The discovery of a highly conserved cellular machinery that can regulate gene expression in response to double-stranded RNA may revolutionize mammalian virology. This revolution promises not only a deeper understanding of host-pathogen interactions and a novel set of experimental tools to explore the mechanism of viral replication but may also yield new therapeutic approaches. Even though the field of RNA silencing (or RNA interference [RNAi]) as applied to mammalian viruses is barely a year old, there is already enough material to appreciate its importance and to discuss its implications. Since an impressive number of excellent reviews on RNAi have been published recently (25, 27, 62), here we emphasize mammalian RNA silencing as it concerns one of its (presumably) natural targets, viruses.
RNAi IN PLANTS AND INVERTEBRATES
RNAi, also known as RNA silencing or posttranscriptional gene silencing, is a process that responds to double-stranded RNA (dsRNA) by silencing gene expression in a sequence-specific manner. The RNAi machinery uses dsRNA duplexes as guides to target and destroy specific cellular or viral RNAs. RNA silencing was initially described in plants more than a decade ago (reviewed in reference 62). Overexpression of transgenes in plants can result in cosuppression of homologous endogenous genes (43, 63). Also, infection by plant RNA viruses can be restricted or prevented by the artificial expression of pieces of the viral genomic RNA. This resistant state appears to be mediated by a cytoplasmic activity that targets specific RNAs for inactivation (38). It was thus proposed that suppression of endogenous genes or resistance to RNA viruses could arise from a sequence-specific RNA degradation system. Similar mechanisms were invoked to explain the phenomenon of cross-protection in which a nonpathogenic strain of plant virus elicits resistance to a related pathogenic virus (reviewed in reference 53). Furthermore, it was observed that cellular genes carried by viruses could lead to decreased expression of cognate endogenous host genes, an occurrence that was suggested to be mediated by the same mechanism responsible for virus resistance and which was named virus-induced gene silencing (33).
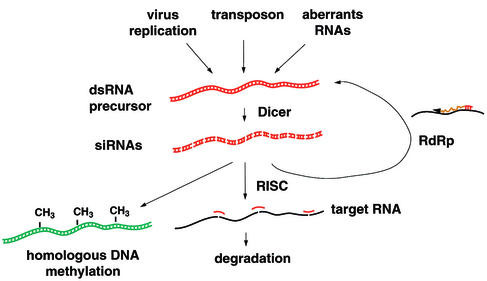
The critical role of dsRNA in initiating a process of sequence-specific mRNA degradation was first described for the nematode Caenorhabditis elegans by the artificial introduction of dsRNA (19). This process was called RNAi and has since been observed in a variety of eukaryotes (31, 44, 67). The emerging view from these studies is that the central player in the RNA silencing pathways is dsRNA, which acts as a trigger and/or intermediate of the process (Fig. 1). Thus, RNA silencing can be triggered by viruses or transposons that generate dsRNA during their replication or artificially by the introduction of synthetic dsRNA. It has also been proposed in plants that “aberrant RNAs” are capable of initiating RNA silencing responses (65). However, these aberrant RNAs have not been well characterized and are likely to contain dsRNA structures or motifs. The initial dsRNA is then cleaved into small, interfering dsRNAs (siRNAs), 21 to 25 nucleotides long, by a protein complex containing Dicer and possibly homologues of the C. elegans genes rde-4, rde-1, and drh-1/2 (58). The siRNAs in turn are incorporated into the RNA-induced silencing complex (RISC), and they are unwound presumably to act as guides to direct the RNA degradation machinery to the target RNAs (41). The RISC monitors the sequence of cytoplasmic RNAs and is able to cleave the cognate target RNA in a sequence-specific, siRNA-dependent manner (41). Although not all the components of the RISC have been identified, it appears that one or more members of the Argonaute gene family, such as the eukaryotic initiation factor 2C proteins, form part of the complex (41). The RNA silencing machinery also appears to be able to amplify the degradation signal. It has been proposed for invertebrates that siRNAs can function as primers that are extended on the targeted RNA by an RNA-dependent RNA polymerase (RdRp) (39, 54). This amplification has not yet been observed in mammalian systems.
FIG. 1.
