Abstract
The three-dimensional structure of a thermostable β-glycosidase (GlyTn) from the thermophilic eubacterium Thermus nonproteolyticus HG102 was determined at a resolution of 2.4 Å. The core of the structure adopts the (βα)8 barrel fold. The sequence alignments and the positions of the two Glu residues in the active center indicate that GlyTn belongs to the glycosyl hydrolases of retaining family 1. We have analyzed the structural features of GlyTn related to the thermostability and compared its structure with those of other mesophilic glycosidases from plants, eubacteria, and hyperthermophilic enzymes from archaea. Several possible features contributing to the thermostability of GlyTn were elucidated.
Glycosyl hydrolases catalyze the hydrolytic cleavage of the glycosidic bonds between two or more carbohydrates and between carbohydrate and noncarbohydrate moieties. They are ubiquitous enzymes that have been isolated and characterized from various organisms (archaea, eubacteria, and eukarya). The glycosyl hydrolases have been classified into over 80 families according to their amino acid sequence homology and structural similarities rather than substrate selectivity. Some of the families can be grouped into “clans,” because the folds of their proteins are better conserved than their sequences (23). Families 1, 2, 5, 10, 17, 26, 30, 35, 39, 42, 51, 53, 59, 72, 79, and 86 are grouped into a superfamily, clan GH-A. Members of this superfamily adopt a (βα)8 barrel fold (23). The β-glycosidases in family 1 constitute a major group among glycosyl hydrolases. They are characterized by broad substrate specificities, which make them potential tools for several applications (24). In this regard, β-glycosidases from thermophilic sources are particularly attractive because of their biotechnological advantages for many stabilized biocatalysts. Furthermore, study of these β-glycosidases may contribute to a better understanding of the structure-function relationships of thermophilic enzymes by comparisons of their properties with those of mesophilic enzymes (47).
The thermostable β-glycosidase GlyTn was produced by the thermophilic eubacterium Thermus nonproteolyticus HG102, which was isolated from a hot spring in Guangdong Province, southern China (8). The gene coding for GlyTn in T. nonproteolyticus HG102 (GenBank accession number AF225213) has been cloned and expressed in Escherichia coli, and the recombinant enzyme was characterized (21). GlyTn belongs to glycosyl hydrolase family 1. It has a broad β-glycosidase activity, and analysis of its substrate specificity revealed that it prefers β-d-glucoside and β-d-fucoside to β-d-galactoside and β-d-mannoside. GlyTn also has transglycosidic activity at high temperature. This enzyme shows optimum activity at 90°C and pH 5.6, with a half-life of 2.5 h at 90°C (21). Besides the β-glycosidase gene in T. nonproteolyticus HG102, findings for other two β-glycosidase genes from Thermus sp. strain Z-1 (GenBank accession number AB034947) and Thermus thermophilus (GenBank accession number Y16753) have also been reported (14, 46). These three β-glycosidases share high levels of sequence similarity, with nearly 95% of the residues identical.
The structures of β-glucosidase from white clover (Protein Data Bank [PDB] code 1CBG) and 6-phospho-β-galactosidase from Lactococcus lactis (PDB code 1PBG) were reported in 1995 (4, 53). Other structures of family 1 β-glycosidases reported include those of myrosinase from Sinapis alba (PDB code 1MYR), β-glycosidase from Sulfolobus solfataricus (1GOW), β-glucosidase from Bacillus polymyxa (1BGA), β-glycosidase from Thermosphaera aggregans (1QVB), β-glucosidase from Bacillus circulans (1QOX), and β-glucosidase from maize (1E1E) (1, 7, 9, 11, 18, 43). All the structures have the same basic (βα)8 barrel fold. The two hyperthermophilic structures 1GOW and 1QVB were from archaea.
The crystal structure of GlyTn from T. nonproteolyticus HG102 described here was determined at a resolution of 2.4 Å. To our knowledge, this is the first β-glycosidase structure determined from a thermophilic eubacterium. It adopts a (βα)8 barrel fold. The model of GlyTn was compared with other mesophilic structures from plants and eubacteria and hyperthermophilic structures from archaea to elucidate the possible basis of its thermostability.
MATERIALS AND METHODS
Expression, purification, and crystallization of GlyTn.
The cloning, expression, purification, and crystallization of GlyTn were performed with the methods described before (22).
Data collection and structure refinement.
Using a Weissenberg camera (42), diffraction data were collected on a beamline BL-6B experimental station (Photon Factory, Ibaraki, Japan). The data were processed with DENZO and SCALEPACK software (36), and the statistics are listed in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value |
|---|---|
| Temperature (K) | 100 |
| Space group | P212121 |
| Unit cell parameters (Å) | |
| a | 66.7 |
| b | 94.8 |
| c | 176.7 |
| Resolution range (Å) | 20.0-2.4 |
| No. of unique reflections | 41,963 |
| Completeness (outer shell) (%) | 94.0 (73.4) |
| I/σ(I) (outer shell) | 9.9 (2.2) |
| Rmerge for all data (outer shell) (%) | 18.2 (32.7) |
| R-factor/Rfree | 23.0 (27.2) |
| rms deviations from ideal geometry | |
| Bond length (Å) | 0.007 |
| Bond angle (°) | 1.38 |
| Average temperature factors (Å2) | |
| Main chain atoms | 29.3 |
| Side chain atoms | 30.6 |
| Water molecules | 31.4 |
The structure was determined by the molecular replacement method using the program Molrep in the CCP4 suite (9a). The positions of the two GlyTn molecules in the asymmetric unit were found with the model of Bacillus polymyxa β-glucosidase structure (PDB code 1BGA) (43). Using the maximum-likelihood simulated annealing protocol and restraining the noncrystallographic symmetry, the initial model was refined with the program CNS (6). The proper GlyTn residues were built into the σA-weighed 2|Fo|-|Fc| electron density map with program O (25). After several rounds of refinement and model building, the Rfree and R factors dropped to 31.5 and 28.0% in the resolution range of 20.0 to 2.4 Å and there were two regions with poor densities: N terminal 1 to 4 and C terminal 431 to 436. The removal of the noncrystallographic symmetry in the following refinement was validated by the decrease of the Rfree value. Water molecules were added to the model at locations with |Fo|-|Fc| densities higher than 3σ and hydrogen-bonding stereochemistry. Inclusion of individual temperature factors was validated by a substantial decrease in the value of Rfree. At the end of refinement, the crystallographic R factor was 23.0%, with an Rfree value of 27.2%. The stereochemistry of the final model was analyzed with PROCHECK (29).
