Abstract
The crystal structures of the ζ-crystalline-like soluble quinone oxidoreductase from Thermus thermophilus HB8 (QORTt) and of its complex with NADPH have been determined at 2.3- and 2.8-Å resolutions, respectively. QORTt is composed of two domains, and its overall fold is similar to the folds of Escherichia coli quinone oxidoreductase (QOREc) and horse liver alcohol dehydrogenase. QORTt forms a homodimer in the crystal by interaction of the βF-strands in domain II, forming a large β-sheet that crosses the dimer interface. High thermostability of QORTt was evidenced by circular dichroic measurement. NADPH is located between the two domains in the QORTt-NADPH complex. The disordered segment involved in the coenzyme binding of apo-QORTt becomes ordered upon NADPH binding. The segment covers an NADPH-binding cleft and may serve as a lid. The 2′-phosphate group of the adenine of NADPH is surrounded by polar and positively charged residues in QORTt, suggesting that QORTt binds NADPH more readily than NADH. The putative substrate-binding site of QORTt, unlike that of QOREc, is largely blocked by nearby residues, permitting access only to small substrates. This may explain why QORTt has weak p-benzoquinone reduction activity and is inactive with such large substrates of QOREc as 5-hydroxy-1,4-naphthoquinone and phenanthraquinone.
An open reading frame, whose sequence indicates coding of a protein belonging to the medium-chain dehydrogenase/reductase superfamily (24), was found in the genome of the gram-negative eubacterium Thermus thermophilus HB8 (37). The protein has a nucleotide-binding fingerprint motif, AXXGXXG, and is expected to bind NADH or NADPH. The medium-chain dehydrogenase/reductase superfamily has such enzymes as ζ-crystallin (5, 11, 25-28, 32), the quinone oxidoreductase from Escherichia coli (QOREc) (6, 7, 18, 31), and horse liver alcohol dehydrogenase (LADH) (3, 4, 8, 9), all of which catalyze NADH- or NADPH-dependent redox reactions of various substrates. This superfamily consists of two subfamilies: dehydrogenases and reductases. The former, represented by LADH, require zinc ions for activity, whereas reductases do not necessarily require metal ions (6). The zinc-binding residues of LADH are replaced by other residues or deleted in the protein of T. thermophilus, indicating that the protein is a reductase. In fact, because the product of the open reading frame from T. thermophilus HB8 has NADPH-dependent p-benzoquinone reduction activity, we have called it quinone oxidoreductase (QORTt) (29).
QORTt has weaker p-benzoquinone reduction activity than ζ-crystallin and no activity for 1,2-naphthoquinone and phenanthraquinone, good ζ-crystallin substrates. ζ-Crystallin is an NADPH-dependent quinone oxidoreductase, a major eye lens protein in such vertebrates as guinea pigs and camels. It is a soluble enzyme and is distinct from membrane-bound quinone oxidoreductase, the large complex in the respiratory chain (36). It is also distinct from the mammalian quinone oxidoreductase called DT-diaphorase, a flavin adenine dinucleotide-containing enzyme that catalyses NAD(P)H-dependent two-electron reduction of quinones (10, 17). ζ-Crystallin reduces such naturally occurring quinones as 1,2-naphthoquinone and phenanthraquinone but is inactive with menadione, ubiquinone, and vitamins K1 and K2 (25, 26, 28). The physiological function of ζ-crystallin is speculated to be detoxification or the metabolism of a quinone (5, 11, 27, 32). Genes encoding ζ-crystallin homologs are widely distributed from bacteria to higher plants and animals, but the only well characterized one is the P1 ζ-crystallin from Arabidopsis thaliana (19, 20). The P1 gene is induced by various oxidative stress treatments in A. thaliana and confers tolerance toward oxidative stress to yeast when it is introduced into the yeast, suggesting that the enzyme is involved in an antioxidative mechanism in plants. The enzyme was shown to reduce not only quinones but also diamides and 2-alkenals which may cause cytotoxic effects. The reason why QORTt and ζ-crystallin differ in substrate specificity is not known.