Schematic representation of the molecular mechanism of the RNA silencing pathway. The process can be triggered by viral infection, transposons, or aberrant RNAs, all of which produce some form of dsRNA. Long dsRNA precursors are processed by Dicer to siRNAs. siRNAs associate with the RISC, which in turn targets and induces degradation of specific RNAs. The siRNAs can be amplified by a cellular RdRp. The RNA silencing system has also been associated with DNA methylation.
It is now generally accepted that RNA silencing is a major antiviral defense mechanism in plants (3a, 17a, 48, 50a). Although the natural trigger for RNA silencing in virally infected cells is presently unknown, because RNA viruses replicate through dsRNA intermediates it is possible that these replication intermediates serve to initiate the RNA silencing response. Even DNA viruses may produce dsRNAs as by-products of bidirectional transcription from their genome (15, 34). Several lines of evidence support the concept that RNA silencing is an antiviral mechanism in plants. First, as described above, natural infection by plant viruses elicits strong gene silencing (38). This observation indicates that the viral genomic RNA or intermediates of replication are used effectively by silencing machinery to trigger RNA silencing. Second, viral replication can be efficiently suppressed by experimentally induced RNA silencing (38, 66). Third, plant viruses encode a variety of inhibitors of the RNA silencing machinery (5, 11, 30, 64). The fact that viruses have acquired mechanisms to interfere with RNA silencing suggests that, for successful replication in nature, viruses need to overcome the plant's antiviral defenses. Fourth, some components of the RNA silencing machinery, like the RdRp, are upregulated by viroid and virus infection (6, 51). Perhaps the most persuasive indication that RNA silencing is an important antiviral defense mechanism in plants is the observation that mutations in genes that encode the RNA silencing machinery, like sgs2 (also called sde1), sgs3, and sde3, result in enhanced susceptibility to virus infection (16, 42). Taken together, these are strong indications that RNAi is an antiviral mechanism in plants.
RNA silencing also appears to contribute to antiviral defense in invertebrates. Preinfection of cells or whole mosquitoes with Sindbis virus carrying dengue virus genome fragments was shown to inhibit dengue virus replication (1, 20, 46). Although these results are reminiscent of the cross-protection phenomenon seen in plants (53), these experiments could not distinguish whether the inhibition was produced by RNA silencing, RNA antisense activity, or dominant-negative effects, such as those induced by defective interfering RNAs. However, a recent study demonstrated that it is indeed possible to inhibit dengue virus production by transfection of dsRNA in mosquito cells; in contrast, a single-stranded RNA control was not effective (12). Furthermore, cells transfected with a plasmid designed to express an inverted-repeat RNA derived from the dengue virus genome were resistant to virus infection and accumulated short species of dsRNAs (siRNAs) (2).
Important evidence supporting a physiological antiviral role for RNAi in invertebrates was obtained by studies of flock house virus (FHV) and its interaction with the RNA silencing machinery in Drosophila cells (37). This nodavirus infects insects but can also replicate in plant and mammalian cells. FHV infection results in accumulation of siRNAs specific for the viral genome. These siRNAs are able to promote the specific degradation of artificially introduced viral RNA. Interestingly, FHV protein B2 can block RNA silencing in both plant and invertebrate cells. B2 can functionally replace the 2b protein of cucumber mosaic virus, which belongs to a group of well-known suppressors of RNA silencing. Notably, FHV-induced RNA silencing was prevented by expression of B2 or depletion of AGO2 (a putative RISC component of the Argonaute family). These experiments argue for a role of RNA silencing as an adaptive antiviral defense in invertebrates. They also raise the question of how RNAi acts in response to natural viral infection and whether, as observed in plants, RNAi in insects can induce a systemic protection against viruses.
siRNAs AS AN ANTIVIRAL MECHANISM IN MAMMALIAN CELLS
Is there a mammalian antiviral RNAi counterpart? It was originally thought that mammalian cells were unlikely to possess an active RNA silencing machinery (19). This skepticism was derived from the fact that mammalian cells respond to the presence of dsRNA with a general, nonspecific shutdown of translation, mediated primarily by protein kinase R (PKR) and RNase L. Indeed, this response is considered an important innate antiviral mechanism that is upregulated by interferon and contributes to controlling viral infection in a nonspecific fashion (36, 55). Furthermore, mammals have evolved a sophisticated immune system based on protein recognition that protects them against infection in a highly specific manner. This system is not present in plants and invertebrates and thus could have functionally replaced the RNA silencing system. Yet it would seem that such a versatile antiviral system as RNA silencing might be conserved during evolution, since viruses (as well as other molecular parasites, like transposons) probably maintain an unrelenting selective pressure on their hosts.