Analysis of parameters affecting protein thermal stability. (i) Secondary structure content.
The α-helix content of a protein is the percentage of residues that show α-helical conformations. The β-sheet content is the percentage of residues that show β-sheet conformations. The corresponding dictionary of protein secondary structure file (26) was used to identify the residues with α-helical and β-sheet conformations.
(ii) Surface areas and cavities.
The total and hydrophobic accessible surface areas (ASA) were calculated with the program NACCESS (S. J. Hubbard and J. M. Thornton, NACCESS computer program, Department of Biochemistry and Molecular Biology, University College London, London, United Kingdom, 1993) with a probe radius of 1.4 Å. An output file containing summed atomic ASA over each residue was used. The number and volume of cavities were calculated with the program VOIDOO (26a).
(iii) Hydrogen bonds.
The WHATIF program was used to identify all hydrogen bonds in the structures (51).
(iv) Ion pairs.
Ion contacts were evaluated with the program CONTACT in CCP4 suite (9a). The ion pair was inferred when Asp or Glu side chain carbonyl oxygen atoms were found to be within 4.0 Å from the nitrogen atoms in Arg, Lys, and His side chains (3).
PDB accession code
The coordinates of the structure and the structure factor file have been deposited in the Protein Data Bank (PDB) under accession code 1NP2.
RESULTS AND DISCUSSION
Quality of the model.
The final model contains two molecules (A and B) in the asymmetric unit, including 6,824 nonhydrogen protein atoms and 334 water molecules. In both molecules A and B, the N-terminal 1 to 4 and C-terminal 431 to 436 residues were not defined in the election density maps, which means that these regions are disordered. The final R factor was 23.0% for reflections in the resolution range of 20.0 to 2.4 Å. The Rfree value for 5% of the total reflections was 27.2%. The model has good stereochemistry, with root mean square (rms) deviations of 0.007 Å on bond length and 1.38° on bond angle (Table 1).
Structural description and comparison with other family 1 β-glycosidases.
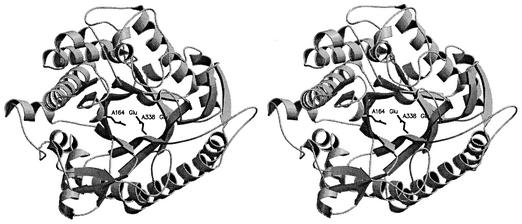
Because the equivalent 426 Ca atoms of molecules A and B are in good agreement (rms value of 0.53 Å) after superimposition, the following description and comparison were based on molecule A. GlyTn adopts the expected topology of a single (β/α)8 barrel fold (TIM barrel) (Fig. 1). Additional secondary structure units were inserted into the connections between the β-strand and α-helix in the (β/α) repeat (Fig. 2). The longest connection (from residue 14 to 54 between β-strand 1 and α-helix 1) contains several turns and two helices. The connections between (β/α) repeats are rather short, with the exception of a 30-residue connection between α-helix 5 and β-strand 6, and there is a short α-helix in the connection. Compared with the more compact bottom half of the barrel (β-strand N-terminus direction), the top half (β-strand C-terminus direction) is loose and four loops on that side form the gate to the active site. These four loops are composed of residues 36 to 54, 175 to 184, 292 to 315, and 386 to 403.
FIG. 1.
The overall fold structure of GlyTn. This figure and Fig. 3 were made with the programs MOLSCRIPT (28) and RASTER3D (33).
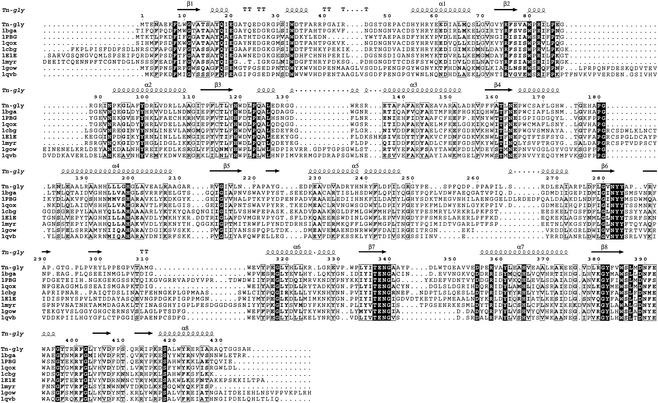
FIG. 2.
Sequence alignments of the nine β-glycosidases from family 1 with known structures. The figure was produced with ESPript (17).
The three-dimensional structure of GlyTn, which is the first such structure determined from thermostable eubacteria, shows high similarities in overall structure with eight other members of the β-glycosidases of family 1, although the sequence identities between GlyTn and those eight members range from 26 to 47%. The TIM barrel folds in these structures were highly conserved (Table 2). The main differences among them are at the level of the connections that link the β-strand and α-helix in the β/α unit. It was also evident at the level of amino acid sequence when the sequences were aligned (Fig. 2). All eight family 1 β-glycosidases with known structures were organized as dimers (1E1E, 1CBG, 1MYR, and 1PBG), tetramers (1QVB and 1GOW) or octamers (1BGA and 1QOX). GlyTn exists in the form of monomeric enzyme, which has been demonstrated by the molecular-mass estimation of about 50 kDa on the native enzyme by a gel filtration method (21). The surface area that is accessible to solvent (1.4 Å probe radius) of one GlyTn molecule is about 16,000 Å2. The surface area buried on the interface of the two molecules A and B in the asymmetric unit is about 1,200 Å2 (600 Å2 per molecule). It corresponds to about 3.7% of the solvent-exposed surface of one molecule.