An interesting problem in relation to enzyme catalysis is how conformational change of the protein occurs when the cofactor or substrate binds to the targeted protein (30, 35). LADH shows conformational change induced by coenzyme binding (4, 9). When the coenzyme or its analog binds to the cleft between the domains, the catalytic domain rotates 7.5° relative to the coenzyme-binding domain. This results in the closure of the coenzyme-binding cleft, and the active site is shielded from the solution. Increased hydrophobicity of the active site has been suggested to facilitate hydride transfer from alcohol to NAD+ (9). In the reductase subfamily, the crystal structure of the quinone oxidoreductase from E. coli (QOREc) in complex with NADPH was determined at 2.2-Å resolution (6, 7, 18, 31). QOREc, as QORTt, is an NADPH-dependent quinone oxidoreductase that has no metal ion. Our interest is how domain movement and/or any other structural changes occur when NADPH binds to quinone oxidoreductases. No structural information about this enzyme is available, however, because the structure of apo-QOREc is unknown. In addition, QORTt is expected to be thermostable because T. thermophilus HB8 is an extremely thermophilic bacterium.
To shed light on its structure-function relationship we determined the crystal structures of QORTt in the absence and presence of NADPH at resolutions of 2.3 and 2.8 Å, respectively. We report the structural basis for its substrate specificity and discuss the conformational change that occurs in QORTt upon coenzyme binding.
MATERIALS AND METHODS
Expression, purification, and crystallization of SeMet-labeled protein.
Native QORTt was expressed and purified as described elsewhere (29). To express the protein labeled with selenomethionine (SeMet), the methionine auxotrophic strain B834(DE3) (Novagen) transformed with the expression plasmid was precultured in Luria-Bertani medium supplemented with ampicillin. At mid-log phase, a portion (10 ml) was transferred to 1 liter of minimal medium containing SeMet and several vitamins and then incubated at 37°C. Cells were harvested, and the SeMet-labeled protein was purified in the same manner as the native protein, giving 13 mg of protein (0.4 mg of protein/g of cells).
The SeMet-labeled protein was crystallized under the same conditions as the native protein. Crystals of the native QORTt in complex with NADPH were grown as described previously (29). Because the electron density for the bound NADPH was broad, indicative of low occupancy, the NADPH concentration in the cocrystallization was increased from 14 to 25 mM.
Data collection.
For the cryogenic experiment, the SeMet-labeled QORTt crystal was soaked in Paratone-N (Hampton Research) and the QORTt-NADPH complex crystal was soaked in a solution of 150 mM NaCl, 20 mM Tris-HCl (pH 8.0), 0.4 M ammonium dihydrogen phosphate, 30% glycerol, and 50 mM NADPH. Each crystal was mounted on a cryo-loop and then flash-cooled in a nitrogen gas stream at 100 K. X-ray diffraction data were collected with a Mar charge-coupled device and synchrotron radiation at BL41XU, SPring-8. Multiwavelength anomalous dispersion (MAD) data for the SeMet-labeled crystal were collected for three wavelengths (peak, 0.9791 Å; edge, 0.9793 Å; remote, 0.9000 Å) at the crystal detector distance of 200 mm. Intensity data for the QORTt-NADPH complex crystal within the 180° rotation range were collected at λ = 1.000 Å and a camera distance of 180 mm. To measure the wide range of diffraction intensities, two data sets were collected with different oscillation angles and exposure times.
All data were processed with the program package HKL2000 (23). Both the apo-QORTt and the QORTt-NADPH complex crystals belong to the hexagonal space group P6122. The unit cell parameters were a = b = 77.7 Å and c = 236.6 Å for the apo-QORTt crystal and a = b = 77.6 Å and c = 235.6 Å for the QORTt-NADPH complex crystal. Data collection statistics are given in Table 1.
TABLE 1.