Indeed, the recent description of RNAi in mammalian cells proved that the RNA silencing machinery is conserved in mammals (9, 12, 57, 61, 67). These studies showed that while the PKR-RNase L pathway is activated by long dsRNA in most cells, siRNAs (21 nucleotides long) are readily recognized by a mammalian RISC and mediate significant reduction in the levels of a specific target mRNA. These observations led several groups to explore the interaction between the RNA silencing machinery and mammalian viruses.
The initial experiments centered on a simple question. Are mammalian viruses susceptible to RNAi? While potentially any RNA can be subject to degradation by the RNAi machinery, it was not clear whether viral RNAs could be effectively targeted. Virus genomes are often protected by a proteinaceous structure (dsRNA viruses), by nucleoproteins and matrix layers (negative-stranded RNA viruses), or by association with cellular membranes during replication (positive-stranded RNA viruses). In addition, a large part of the newly synthesized RNA is often rapidly encapsidated in new virus particles. Thus, the recent discovery that siRNAs can block replication of several different types of mammalian viruses is significant, especially given its therapeutic implications. siRNA has been shown to inhibit production of two retroviruses, the human immunodeficiency virus (HIV) (13, 14, 26, 28, 35, 45, 48, 56) and Rous sarcoma virus (26); a negative-stranded RNA virus, respiratory syncytial virus (10); and a positive-stranded RNA virus, poliovirus (21), and to inhibit the gene expression of a DNA virus, human papillomavirus (29). In all these reports, cells transfected with siRNA corresponding to the viral genome induced a clear reduction in virus production. In general, a decrease in titer of 10- to 200-fold was observed (14, 21, 26, 28, 45). All these studies demonstrate that accumulation of viral RNA in infected cells was significantly reduced by siRNAs, indicating that viral RNA can be targeted by the RNA silencing machinery.
The action of siRNAs appears to be independent of the nonspecific PKR-RNase L responses. Thus, control siRNAs of unrelated sequences failed to inhibit virus production (13, 14, 21, 26, 28, 45, 56). Also, siRNA transfection did not induce phosphorylation of PKR (10, 28), a signal of interferon-mediated activation of the innate defense system (55). Furthermore, transfection of siRNAs into mouse embryonic fibroblasts deficient in both PKR and RNase L was effective in inhibiting virus replication (21). In addition, the antiviral siRNAs did not result in interferon production, as viral resistance could not be induced by culturing cells in medium containing supernatant from siRNA-transfected cells (21). These studies suggest that the classical interferon and nonspecific dsRNA pathways are not part of the specific RNA silencing mechanism.
An important question raised by these recent studies concerns the duration and amplification of the siRNA signal and effect. The ability of siRNA-transfected cells to resist virus infection was maintained over the course of several days (21, 28). The duration of the effect could either indicate that the interference state persists for a few days or, alternatively, that the siRNAs could be slowly released from the cell-associated transfection mixture. Assuming that cells can indeed maintain the silencing state for several days, a remarkable parallel to the protein-based immune system can be drawn, whereby the RNA silencing system induces a specific response which can last for at least a few days. Perhaps the system is capable of establishing a rudimentary form of memory. It would be interesting to determine whether siRNAs are preserved freely in the cytoplasm or are associated with components of the RNAi machinery in order to resist degradation. Perhaps, transfected siRNAs are able to associate with the RISC or even with the hypothetical RdRp and in this manner not only prolong their half-lives but also amplify the signal over a certain period of time.