TABLE 2.
Structural comparisons for TIM barrel folds
| Enzyme | rms value (Å)a | Sequence identity (%) |
|---|---|---|
| 1BGA | 0.99 | 45 |
| 1PBG | 1.32 | 34 |
| 1QOX | 1.16 | 47 |
| 1CBG | 1.30 | 35 |
| 1E1E | 1.29 | 28 |
| 1MYR | 1.14 | 28 |
| 1GOW | 1.31 | 27 |
| 1QVB | 1.49 | 26 |
The rms values were calculated by a least-squares fit after superimposition of the 184 Ca atoms in the TIM barrel folds.
Active site.
Enzymatic hydrolysis of glycosidic bonds can be performed via two major mechanisms, giving rise to either an overall retention, or an overall inversion, of the anomeric configuration (12). Two critical residues (a proton donor and a nucleophile or base) are required for this reaction (12). Catalysis by retaining family 1 β-glycosidases proceeds via a double-displacement mechanism, and the active site contains a pair of carboxylic acids that are about 5.5 Å apart. The two motifs T(F/L/M)NE(P/L/I) and -(I/V)TENG (involved in glycone binding and enzymatic hydrolysis of glycosidic bonds within the active site) are highly conserved (55). In the structure of GlyTn, the acid catalyst Glu164 in motif -TLNEP and the nucleophilic Glu338 in motif -ITENG were found at β-strands 4 and 7, respectively (Fig. 1, 2). The distance between the Cδ atoms of the two residues is 5.25 Å, which is consistent with the properties of retaining β-glycosidases.
Structural basis for thermal stability.
Structural comparisons between thermophilic proteins from organisms living under extreme conditions and their mesophilic counterparts have been utilized to discover the possible thermostabilizing factors. The contributions of parameters (including ion pairs, hydrogen bonding, secondary structure, cavities, surface areas, amino acid composition, and flexibility) to the protein stability have been analyzed extensively (45, 48, 49). It seems that the thermal stability cannot be explained by a unique mechanism. Each thermostable protein uses one or a combination of the mechanisms elucidated in comparative studies to maintain its structure at high temperature.
The optimal growing temperature of T. nonproteolyticus HG102 (producing GlyTn) is 65°C (8), while the two hyperthermophilic archaea S. solfataricus and T. sphaera aggregans (producing 1GOW and 1QVB, respectively) can grow at temperatures as high as 87 and 90°C (9, 54). GlyTn displays optimal activity with a half-life of 2.5 h at 90°C. Enzyme 1GOW shows stronger thermostability, with a half-life of 48 h at 85°C (34); similarly, 1QVB can retain 95% of its activity after incubation at 80°C for 130 h (9). Other mesophilic glycosidases with known structures are from eubacteria or plants growing at normal temperature and should show weaker thermostability than GlyTn. For 1QOX, 80% of its activity remained after being heated at 50°C for 15 min in phosphate buffer, while 1% was left after 15 min at 60°C (37). The eubacterium B. circulans producing 1QOX cannot grow well at temperatures higher than 40°C (54). The half-life of 1BGA produced by B. polymyxa (whose highest growing temperature is 40°C [54]) is only 3.6 min at 48°C (31). Structural comparisons between GlyTn and these mesophilic and hyperthermophilic β-glycosidases would help to elucidate the possible structural determinants of its thermostability.
Amino acid composition and the stabilization of secondary structure.
The amino acid compositions of the nine family 1 β-glycosidases with known three-dimensional structures are listed in Table 3. The glycosidases 1GOW and 1QVB from hyperthermophilic archaea do not show significant changes in amino acid composition compared to the other six glycosidases from mesophiles. For GlyTn, the significant changes were (i) high content of Ala residues, (ii) high content of Pro residues, (iii) low content of thermolabile residues, and (iv) high Arg/Lys ratio. Besides the high content of Ala in helices, the number of β-branched residues decreased significantly in GlyTn (Table 4). The secondary structure contents of the nine glycosidases are very similar (Table 3), but two significant amino acid composition changes in GlyTn (increased content of Ala and decreased content of β-branched residues) are both helpful for the stabilization of α-helices. The α-helices can be stabilized by the introduction of residues with a high level of helix-forming propensity, such as Ala. The level of Ala content in the α-helices of GlyTn (17.9%) is much higher than those in mesophilic glycosidases and hyperthermophilic enzymes from archaea. The β-branched residues Ile, Val, and Thr were found to destabilize helices (38, 40). Their effects were ascribed to conformational entropy loss upon transfer from the extended conformation to the helix (10). The percentage of Val, Ile, and Thr in the helices of GlyTn is 9.5%, which is much lower than those of other glycosidases. Facchiano et al. had also found that the helices of thermophilic proteins contain a lower percentage of β-branched residues than their mesophilic equivalents (16). These two features that may contribute to the thermostability of GlyTn were not observed in the hyperthermophilic glycosidases from archaea.
TABLE 3.