Data collection statistics
| Type of crystal | Wavelength (Å) | Resolution (Å) | No. of measured reflections | No. of unique reflections | Completeness (%)a | Rmerge (%)a,b |
|---|---|---|---|---|---|---|
| SeMet apo-QORTt | ||||||
| Peak | 0.9791 | 2.45 | 297,872 | 15,986 | 97.6 (76.1) | 6.1 (12.7) |
| Edge | 0.9793 | 2.45 | 298,747 | 16,013 | 97.5 (75.5) | 5.3 (15.5) |
| Remote | 0.9000 | 2.25 | 383,738 | 20,471 | 97.6 (76.6) | 7.3 (14.8) |
| QORTt-NADPH | 1.000 | 2.8 | 278,044 | 11,091 | 99.9 (100.0) | 11.6 (20.5) |
Values in parentheses are for the outermost shell.
Rmerge = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where 〈I(h)〉 is the average intensity over equivalent reflections.
Structure determination and refinement.
The structure of apo-QORTt was solved by the use of MAD data and the CNS program package (2). Two selenium atom sites were determined from the difference Patterson map, and the subsequent difference Fourier map located the remaining three selenium atoms. Density modification was applied to the electron density derived from the MAD phases. One of two sets of phases (two possible enantiomers) produced an interpretable electron density map. The overall figure of merit was 0.73 for 27,483 reflections in the resolution range of 44 to 2.5 Å.
The apo-QORTt model was built with the aid of the amino acid sequence and the program O (13). Models of the secondary structure elements were fitted manually to the electron density map and then connected by loops. A segment (residues 218 to 224) whose electron density was not clear was excluded from the model, and residues whose side chains were not clear were replaced by alanine. The structure was revised by adjusting the model and by simulated annealing and individual temperature factor refinements, in which remote data of the SeMet-labeled crystal were used. Finally, 104 water molecules and one sulfate anion were included in the refinement. The R and Rfree values for the apo-QORTt were 22.4 and 24.9%, respectively.
Because the structure of the QORTt-NADPH complex was isomorphous with that of apo-QORTt, rigid body, simulated annealing, and individual temperature factor refinements were applied to the apo-QORTt model by using diffraction data of the QORTt-NADPH complex. Electron densities for several residues in the segment invisible in the apo-QORTt, as well as for the NADPH molecule, were found in the Fo-Fc map, to which the model was fitted. After several cycles of refinement and minor manual revisions, 36 water molecules were added to the model. Residues 222 to 224 were not included, and 26 residues were replaced by alanine because their electron densities were not clear. The R and Rfree values were 21.9 and 25.6%, respectively, for the QORTt-NADPH complex. Refinement statistics are given in Table 2.
TABLE 2.
Refinement statistics
| Parameter | apo-QORTt | QORTt-NADPH complex |
|---|---|---|
| Resolution range (Å) | 50-2.3 | 50-2.8 |
| No. of reflections in working/no. in test set | 31932/3446 | 9772/1109 |
| % of reflections of working set/test set | 89.9/9.7 | 88.1/10.0 |
| R factor (%)a | 22.4 | 21.9 |
| Rfree (%)b | 24.9 | 25.6 |
| No. of atoms | ||
| Protein | 2,143 | 2,127 |
| NADPH | 48 | |
| Sulfate | 5 | |
| Solvent | 104 | 36 |
| Avg B factor (Å2) | 27.8 | 35.2 |
| Root mean square deviations of geometry from ideal values | ||
| Bonds (Å) | 0.006 | 0.009 |
| Angles (°) | 1.4 | 1.8 |
| Dihedrals (°) | 24.5 | 24.3 |
| Impropers (°) | 1.0 | 3.7 |
| Coordinate error (Å) for σA(cross- validated σA) | 0.20 (0.27) | 0.33 (0.55) |
R factor = Σ∥Fo| − |Fc∥/Σ|Fo|.
Rfree is the R factor computed for the test set of reflections omitted from the refinement process (1).
CD spectroscopy.
Circular dichroic (CD) spectra of QORTt were measured at a protein concentration of 10 μM in a 0.1-cm-path cell in a Jasco J-720W spectropolarimeter. QORTt was dissolved in a solution of 150 mM NaCl, 20 mM Tris-HCl (pH 8.0), and 1 mM dithiothreitol. To monitor its thermostability, the molar ellipticity at 222 nm was measured by changing the temperature from 15 to 95°C by heating at a rate of 1.0°C/min.