Another important question is whether viral nucleoproteins and/or replication strategies play a role in shielding the viral genome from the RNAi machinery (discussed in reference 3). At this stage, the question has mainly been addressed for HIV. Since different studies have used different siRNAs, cell types, viral strains, multiplicities of infection, and times between transfection and infection, the answer is not yet conclusive. However, there is a consensus that expression of HIV RNA from proviral DNA can be effectively inhibited by RNAi (26, 28, 35, 45). What is unclear at this time is whether RNAi can target the incoming viral RNA while in transit to the nucleus, when it is still associated with nucleocapsid proteins. Using PCR to detect integrated viral genomes, Hu and colleagues (26) found that the number of integrated proviruses is identical in HIV siRNA- and control siRNA-treated cells, suggesting that the incoming viral RNA is inaccessible to RNAi. In contrast, Coburn and Cullen's study, using hybridization to detect viral genomic DNA in infected cells, revealed a difference between antiviral siRNA-treated and control samples (14). Consistent with this result, Capodici et al. observed a clear reduction in the amount of HIV DNA that accumulated in cells treated with siRNAs (13) and Jacque et al. found a significant difference in HIV genomic RNA in siRNA-treated cells as early as 1 h after infection, indicating that incoming RNA can be targeted by RNAi (28). A critical difference between these studies is that they target different sequences within the HIV genome. This difference can account for the discrepant results since the nucleocapsid of the incoming virus may preferentially shield some regions of the RNA. For negative-stranded RNA viruses, it appears that Rous sarcoma virus mRNA is susceptible to siRNA targeting, but genomic RNA, which is known to be coated by a nucleoprotein, is not (10).
Since all the studies on antiviral activity of RNAi have employed defined, short RNA sequences (siRNAs or short hairpin RNAs [shRNAs]), they allow evaluation of the tolerance of the RNAi machinery to mismatches. It appears that mismatches between the siRNA and its viral target generally are not well tolerated. One mismatch located approximately in the center of the siRNA nearly abolishes silencing of poliovirus (L. Gitlin and R. Andino, unpublished data). However, certain anti-HIV siRNAs carrying a single-nucleotide mismatch can be as effective as those with sequences that perfectly match the target RNA (28). The tolerance of RNA silencing to mismatches is important because it relates to the ability of viruses to escape inhibition by RNA silencing. If the RNAi machinery can accommodate mismatches, it would be more difficult for the virus to escape the action of siRNA. The fact that a single point mutation yielded an RNAi-resistant virus raises an important challenge for the use of short, defined siRNAs in therapeutic approaches. On the other hand, a “natural” RNA silencing response that targets multiple viral sequences would be harder to evade, as escape would require extensive alterations in the viral genome.
The viruses which are least likely to escape from siRNAs are the DNA viruses, due to their lower mutation frequency. Among them, an important therapeutic target is papillomaviruses, because it appears that the constitutive expression of viral proteins E6 and E7 is required for carcinogenic growth (18). Recent results with siRNAs targeting E6 and E7 in cultured cells suggested that E6 downregulation by siRNAs can lead to cell growth suppression, while E7 downregulation results in apoptosis (29).
A striking conclusion from one of the recent studies is that siRNAs can clear viral infection without causing any visible harm to the infected cell (21). It has been a long-standing assumption in immunology that clearance of virus from the mammalian host requires destruction of infected cells (22), either by the action of the immune system or by apoptosis induced by the virus. This idea has been challenged in studies that observed reduction and even clearance of viral genomes in the absence of significant cytopathology (23, 32). Although the mechanism that induced viral clearance in these systems is unknown, it is tempting to speculate that it could be mediated by RNA silencing. Indeed, siRNA can effectively clear poliovirus from infected HeLa cells (21). A recent study has also demonstrated that siRNAs can be effective in clearing hepatitis C virus replicons (50). These experiments highlight the potential of RNA silencing as an agent of noncytopathic viral clearance.
FUTURE DIRECTIONS
These initial studies of the effect of RNA silencing on viral replication in mammalian cells open two major avenues of investigation. First, it will be important to understand the role (if any) of RNA silencing in providing an antiviral defense against natural infections in mammalian systems. Second, these results raise the exciting yet challenging opportunity to develop therapeutic approaches employing RNAi.
Is RNA silencing an antiviral system?
Even though synthetic siRNA transfected into mammalian cells can inhibit viral replication, it is not yet clear whether RNA silencing plays a physiological role as an antiviral system. For instance, it is unclear whether RNA silencing can be elicited naturally during viral infection. Since the presumably natural precursors of siRNAs, long dsRNA intermediates of RNA replication, seem to cause a general translation shutoff in mammalian cells, it is not yet clear how specific RNAi responses could be initiated during the course of a natural infection. Although the recent findings using siRNAs discussed here represent an important step toward determining the role and potential of RNAi as an antiviral mechanism, they are based on artificial experimental designs.