Amino acid compositions and secondary structures
| Enzyme | No. (%) of amino acids
|
Total no. of amino acids | % Charged amino acids | % Hydrophobic amino acids | % Pro amino acid | % Ala amino acid | % Thermolabile amino acids | Arg/Lys ratio | % α-Helices | % β-Sheets | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ala | Cys | Asp | Glu | Phe | Gly | His | Ile | Lys | Leu | Met | Asn | Pro | Gln | Arg | Ser | Thr | Val | Trp | Tyr | ||||||||||
| GlyTn | 56 (12.84) | 2 (0.46) | 22 (5.05) | 34 (7.80) | 19 (4.36) | 40 (9.17) | 12 (2.75) | 14 (3.21) | 4 (0.92) | 44 (10.09) | 4 (0.92) | 9 (2.06) | 35 (8.03) | 7 (1.61) | 42 (9.63) | 13 (2.98) | 16 (3.67) | 26 (5.96) | 13 (2.98) | 24 (5.50) | 436 | 23.4 | 48.4 | 8.0 | 12.8 | 5.0 | 10.5 | 38.5 | 15.4 |
| 1BGA | 25 (5.59) | 5 (1.12) | 28 (6.26) | 27 (6.04) | 23 (5.15) | 39 (8.72) | 17 (3.80) | 30 (6.71) | 8 (1.79) | 31 (6.94) | 12 (2.68) | 25 (5.59) | 18 (4.03) | 24 (5.37) | 25 (5.59) | 21 (4.70) | 22 (4.92) | 29 (6.49) | 15 (3.36) | 23 (5.15) | 447 | 19.7 | 40.9 | 4.0 | 5.6 | 14.8 | 3.1 | 38.9 | 16.1 |
| 1PBG | 33 (7.05) | 2 (0.43) | 40 (8.55) | 37 (7.91) | 25 (5.34) | 38 (8.12) | 19 (4.06) | 28 (5.98) | 34 (7.26) | 28 (5.98) | 8 (1.71) | 22 (4.70) | 20 (4.27) | 10 (2.14) | 14 (2.29) | 15 (3.21) | 20 (4.27) | 28 (5.98) | 10 (2.15) | 37 (7.91) | 468 | 26.7 | 38.5 | 4.3 | 7.1 | 9.0 | 0.4 | 40.6 | 15.8 |
| 1QOX | 30 (6.68) | 5 (1.11) | 34 (7.57) | 27 (6.01) | 16 (3.56) | 46 (10.24) | 13 (2.90) | 29 (6.46) | 17 (3.79) | 41 (9.13) | 14 (3.12) | 20 (4.45) | 19 (4.23) | 11 (2.45) | 21 (4.68) | 22 (4.90) | 17 (3.79) | 21 (4.68) | 16 (3.56) | 30 (6.68) | 449 | 22.0 | 41.4 | 4.0 | 6.7 | 11.1 | 1.2 | 39.0 | 16.0 |
| 1CBG | 36 (7.35) | 5 (1.02) | 31 (6.33) | 24 (4.90) | 31 (6.33) | 37 (7.55) | 13 (2.65) | 23 (4.69) | 32 (6.53) | 44 (8.98) | 9 (1.84) | 28 (5.71) | 26 (5.31) | 9 (1.84) | 26 (5.31) | 30 (6.12) | 20 (4.08) | 21 (4.29) | 12 (2.45) | 33 (6.73) | 490 | 23.1 | 41.2 | 5.3 | 7.4 | 10.4 | 0.8 | 39.2 | 15.5 |
| 1E1E | 29 (5.66) | 5 (0.98) | 35 (6.84) | 31 (6.05) | 26 (6.33) | 42 (8.20) | 10 (1.95) | 28 (5.47) | 33 (6.45) | 39 (7.62) | 13 (2.54) | 34 (6.64) | 32 (6.25) | 11 (2.15) | 22 (4.30) | 33 (6.45) | 24 (4.69) | 21 (4.10) | 12 (2.34) | 32 (6.25) | 512 | 23.6 | 39.1 | 6.3 | 5.7 | 12.3 | 0.7 | 36.1 | 15.2 |
| 1MYR | 23 (4.59) | 8 (1.60) | 38 (7.58) | 20 (3.99) | 27 (5.39) | 46 (9.18) | 12 (2.40) | 32 (6.39) | 25 (4.99) | 35 (6.99) | 8 (1.60) | 32 (6.13) | 26 (5.19) | 16 (3.19) | 18 (3.59) | 34 (6.79) | 34 (6.79) | 20 (3.99) | 10 (2.00) | 37 (7.39) | 501 | 20.2 | 36.1 | 5.2 | 4.6 | 12.8 | 0.7 | 38.1 | 17.0 |
| 1GOW | 28 (5.73) | 1 (0.20) | 32 (6.54) | 30 (6.13) | 23 (4.70) | 40 (8.18) | 13 (2.66) | 20 (4.09) | 23 (4.70) | 36 (7.36) | 11 (2.25) | 30 (6.13) | 26 (5.32) | 11 (2.25) | 32 (6.54) | 32 (6.54) | 20 (4.09) | 30 (6.13) | 17 (3.48) | 34 (6.95) | 489 | 23.9 | 39.0 | 5.3 | 5.7 | 10.8 | 1.4 | 34.7 | 17.4 |
| 1QVB | 29 (6.03) | 2 (0.42) | 29 (6.03) | 33 (6.86) | 22 (4.57) | 35 (7.28) | 12 (2.49) | 23 (4.78) | 29 (6.03) | 42 (8.73) | 11 (2.29) | 27 (5.61) | 30 (6.24) | 19 (1.87) | 24 (4.99) | 27 (5.61) | 11 (2.29) | 41 (8.52) | 17 (3.53) | 28 (5.82) | 481 | 23.9 | 44.7 | 6.2 | 6.0 | 10.2 | 0.83 | 36.2 | 14.6 |
Charged residues, Arg, Lys, Asp, and Glu; hydrophobic residues, Ala, Val, Ile, Leu, Trp, Phe, Met, and Pro; thermolabile residues, Gln, Asn, Cys, and Met.
TABLE 4.