Protein Data Bank accession codes.
The atomic parameters and observed structure factors have been deposited in the RCSB Protein Data Bank (apo-QORTt, 1IZ0; QORTt-NADPH complex, 1IYZ).
RESULTS AND DISCUSSION
Overall structure.
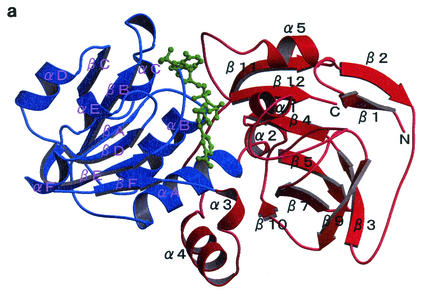
A ribbon model of the QORTt monomer is shown in Fig. 1a, and the amino acid sequence and secondary structure elements are shown in Fig. 1b. QORTt consists of two domains: domain I (residues 1 to 110 and 244 to 302) and domain II (residues 111 to 243). Domain I has five α-helices and 10 β-strands that form three β-sheets: βI (β4, β5, β7, β10, β11, and β12), βII (β3, pseudo-β8, and β9), and βIII (β1 and β2). Domain II has six α-helices and six β-strands that form one β-sheet (βIV). Domain II is composed of two βαβαβ units connected by αD. The first βαβ unit (βA, αB, and βB) has the AXXGXXG motif and is involved in NADPH binding.
FIG. 1.
Structure of QORTt. (a) Ribbon diagram with secondary structure assignments of the QORTt monomer. Domain I is shown in red, and domain II is shown in blue. NADPH (green) is between domains I and II. Secondary structure elements of QORTt were assigned with the program PROCHECK (16). (b) Amino acid sequence of QORTt expected from the DNA sequence together with secondary structure assignments. Cylinders indicate α-helices, and arrows indicate β-strands. All the figures in this paper were prepared by the programs O (13), Molray (12), MOLSCRIPT (15), and Raster3D (21).
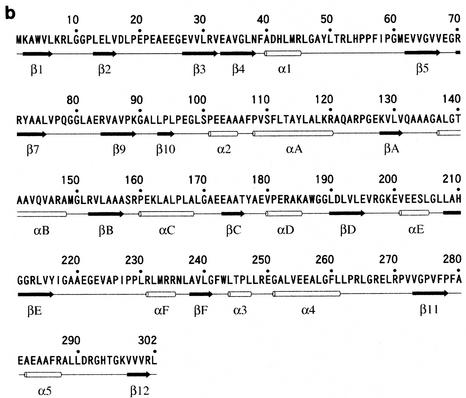
Although QORtt has very weak sequence identity to QOREc (27%) and LADH (23%), its overall structure is similar to their structures. Domain I of QORTt corresponds to the catalytic domain of LADH, and domain II corresponds to the coenzyme-binding domain (8). As the spatial arrangement of the secondary structure elements of QORTt basically coincides with that of those enzymes, the α-helices and β-strands in QORTt were so labeled to be consistent with those of the two proteins. Superimposition of Cα-traces of the QORTt-NADPH and QOREc-NADPH complexes and of the QORTt-NADPH complex and apo-LADH are shown in Fig. 2. Root mean square deviation between QORTt and QOREc is 1.7 Å for 275 Cα atoms, and between QORTt and LADH it is 2.0 Å for 259 Cα atoms.
FIG. 2.
Superimposition of Cα traces of the QORTt-NADPH complex and the QOREc-NADPH complex (Protein Data Bank code 1QOR) (a) and the QORTt-NADPH complex and apo-LADH (Protein Data Bank code 8ADH) (b). QORTt is shown in green, and QOREc and LADH are shown in blue. Segments in QOREc and LADH that are absent in QORTt are highlighted in red. Red spheres in panel b indicate the zinc atoms in LADH. For clarity, coenzymes are not shown.
Thermostability.