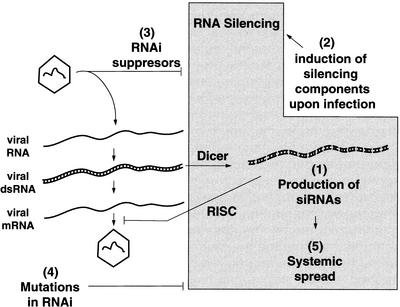
We propose that establishing RNAi as a natural antiviral defense in mammals will require addressing five critical issues that are schematically described in Fig. 2.
FIG. 2.
Is RNA silencing a natural antiviral defense system in mammals? Establishing RNAi as a natural antiviral defense system in mammals will require addressing the five critical questions which are schematically presented in the figure. See text for details.
Issue 1: Are siRNAs generated during the course of a natural infection? To generate siRNAs from long dsRNA replication intermediates necessitates functional Dicer expression. Intriguingly, there is a difference in the response to dsRNA by embryonic stem cells and differentiated cell lines. Differentiated cell lines express PKR and RNase L, and apparently only low levels of Dicer (9), and thus dsRNA induces a general translation shutoff. In contrast, embryonic carcinoma cells process dsRNA to induce sequence-specific silencing (9). One possibility is that the antiviral/antitransposon function of RNA silencing is limited to embryos. Alternatively, certain cell types in the adult may be able to process and disseminate long dsRNA, while most other cells are only capable of reacting to fully processed siRNAs. However, since undifferentiated embryonic carcinoma cell lines are susceptible to viral infection (59), there may be unidentified regulatory mechanisms that control RNA silencing in embryonic cells in vivo. Otherwise, viruses may have found ways to overcome silencing or the RNAi machinery in embryonic cell lines may lack additional components required for antiviral function.
Issue 2: Are RNAi components upregulated during viral infection? The upregulation of RNAi components during viral infection would support the idea that this system plays a natural role in antiviral defense. The recent finding that porcine reproductive and respiratory syndrome virus infection induces upregulation of the RNA helicase RHIV-1 (70), a homologue of drh-1 helicase from C. elegans apparently required for dsRNA processing (58), may support this idea. However, it is possible, as suggested by the authors, that upregulation of RHIV-1 plays a role during virus replication rather than in antiviral defense. A related issue is the relationship between RNA silencing and the interferon system. Although it has been assumed that they are quite distinct and independent from each other, the relationship between these systems remains to be analyzed. Since many components of the RNAi system appear to participate in micro-RNA processing (4), it will also be interesting to elucidate the overlap between RNAi and micro-RNA processing. Understanding the regulation and interplay of these two systems, especially in a context of a viral infection may have important consequences for the adaptation of RNAi for therapeutic purposes.
Issue 3: Have viruses evolved mechanisms to suppress or escape an RNAi response? Two types of mechanisms of RNAi evasion can be envisioned. First, as mentioned before, it is possible that viruses protect their RNAs by sequestering them in viral particles or replication complexes. However, because viruses need to express their genomic information by translating their RNAs and since mRNAs are susceptible to RNAi, it is possible that the RNA silencing machinery can target and reduce the expression of viral protein, thus inhibiting viral replication. Assuming the viral dsRNA intermediates are shielded from Dicer, how is the RNAi response initiated? One possibility is that dsRNA leaks out from the shielded compartment inside the infected cells or even from the remnants of cells lysed by the virus. A second type of evasion mechanism could rely on virus-encoded proteins that directly inhibit specific steps of RNA silencing. So far, there have been no reports of such activities encoded by mammalian viruses, but as the study of RNAi in mammalian cells develops, it is likely that information about virus-encoded RNAi suppressors will be obtained.
Issue 4: Do mutations or deletions in mammalian homologues of RNAi components render cells or animals more susceptible to viral infection? Both genetic and biochemical approaches have already identified several key components of the mammalian RNAi machinery (including Dicer and the RISC) (8, 41, 24), and more are likely to emerge in the near future. It is possible that knockouts of genes encoding RNA silencing components will be lethal, considering that they may participate in several RNA-based gene expression regulatory systems, as shown for DICER in micro-RNA processing (4). However, it may be possible to obtain cell lines deficient in specific components of the RNA silencing machinery to address whether defects in the RNAi pathway increase susceptibility to virus infection.