Amino acid composition in the α-helices
| Amino acid | No. of amino acids
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| GlyTn | 1BGA | 1PBG | 1QOX | 1CBG | 1E1E | 1MYR | 1GOW | 1QVB | |
| Ala | 30 | 12 | 23 | 17 | 18 | 12 | 7 | 16 | 20 |
| Cys | 1 | 2 | 2 | 2 | 1 | 2 | 3 | 0 | 0 |
| Asp | 8 | 13 | 17 | 16 | 12 | 12 | 16 | 8 | 9 |
| Glu | 17 | 11 | 20 | 13 | 14 | 17 | 10 | 11 | 13 |
| Phe | 6 | 9 | 9 | 7 | 9 | 10 | 11 | 9 | 7 |
| Gly | 6 | 4 | 7 | 5 | 7 | 4 | 9 | 6 | 2 |
| His | 9 | 10 | 12 | 6 | 8 | 7 | 7 | 5 | 6 |
| Ile | 7 | 13 | 12 | 11 | 10 | 11 | 17 | 9 | 7 |
| Lys | 2 | 2 | 15 | 6 | 13 | 16 | 17 | 10 | 14 |
| Leu | 25 | 16 | 15 | 26 | 20 | 20 | 14 | 10 | 12 |
| Met | 1 | 4 | 4 | 6 | 5 | 5 | 5 | 4 | 5 |
| Asn | 0 | 4 | 5 | 3 | 6 | 10 | 5 | 10 | 10 |
| Pro | 7 | 3 | 3 | 3 | 5 | 6 | 5 | 5 | 6 |
| Gln | 3 | 13 | 6 | 7 | 8 | 6 | 11 | 5 | 4 |
| Arg | 20 | 15 | 6 | 11 | 10 | 7 | 8 | 16 | 11 |
| Ser | 3 | 7 | 6 | 5 | 7 | 9 | 10 | 8 | 8 |
| Thr | 2 | 6 | 3 | 4 | 8 | 7 | 8 | 5 | 2 |
| Val | 7 | 12 | 10 | 9 | 8 | 6 | 9 | 11 | 17 |
| Trp | 4 | 6 | 3 | 6 | 5 | 5 | 5 | 7 | 8 |
| Tyr | 10 | 12 | 12 | 12 | 18 | 13 | 14 | 15 | 13 |
| Total | 168 | 174 | 190 | 175 | 192 | 185 | 191 | 170 | 174 |
| % Ala | 17.9 | 6.9 | 12.1 | 9.7 | 9.4 | 6.5 | 3.7 | 9.4 | 11.5 |
| % β-branched residuesa | 9.5 | 17.8 | 13.2 | 13.7 | 13.5 | 13.0 | 17.8 | 14.7 | 14.9 |
β-branched residues: Ile, Val, and Thr.
Pro residues.
Matthews et al. have proposed that the protein structures can be stabilized by decrease in their entropy of unfolding (32). Pro differs from all other amino acids because the side chain curls back to the preceding peptide-bond nitrogen and forms the five-member pyrrolidine ring. It can adopt only a few configurations and can restrict the configurations allowed for the preceding residue (44); thus, it has the lowest conformational entropy. Replacement of other residues by Pro at suitable positions can enhance protein thermostability (19). It has been noted that Pro has an increased occurrence in thermophilic proteins, especially in loops (5, 30, 52). The Pro content in GlyTn (8.0%) is the highest of all nine glycosidases, and the positional distribution in GlyTn of the total 35 Pro residues was examined. In order of frequency, these Pro residues lie in random coil, helices, sheet, and turn. There are 23 Pro residues occurring in random coils, and the Pro-richest regions are the four loops: 248 to 265, 291 to 297, 301 to 315, and 330 to 333 (Fig. 2). There are six (252, 255, 259, 260, 261, and 263), three (291, 295, and 297), three (302, 303, and 306), and two (331 and 333) Pro residues in these loops. These constrained loops often strengthen the stabilizing interactions between the two adjacent core elements, thus increasing the protein rigidity (48). Of the seven Pro residues occurring in helices, five of them are at the N1 position. They are Pro93, Pro165, Pro228, Pro316, and Pro356. At the N1 position, a kink in the peptide backbone introduced by Pro can thermodynamically stabilize an α-helix (57).
Surfaces and cavities.
The importance of the hydrophobic effect on the stability of proteins has been accepted (13). The calculation of the ASA was used to simplify the complex analytical solution for the hydrophobic interaction (15, 56). It has been suggested that the reduction of hydrophobic ASA contributes to the stability of proteins (2, 20, 27). The charged and hydrophobic surface areas of the nine glycosidases were calculated and are listed in Table 5. From the results, we can see that reduction of hydrophobic surface areas does not contribute to the stability of GlyTn.
TABLE 5.
Surface areas and cavities
| Characteristic | Enzyme
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| GlyTn | 1BGA | 1PBG | 1QOX | 1CBG | 1E1E | 1MYR | 1GOW | 1QVB | |
| Total ASA of the monomer | 16,120 | 16,251 | 17,522 | 15,968 | 19,938 | 18,715 | 18,087 | 18,361 | 18,741 |
| Hydrophobic ASA | 5,139 | 3,856 | 4,241 | 4,337 | 5,110 | 5,180 | 4,003 | 4,353 | 5,625 |
| Charged ASA | 7,723 | 6,090 | 8,434 | 6,733 | 8,734 | 8,098 | 6,456 | 7,942 | 8,830 |
| Hydrophobic ASA/total ASA (%) | 32 | 24 | 24 | 27 | 26 | 28 | 22 | 24 | 30 |
| No. of cavities | 4 | 3 | 3 | 2 | 3 | 6 | 3 | 4 | 5 |
| Vol. of cavities | 38.4 | 46.0 | 86.7 | 19.9 | 56.8 | 92.2 | 23.5 | 54.0 | 71.7 |
A decrease in the number and volume of the internal cavities and pockets has also been indicated as providing thermal stability (45). The nine glycosidases were analyzed for the presence of the cavities to try to ascertain whether they contribute to the thermal stability of these enzymes (Table 5). No significant trend for the decrease of the number and volume of cavities was observed in the thermostable GlyTn, 1GOW, and 1QVB.
Hydrogen bonds.
Hydrogen bonding was compared among the nine glycosidases (Table 6). The hydrogen bonds were divided into three classes: main chain-main chain (MM H-bonds), main chain-side chain (MS H-bonds), and side chain-side chain (SS H-bonds). There is no significant difference in the number of hydrogen bonds among mesophilic, thermophilic, and hyperthermophilic enzymes.
TABLE 6.