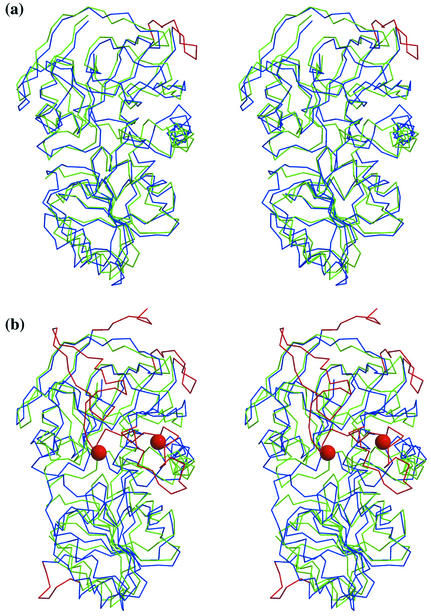
The thermostability of QORTt was investigated by CD spectroscopy. Its CD spectrum (Fig. 3, inset) has two negative peaks, at 208 and 222 nm, characteristic of an α-helix. Thermal denaturation of QORTt monitored at 222 nm (Fig. 3) occurred at about 80°C, and therefore, it is indeed a highly thermostable enzyme. A notable structural difference between QORTt and QOREc is that a surface loop in QOREc (residues 69 to 81) is absent in QORTt (Fig. 2 a). That region has strand β6 and its neighboring loop, corresponding to residues 73 to 85 of LADH; therefore, it may not affect enzyme activity or dimer association. In QOREc and LADH, the surface regions that are absent in QORTt indicate higher B factor values than the remaining regions. Similar deletions are reported in other thermostable proteins (14, 34) and are believed to be a source of thermostability. The other source of QORTt thermostability may be its high proline content (7.95%) (22, 33).
FIG. 3.
Thermal denaturation curve of QORTt monitored at 222 nm. Inset, the CD spectrum of QORTt.
Dimer structure.
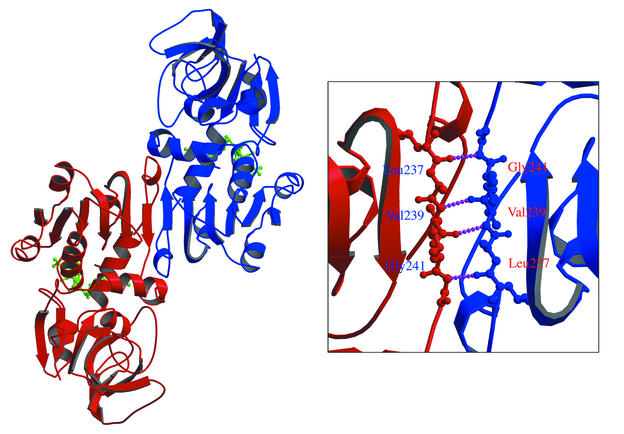
A ribbon diagram of the QORTt dimer is shown in Fig. 4. Gel filtration results indicate that in solution QORTt molecules exist as dimers (29). Each crystallographic asymmetric unit has one QORTt molecule, and the two QORTt molecules are close together at the crystallographic two-fold axis. The dimer shown in Fig. 4 therefore corresponds to the dimer in solution. Its crystal structure shows that any other subunit association is unlikely. The two QORTt subunits interact in domain II, in which several hydrogen bonds between the main chains of the βF strands connect the subunits, forming a 12-strand β-sheet that crosses the dimer interface. The mode of subunit interaction in QORTt is identical to that in QOREc (31).
FIG. 4.
Structure of the QORTt dimer. Inset, close-up view of the interface. Residues of βF strands are shown with ball-and stick models. Dotted lines indicate hydrogen bonds.
NADPH-binding site.