Issue 5: Can RNAi in one infected cell trigger a systemic antiviral response? In C. elegans, the injection of dsRNA in one region of the worm triggers its spread to many different tissues, including the gonads (19). It is not known how the dsRNA exits the cell in which it was originally produced, how it is transported through the organism, and how it is taken up by distant target cells. It will thus be important to address whether RNA silencing in mammals also induces a systemic response.
Is it possible to manipulate the RNAi system to develop therapeutic approaches?
Present data has generated much hope for the use of RNAi as a novel antiviral therapy. However, establishing RNAi as a viable therapeutic approach requires resolving at least three major issues.
(i) Persistence of the RNAi inhibitory effect.
As mentioned above, an inhibitory effect in cell culture is observed for only a few days after transfection of siRNAs. This relatively short duration of the RNA inhibitory effect has been circumvented by constitutively expressing shRNAs from RNA polymerase III promoters, with transcription initiated and terminated at precise sites (for a review, see reference 61) or as polymerase II transcripts (68, 69). Although shRNAs have not yet been extensively employed, it appears that the system is effective in a variety of cell types. Intriguingly, it appears that Dicer is responsible for processing shRNA into siRNAs (47), suggesting that this artificial system employs the RNAi processing apparatus to channel the shRNA into the correct pathway. Similar approaches may be extended to solve the persistence issue.
(ii) Delivery.
A major problem to be addressed is how to efficiently deliver the shRNA-expressing plasmids and how to target specific cell types. Perhaps a reasonable approach will be to employ viral vectors. Initial reports indicate that retroviral (7, 17) and adenoviral (68) vectors are capable of carrying shRNAs and inducing RNAi in target cells. Nonetheless, it is expected that this approach will encounter benefits and limitations similar to those observed in gene therapy methodologies.
(iii) Viral escape.
As mentioned above, viruses are likely to evade any given siRNA by mutations of the target sequences. Therefore, it may be important to produce multiple siRNAs, focusing on the conserved regions of the viral genome. However, this strategy has not yet been evaluated and it is possible that viruses are able to find additional ways to evade a strategy based on targeting multiple sites in the viral genome. An alternative method to circumvent the high rate of viral mutation may be to target a cellular protein required for viral replication. For example, depletion of CD4 using siRNAs has led to a decrease in the infectivity of HIV (45). These experiments suggest that targeting CCR5 (which is mutated in some individuals that are resistant to HIV infection [40]) may be a successful course in protection from AIDS. In a very recent report, CCR5 was targeted by siRNAs. Blocking CCR5 expression resulted in substantial protection for the lymphocyte populations susceptible to HIV-1 infection (49). However, the biological plasticity of viruses may find ways to overcome this strategy and, thus, targeting a cellular factor may not guarantee complete protection from viral infection (52, 60). Hence, it is possible that the genetic variability of RNA viruses may render targeting a single cellular factor ineffective. Therefore, targeting several host cell factors involved in viral replication may be required, an approach designed as an RNAi equivalent to the multi-drug antiretrovirus therapy HAART, whose success is based on the simultaneous targeting of multiple viral proteins.
In hindsight, one would expect researchers to have predicted the existence of a nucleic acid-based immune system earlier, by analogy to the protein-based immune system of mammals. After all, if life began as an RNA world it would have been appropriate to evolve an adaptive system that recognized foreign nucleic acids early on. The discovery of this highly sophisticated system stresses the advantages of being both adaptive and specific in combating invaders. It also stresses the fact that there is more to RNA silencing and virology in general than we can yet fathom. The years to come promise to bring interesting and unanticipated results regarding the mechanism and therapeutic applications of this fascinating system.