Ion pairs and hydrogen bonds
| Characteristic | Enzyme
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| GlyTn | 1BGA | 1PBG | 1QOX | 1CBG | 1E1E | 1MYR | 1GOW | 1QVB | |
| All protein-protein H-bonds | 376 | 450 | 445 | 444 | 459 | 432 | 494 | 495 | 433 |
| MM H-bonds | 254 | 285 | 284 | 283 | 283 | 263 | 293 | 285 | 275 |
| MS H-bonds | 61 | 100 | 85 | 92 | 101 | 99 | 121 | 119 | 91 |
| SS H-bonds | 61 | 65 | 76 | 69 | 75 | 70 | 80 | 91 | 67 |
| No. of hydrogen bonds per residue | 0.88 | 1.01 | 0.98 | 0.99 | 0.94 | 0.88 | 0.99 | 1.01 | 0.90 |
| No. of ion pairs | 44 | 32 | 45 | 34 | 27 | 32 | 23 | 47 | 32 |
| No. of ion pairs per residue | 0.10 | 0.07 | 0.10 | 0.08 | 0.06 | 0.07 | 0.05 | 0.10 | 0.07 |
| No. of ion pairs formed by Arg/Lys/His | 35/3/6 | 19/5/8 | 17/20/8 | 21/7/6 | 12/11/4 | 14/13/5 | 13/7/3 | 33/3/11 | 23/3/6 |
| No. of ion pairs formed by Glu/Asp | 23/21 | 15/17 | 23/22 | 11/23 | 10/17 | 14/18 | 9/14 | 19/28 | 14/18 |
| % of ion pairs including Arg | 80 | 59 | 38 | 62 | 44 | 44 | 57 | 70 | 72 |
| % of ion pairs occurring in ion network with three or more charged residues | 68 | 47 | 60 | 56 | 41 | 28 | 17 | 62 | 47 |
Ion pairs.
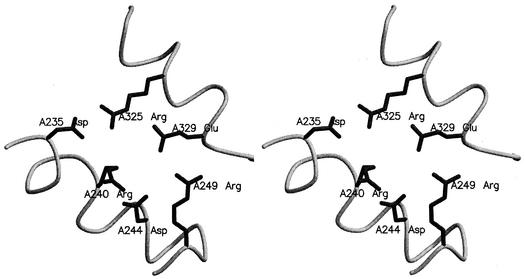
Earlier work by Perutz suggested that electrostatic interactions represent a significant stabilizing factor in folded protein (39). After comprehensive studies of proteins with mesophilic and thermophilic structures in the PDB, Szilagyi and Zavodszky proposed that the only parameter with a consistent and significant contribution to thermal stability is the number of charged pairs and ion pair networks (45). An extensive analysis of ion pairs in the nine glycosidases was carried out, and the results are listed in Table 6. The number of ion pairs per residue was 0.10 for thermophilic GlyTn, 1GOW, and mesophilic 1PBG, a number which was the highest among the nine glycosidases. For thermophilic 1QVB, the number was 0.07, not departing significantly from the value of other mesophilic enzymes. After analyzing the constitutions of ion pairs in the nine enzymes, it was found that the ion pairs formed by Arg had the largest percentage for the three thermophilic enzymes. The percentages of ion pairs with Arg were 80, 70, and 72% in GlyTn, 1GOW, and 1QVB, respectively. In GlyTn, the high percentage of ion pairs with Arg is related to its extremely high Arg/Lys ratio. The Arg-forming ion pairs are over 70% of all of the ion pairs in 1GOW and 1QVB, although the Arg/Lys ratios are 1.4 and 0.8. The strong preference for utilization of Arg in ion pairs is determined by its properties. Arg is better at maintaining ion pairs at elevated temperature, because it has a higher pKa than Lys. The pKa value typically drops with increasing temperature (35, 50). It might also be due to its δ-guanido and longer side chain, which offer a wider range of possible ion pairs than those of Lys (35). In GlyTn, 68% of the ion pairs occur as part of multiple ion pair networks involving three or more charged residues. Many of the ion pair networks help to cross-link noncontiguous parts of the structure at the protein surface. One network (involving six charged residues, Asp235, Arg240, Asp244, Arg249, Arg325, and Glu329) acts as electrostatic links between two helices, α5 and α6 (Fig. 3). The other six-residue-forming ion pair network, which links the two connections β7-α7 and β8-α8, includes Asp345, Asp355, Arg358, Asp389, Arg400, and Lys416.
FIG. 3.
Stereo figures of an ion pair network acting as electrostatic links between two helices, α5 and α6.
Reduction in thermolabile residues.
Of the 20 amino acids, Asn, Gln, Cys, and Met can be classified as thermolabile due to their tendency to undergo deamidation or oxidation at high temperature and therefore may be naturally discriminated against in thermostable proteins (41). In GlyTn, the level of Asn, Gln, Met, and Cys is 5.0%, which is much lower than those of other glycosidases. The difference mainly comes from the decrease of Asn, Gln, and Met content, and the difference of Cys is not significant because the content of Cys in all the nine glycosidases is very low. From the structural view, there are seven occurrences of the Gln residues in 1BGA changing to Glu or Asp that form an ion pair in GlyTn. Three Asn residues in 1BGA are changed to Glu or Arg that form an ion pair. Six Gln and Asn residues in 1BGA are changed to Pro that locate in loops of GlyTn.
Conclusion.
The crystal structure of β-glycosidase GlyTn from thermophilic T. nonproteolyticus HG102 has been determined at a resolution of 2.4 Å. Analysis of its primary structure and the positions of two Glu residues in the active center indicate that it belongs to the glycosyl hydrolase of retaining family 1. Of all the structural features analyzed, those that may contribute to the thermostability of GlyTn are increased stabilization of the α-helices, high content of Pro residues and the resulting restricted flexibility of loops, increased number of ion pairs and high percentage of Arg occurrence in them, and a reduction in thermolabile residues.