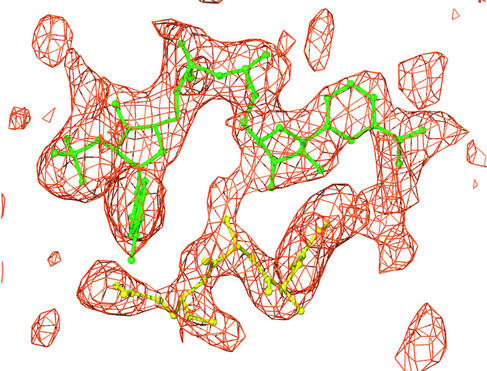
NADPH is located in the cleft between domains I and II. The Fo-Fc electron density map around NADPH is shown in Fig. 5. Previously, cocrystals of QORTt and NADPH were prepared at a concentration of 14 mM (29). The resulting electron density for NADPH and residues invisible in the apo-QORTt map became visible but was still broad, indicating that NADPH is partially occupied and that the residues are partly fixed (data not shown). The electron density for the corresponding region of the present crystal, obtained in the presence of 25 mM NADPH, was much clearer.
FIG. 5.
Fo-Fc map around NADPH (contour level > 2σ). Models of NADPH (green) and the disordered region in apo-QORTt (yellow) are overlaid on the map.
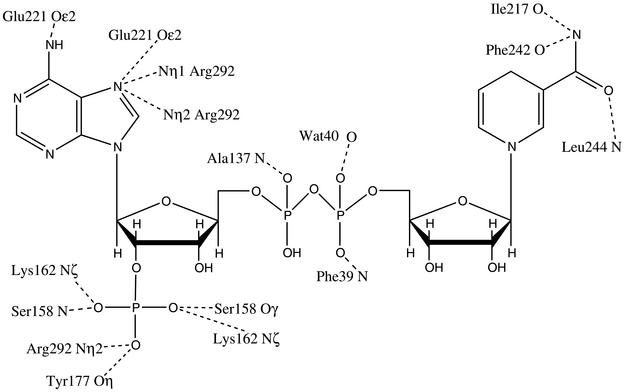
Possible hydrogen bonds involving NADPH are shown in Fig. 6. The adenine ring is sandwiched between the main chains of Ala220 and Glu221 and the side chain of Arg292. Ala220 and Glu221 are disordered in apo-QORTt. Interactions between these residues and the adenine ring may fix the conformation of the disordered segment. In fact, several hydrogen bonds exist between these residues and the adenine ring. The amino group of the adenine ring is exposed to solvent. The 2′-phosphate group is surrounded by polar and positively charged side chains (Ser158, Lys162, Tyr177, and Arg292). These residues provide a positively charged hydrophilic pocket which the 2′-phosphate group of adenine ribose enters. A sulfate anion is located in the pocket of apo-QORTt, indicating that the pocket accepts a negatively charged molecule. The positively charged pocket may accept NADPH more readily than NADH, which has no 2′-phosphate group. In LADH, which binds NAD+ as a coenzyme, Ser158, Tyr177, and Arg292 in QORTt are replaced by the nonpolar residues Ile224, Pro243, and Gly365, respectively.
FIG. 6.
Possible hydrogen bonds (less than 3.2 Å) between NADPH and QORTt. Hydrogen bonds are shown by broken lines. The distances (in angstroms) between NADPH and QORTt are as follows: Leu224 to 7N, 2.89; Ile217O to N7N, 2.71; Phe242O to N7N, 2.91; Wat40 to O1PN, 2.80; Phe39N to O2PN, 2.86; Ala137N to O1PA, 2.70; Ser158N to OP1, 2.96; Tyr177Oη to OP2, 2.87; Arg292Nη2 to OP2, 2.53; Ser158Oγ to OP3, 2.66; Lys162Nζ to OP3, 2.87; Glu221Oɛ2 to N6A, 2.68; Glu221Oɛ1 to N7A, 3.06; Arg292Nη1 to N7A, 3.17; Arg292Nη2 to N7A, 3.06.
The region from Gly218 to Val224 is invisible in the electron density map of apo-QORTt, indicating that this region is flexible. Most residues of Gly218-Glu221 become ordered when NADPH is bound. The loop, located near NADPH, is on the surface of the protein molecule and covers NADPH. This loop region apparently functions as a flexible lid of the NADPH-binding site. Although there is such structural difference between apo-QORTt and the QORTt-NADPH complex, unlike with LADH, it causes no interdomain movement in QORTt on NADPH binding.
Substrate-binding site.