ADDENDUM IN PROOF
While this paper was under review, the following articles dealing with siRNA application against an orthomyxovirus, a herpesvirus, and hepatitis C virus replicons appeared: Q. Ge, M. T. McManus, T. Nguyen, C. H. Shen, P. A. Sharp, H. N. Eisen, and J. Chen, Proc. Natl. Acad. Sci. USA 100:2718-2723, 2003; Q. Jia and R. Sun, J. Virol. 77:3301-3306, 2003; J. A. Wilson, S. Jayasena, A. Khvorova, S. Sabatinos, I. G. Rodrigue-Gerbais, S. Arya, F. Sarangi, M. Harris-Brandts, S. Beaulieu, and C. D. Richardson, Proc. Natl. Acad. Sci USA 100:2783-2788, 2003; and S. B. Kapadia, A. Brideau-Andersen, and F. V. Chisari, Proc. Natl. Acad. Sci. USA 100:2014-2018, 2003.
Acknowledgments
We are grateful to Judith Frydman and members of the Andino laboratory for useful comments on the manuscript.
This work was supported by funds provided by Public Health Service grant AI40085 to R.A.
REFERENCES
- 1.Adelman, Z. N., C. D. Blair, J. O. Carlson, B. J. Beaty, and K. E. Olson. 2001. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol. Biol. 10:265-273. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, Z. N., I. Sanchez-Vargas, E. A. Travanty, J. O. Carlson, B. J. Beaty, C. D. Blair, and K. E. Olson. 2002. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 76:12925-12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270-1273. [DOI] [PubMed] [Google Scholar]
- 3a.Al-Kaff, N. S., S> N. Covey, M. M. Kreike, A. M. Page, R. Pinder, and P. J. Dale. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113-2115. [DOI] [PubMed] [Google Scholar]
- 4.Ambros, V. 2001. MicroRNAs: tiny regulators with great potential. Cell 107:823-826. [DOI] [PubMed] [Google Scholar]
- 5.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Astier-Manifacier, S., and P. Cornuet. 1971. RNA-dependent RNA polymerase in Chinese cabbage. Biochim. Biophys. Acta 232:484-493. [DOI] [PubMed] [Google Scholar]
- 7.Barton, G. M., and R. Medzhitov. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. USA 99:14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 9.Billy, E., V. Brondani, H. Zhang, U. Muller, and W. Filipowicz. 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 98:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 Infection by small interfering RNA-mediated RNA interference. J. Immunol. 169:5196-5201. [DOI] [PubMed] [Google Scholar]
- 14.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colby, C., C. Jurale, and J. R. Kates. 1971. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. J. Virol. 7:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalmay, T., R. Horsefield, T. H. Braunstein, and D. C. Baulcombe. 2001. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20:2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devroe, E., and P. A. Silver. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Dougherty, W. G., and T. D. Parks. 1995. Transgene and gene suppression: telling us something new? Curr. Opin. Cell Biol. 7:399-405. [DOI] [PubMed] [Google Scholar]
- 18.Fields, B. N., D. M. Knipe, and P. M. Howley. 1996. Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 19.Fire, A. 1999. RNA-triggered gene silencing. Trends Genet. 15:358-363. [DOI] [PubMed] [Google Scholar]
- 20.Gaines, P. J., K. E. Olson, S. Higgs, A. M. Powers, B. J. Beaty, and C. D. Blair. 1996. Pathogen-derived resistance to dengue type 2 virus in mosquito cells by expression of the premembrane coding region of the viral genome. J. Virol. 70:2132-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 22.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 23.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 24.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 25.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 26.Hu, W., C. Myers, J. Kilzer, S. Pfaff, and F. Bushman. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 12:1301. [DOI] [PubMed] [Google Scholar]
- 27.Hutvagner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12:225-232. [DOI] [PubMed] [Google Scholar]
- 28.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, M., and J. Milner. 2002. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21:6041-6048. [DOI] [PubMed] [Google Scholar]
- 30.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 31.Kennerdell, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, T., and D. E. Griffin. 2000. The role of CD8+ T cells and major histocompatibility complex class I expression in the central nervous system of mice infected with neurovirulent Sindbis virus. J. Virol. 74:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumagai, M. H., J. Donson, G. della-Cioppa, D. Harvey, K. Hanley, and L. K. Grill. 1995. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 92:1679-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, M., and G. G. Carmichael. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. USA 94:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 36.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, H. W., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 38.Lindbo, J. A., L. Silva-Rosales, W. M. Proebsting, and W. G. Dougherty. 1993. Induction of a highly specific antiviral state in transgenic plants—implications for regulation of gene expression and virus resistance. Plant Cell 5:1749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipardi, C., Q. Wei, and B. M. Paterson. 2001. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107:297-307. [DOI] [PubMed] [Google Scholar]