Acknowledgments
The project was supported by the National Key Research Development Project of China (Project No. G1999075601).
REFERENCES
- 1.Aguilar, C. F., I. Sanderson, M. Moracci, M. Ciaramella, R. Nucci, M. Rossi, and L. H. Pearl. 1997. Crystal structure of the β-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus: resilience as a key factor in thermostability. J. Mol. Biol. 271:789-802. [DOI] [PubMed] [Google Scholar]
- 2.Auerbach, G., R. Ostendorp, L. Prade, I. Korndorfer, T. Dams, R. Huber, and R. Jaenicke. 1998. Lactate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima: the crystal structure at 2.1 Å resolution reveals strategies for intrinsic protein stabilization. Structure 6:769-781. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, D. J., and T. M. Thornton. 1983. Ion-pairs in proteins. J. Mol. Biol. 168:867-885. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, T., C. G. Suresh, S. P. Tolley, E. J. Dodson, and M. A. Hughes. 1995. The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure 3:951-960. [DOI] [PubMed] [Google Scholar]
- 5.Bogin, O., M. Peretz, Y. Hacham, Y. Korkhin, F. Frolow, A. J. Kalb (Gilboa), and Y. Burstein. 1998. Enhanced thermal stability of Clostridium beijerinckii alcohol dehydrogenase after strategic substitution of amino acid residues with prolines from the homologous thermophilic Thermoanaerobacter brockii alcohol dehydrogenase. Protein Sci. 7:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Burmeister, W. P., S. Cottaz, H. Driguez, R. Iori, S. Palmieri, and R. B. Henrissat. 1997. The crystal structure of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure 5:663-675. [DOI] [PubMed] [Google Scholar]
- 8.Cai, M. Y., S. J. Yang, J. F. Liu, and J. Wei. 1992. Thermus nonproteolyticus sp. nov., a new species of thermophilic nonsporeformer producing LDH. Acta Microbiol. Sin. 32:233-237. [Google Scholar]
- 9.Chi, Y. I., L. A. Martinez-Cruz, J. Jancarik, R. V. Swanson, D. E. Robertson, and S. H. Kim. 1999. Crystal structure of the β-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability. FEBS Lett. 445:375-383. [DOI] [PubMed] [Google Scholar]
- 9a.Collaborative Computational Project Number 4.1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 10.Creamer, T. P., and G. D. Rose. 1994. Alpha-helix-forming propensities in peptides and proteins. Proteins 19:85-97. [DOI] [PubMed] [Google Scholar]
- 11.Czjzek, M., M. Cicek, V. Zamboni, W. P. Burmeister, D. R. Bevan, B. Henrissat, and A. Esen. 2001. Crystal structure of a monocotyledon (maize ZMGlu1) β-glucosidase and a model of its complex with p-nitrophenyl β-d-thioglucoside. Biochem. J. 354:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, G. J., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853-859. [DOI] [PubMed] [Google Scholar]
- 13.Dill, K. A. 1990. Dominant forces in protein folding. Biochemistry 29:7133-7155. [DOI] [PubMed] [Google Scholar]
- 14.Dion, M., L. Fourage, J. N. Hallet, and B. Colas. 1999. Cloning and expression of a β-glycosidase gene from Thermus thermophilus. Sequence and biochemical characterization of the encoded enzyme. Glycoconj. J. 16:27-37. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg, D., and A. D. McLachlan. 1986. Solvation energy in protein folding and binding. Nature 319:199-203. [DOI] [PubMed] [Google Scholar]
- 16.Facchiano, A. M., G. Colonna, and R. Ragone. 1998. Helix stabilizing factors and stabilization of thermophilic proteins: an X-ray based study. Protein Eng. 11:753-760. [DOI] [PubMed] [Google Scholar]
- 17.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 18.Hakulinen, N., S. Paavilainen, T. Korpela, and J. Rouvinen. 1999. The crystal structure of β-glucosidase from Bacillus circulans sp. alkalophilus: ability to form long polymeric assemblies. J. Struct. Biol. 129:69-79. [DOI] [PubMed] [Google Scholar]
- 19.Hardy, F., G. Vriend, O. R. Veltman, B. van der Vinne, G. Venema, and V. J. Eijsink. 1993. Stabilization of Bacillus stearothermophilus neutral protease by introduction of prolines. FEBS Lett. 317:89-92. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto, H., T. Inoue, M. Nishioka, S. Fujiwara, M. Takagi, T. Imanaka, and Y. Kai. 1999. Hyperthermostable protein structure maintained by intra- and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase. J. Mol. Biol. 292:707-716. [DOI] [PubMed] [Google Scholar]
- 21.He, X. Y., S. Z. Zhang, and S. J. Yang. 2001. Cloning and expression of thermostable β-glycosidase gene from Thermus nonproteolyticus HG102 and characterization of recombinant enzyme. Appl. Biochem. Biotechnol. 94:243-255. [DOI] [PubMed] [Google Scholar]
- 22.He, X. Y., X. Q. Wang, S. J. Yang, W. R. Chang, and D. C. Liang. 2001. Overexpression, purification, crystallization and preliminary crystallographic studies on a thermostable β-glycosidase from Thermus nonproteolyticus HG102. Acta Crystallogr. D 57:1650-1651. [DOI] [PubMed] [Google Scholar]
- 23.Henrissat, B., and G. J. Davies. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637-644. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa, Y., G. C. Look, and C. H. Wong. 1992. Enzyme-catalyzed oligosaccharide synthesis. Anal. Biochem. 202:215-238. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 26a.Kleywegt, G. J. and T. A. Jones. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:178-185. [DOI] [PubMed] [Google Scholar]
