed] [Google Scholar]
- 58.Wassarman, D. A., and J. A. Steitz. 1993. A base-pairing interaction between U2 and U6 small nuclear RNAs occurs in > 150S complexes in HeLa cell extracts: implications for the spliceosome assembly pathway. Proc. Natl. Acad. Sci. USA 90:7139-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyatt, J. R., E. J. Sontheimer, and J. A. Steitz. 1992. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 6:2542-2553. [DOI] [PubMed] [Google Scholar]
- 60.Yean, S. L., and R. J. Lin. 1991. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol. Cell. Biol. 11:5571-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yitzhaki, S., E. Miriami, R. Sperling, and J. Sperling. 1996. Phosphorylated Ser/Arg-rich proteins: limiting factors in the assembly of 200S large nuclear ribonucleoprotein particles. Proc. Natl. Acad. Sci. USA 93:8830-8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, D., N. Abovich, and M. Rosbash. 2001. A biochemical function for the Sm complex. Mol. Cell 7:319-329. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhuang, Y., and A. M. Weiner. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46:827-835. [DOI] [PubMed] [Google Scholar]