- 40.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 41.Martinez, J., A. Patkaniowska, H. Urlaub, R. Luhrmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563. [DOI] [PubMed] [Google Scholar]
- 42.Mourrain, P., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon, J. B. Morel, D. Jouette, A. M. Lacombe, S. Nikic, N. Picault, K. Remoue, M. Sanial, T. A. Vo, and H. Vaucheret. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533-542. [DOI] [PubMed] [Google Scholar]
- 43.Napoli, C., C. Lemieux, and R. Jorgensen. 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 46.Olson, K. E., S. Higgs, P. J. Gaines, A. M. Powers, B. S. Davis, K. I. Kamrud, J. O. Carlson, C. D. Blair, and B. J. Beaty. 1996. Genetically engineered resistance to dengue-2 virus transmission in mosquitoes. Science 272:884-886. [DOI] [PubMed] [Google Scholar]
- 47.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park, W. S., N. Miyano-Kurosaki, M. Hayafune, E. Nakajima, T. Matsuzaki, F. Shimada, and H. Takaku. 2002. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interference. Nucleic Acids Res. 30:4830-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Ratcliff, F. G., S. A. MacFarlane, and D. C. Baulcombe. 1999. Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiebel, W., B. Haas, S. Marinkovic, A. Klanner, and H. L. Sanger. 1993. RNA-directed RNA polymerase from tomato leaves. I. Purification and physical properties. J. Biol. Chem. 268:11851-11857. [PubMed] [Google Scholar]
- 52.Schols, D., J. A. Este, C. Cabrera, and E. De Clercq. 1998. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J. Virol. 72:4032-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sequeira, L. 1984. Cross-protection and induced resistance: their potential for plant disease control. Trends Biotechnol. 2:25-29. [Google Scholar]
- 54.Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish, L. Timmons, R. H. Plasterk, and A. Fire. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465-476. [DOI] [PubMed] [Google Scholar]
- 55.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 56.Surabhi, R. M., and R. B. Gaynor. 2002. RNA interference directed against viral and cellular targets inhibits human immunodeficiency virus type 1 replication. J. Virol. 76:12963-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svoboda, P., P. Stein, H. Hayashi, and R. M. Schultz. 2000. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127:4147-4156. [DOI] [PubMed] [Google Scholar]
- 58.Tabara, H., E. Yigit, H. Siomi, and C. C. Mello. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109:861-871. [DOI] [PubMed] [Google Scholar]
- 59.Teich, N. M., R. A. Weiss, G. R. Martin, and D. R. Lowy. 1977. Virus infection of murine teratocarcinoma stem cell lines. Cell 12:973-982. [DOI] [PubMed] [Google Scholar]
- 60.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tuschl, T. 2002. Expanding small RNA interference. Nat. Biotechnol. 20:446-448. [DOI] [PubMed] [Google Scholar]
- 62.van der Krol, A. R., L. A. Mur, M. Beld, J. N. Mol, and A. R. Stuitje. 1990. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2:291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants-defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 64.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wassenegger, M., and T. Pelissier. 1998. A model for RNA-mediated gene silencing in higher plants. Plant Mol. Biol. 37:349-362. [DOI] [PubMed] [Google Scholar]
- 66.Waterhouse, P. M., M. W. Graham, and M. B. Wang. 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95:13959-13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wianny, F., and M. Zernicka-Goetz. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell. Biol. 2:70-75. [DOI] [PubMed] [Google Scholar]
- 68.Xia, H., Q. Mao, H. L. Paulson, and B. L. Davidson. 2002. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 16:16. [DOI] [PubMed] [Google Scholar]
- 69.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, X., C. Wang, L. B. Schook, R. J. Hawken, and M. S. Rutherford. 2000. An RNA helicase, RHIV-1, induced by porcine reproductive and respiratory syndrome virus (PRRSV) is mapped on porcine chromosome 10q13. Microb Pathog. 28:267-278. [DOI] [PubMed] [Google Scholar]