- 27.Knapp, S., S. Kardinahl, N. Hellgren, G. Tibbelin, G. Schafer, and R. Ladenstein. 1999. Refined crystal structure of a superoxide dismutase from the hyperthermophilic archaeon Sulfolobus acidocaldarius at 2.2 Å resolution. J. Mol. Biol. 285:689-702. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 29.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 30.Li, C., J. Heatwole, S. Soelaiman, and M. Shoham. 1999. Crystal structure of a thermophilic alcohol dehydrogenase substrate complex suggests determinants of substrate specificity and thermostability. Proteins 37:619-627. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Camacho, C., J. Salgado, J. L. Lequerica, A. Madarro, E. Ballestar, L. Franco, and J. Polaina. 1996. Amino acid substitutions enhancing thermostability of Bacillus polymyxa β-glucosidase A. Biochem. J. 314:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews, B. W., H. Nicholson, and W. J. Becktel. 1987. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. USA 84:6663-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrit, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 34.Moracci, M., R. Nucci, F. Febbraio, C. Vaccaro, N. Vespa, F. La Cara, and M. Rossi. 1995. Expression and extensive characterization of a β-glycosidase from the extreme thermoacidophilic archaeon Sulfolobus solfataricus in Escherichia coli: authenticity of the recombinant enzyme. Enzyme Microbiol. Technol. 17:992-997. [DOI] [PubMed] [Google Scholar]
- 35.Mrabet, N. T., A. Van den Broeck, I. Van den brande, P. Stanssens, Y. Laroche, A. M. Lambeir, G. Matthijssens, J. Jenkins, M. Chiadmi, H. van Tilbeurgh, F. Rey, J. Janin, W. J. Quax, J. Lasters, M. D. Maeyer, and D. J. Wosak. 1992. Arginine residues as stabilizing elements in proteins. Biochemistry 31:2239-2253. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 37.Paanilainen, S., J. Hellman, and P. Korpela. 1993. Purification, characterization, gene cloning, and sequencing of a new β-glucosidase from Bacillus circulans subsp. alkalophilus. Appl. Environ. Microbiol. 59:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padmanabhan, S., S. Marqusee, T. Ridgeway, T. M. Laue, and R. L. Baldwin. 1990. Relative helix-forming tendencies of nonpolar amino acids. Nature 344:268-270. [DOI] [PubMed] [Google Scholar]
- 39.Perutz, M. F. 1976. Electrostatic effects in protein. Science 201:1187-1191. [DOI] [PubMed] [Google Scholar]
- 40.Piela, L., G. Nemethy, and H. A. Scheraga. 1987. Conformational constraints of amino acid side chains in α-helices. Biopolymers 26:1273-1286. [DOI] [PubMed] [Google Scholar]
- 41.Russell, R. J., J. M. Ferguson, D. W. Hough, M. J. Danson, and G. L. Taylor. 1997. The crystal structure of citrate synthase from the hyperthermophilic archaeon Pyrococcus furiosus at 1.9 Å resolution. Biochemistry 36:9983-9994. [DOI] [PubMed] [Google Scholar]
- 42.Sakabe, N. 1991. X-ray diffraction data collection system for modern protein crystallography with a Weissenberg camera and an imaging plate using synchrotron radiation. Nucl. Instrum. Methods Phys. Res. A303:448-463. [Google Scholar]
- 43.Sanz-Aparicio, J., J. A. Hermoso, M. Martinez-Ripoll, J. L. Lequerica, and J. Polaina. 1998. Crystal structure of β-glucosidase from Bacillus polymyxa: insights into the catalytic activity of family 1 glycosyl hydrolases. J. Mol. Biol. 275:491-502. [DOI] [PubMed] [Google Scholar]
- 44.Sriprapundh, D. C., D. C. Vieille, and J. G. Zeikus. 2000. Molecular determinants of xylose isomerase thermal stability and activity: analysis by site-directed mutagenesis. Protein Eng. 13:259-265. [DOI] [PubMed] [Google Scholar]
- 45.Szilagyi, A., and P. Zavodszky. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure Fold Des. 8:493-504. [DOI] [PubMed] [Google Scholar]
- 46.Takase, M., and K. Horikoshi. 1989. Purification and properties of a β-glucosidase. Agric. Biol. Chem. 51:559-560. [Google Scholar]
- 47.Thomas, K. N., and R. K. William. 1986. Thermophiles, p. 197-216. In T. D. Brock (ed.), Thermophiles: general, molecular, and applied microbiology. Wiley-Interscience, New York, N.Y.
- 48.Vieille, C., and G. J. Zeikus. 1996. Thermozymes: identifying molecular determinants of protein structural and functional stability. Trends Biotechnol. 14:183-190. [Google Scholar]
- 49.Vieille, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volkin, D. B., and C. R. Middaugh. 1992. The effect of temperature on protein structure, p. 215-247. In T. J. Ahern and C. Manning (ed.), Stability of protein pharmaceuticals, part A. Chemical and physical pathways of protein degradation. Plenum Press, New York, N.Y.
- 51.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe, K., Y. Hata, H. Kizaki, Y. Katsube, and Y. Suzuki. 1997. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 Å resolution: structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 269:142-153. [DOI] [PubMed] [Google Scholar]
- 53.Wiesmann, C., G. Beste, W. Hengstenberg, and G. E. Schulz. 1995. The three-dimensional structure of 6-phospho-β-galactosidase from Lactococcus lactis. Structure 3:961-968. [DOI] [PubMed] [Google Scholar]
- 54.Williams, S. T., M. E. Sharpe, and J. G. Holt (ed.). 1989. Bergey's manual of systematic bacteriology. Williams and Wilkins, Baltimore, Md.
- 55.Wither, S. G. 2001. Mechanism of glycosyl transferases and hydrolases. Carbohydrate Polymers 44:325-337. [Google Scholar]
- 56.Xie, D., and E. Freire. 1994. Molecular basis of cooperativity in protein folding. Thermodynamic and structural conditions for the stabilization of compact denatured states. Proteins 19:291-301. [DOI] [PubMed] [Google Scholar]
- 57.Yun, R. H., A. Anderson, and J. Hermans. 1991. Proline in α-helix: stability and conformation studied by dynamics simulation. Proteins 10:219-228. [DOI] [PubMed] [Google Scholar]