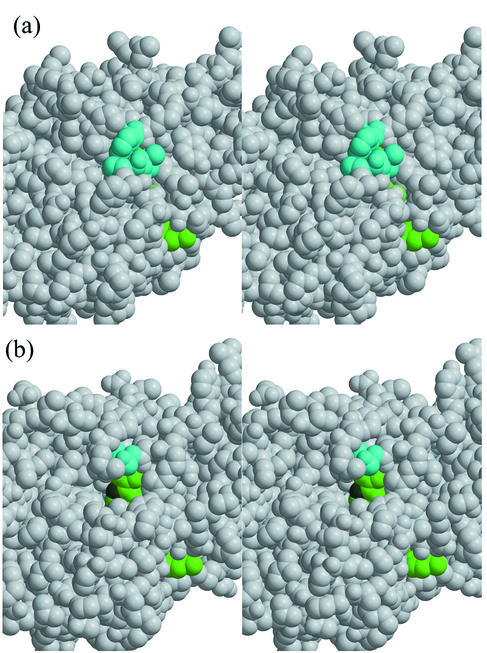
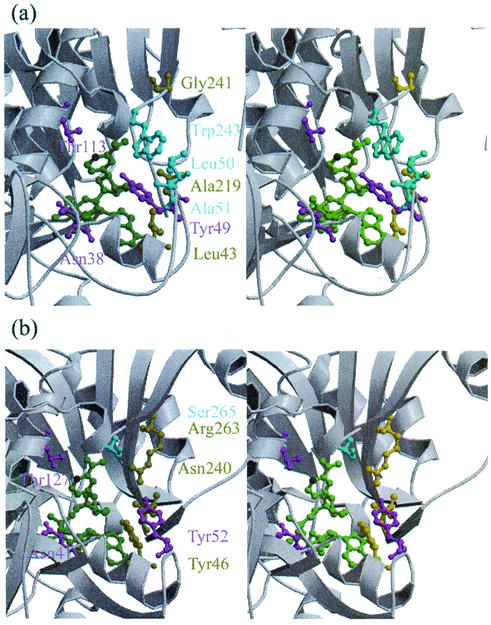
The substrate-binding site is accepted as being close to the C-4 atom in the nicotinamide ring of the NADPH bearing the catalytic hydrogens. Residues Thr113, Leu138, Ile217, and Phe242 in QORTt are close to the B side of the nicotinamide ring, and therefore, the substrate cannot access the nicotinamide ring from the B side. The A side of the nicotinamide ring is accessible from the solvent, indicating that, like LADH, QORTt is an A-side-specific enzyme (3). In the ternary complex of LADH with NAD+ and dimethyl sulfoxide, the substrate analog is located between the A side of the nicotinamide ring and the catalytic domain (9). The corresponding site in QOREc is speculated to be the putative substrate-binding site (31). Unexpectedly, local structures of the putative substrate-binding sites of QORTt and QOREc differ markedly although their overall backbone structures are similar. In QORTt, entrance to this site from the solvent is blocked by residues Leu50, Ala51, and Trp243, whereas in QOREc the substrate-binding site opens more widely toward the solvent (Fig. 7). Accessibility of the substrate-binding site of QORTt is limited, and large substrates cannot reach it unless large conformation change occurs in the protein. This structural feature may explain why QORTt has weak p-benzoquinone reduction activity and is inactive with such large substrates as 5-hydroxy-1,4-naphthoquinone and phenanthraquinone (Y. Shimomura, unpublished data).
FIG.7.
Stereo views around the putative substrate-binding sites of QORTt (a) and QOREc (b). NADPH is shown in green; the C-4 atom that binds active hydrogen atoms is shown in dark green. Leu50, Ala51, and Trp243 in QORTt, which block the substrate-binding site from the solvent, and Ser265 in QOREc, which corresponds to Trp243 in QORTt, are shown in cyan.
Figure 8 compares the characteristic residues around the putative substrate-binding sites of QORTt and QOREc. Residues Asn38, Tyr49, and Thr113, which are highly conserved in QORTt, QOREc, and ζ-crystallin, may be involved in catalysis. Interestingly, features of the putative substrate-binding site of QORTt differ markedly from those of QOREc, even though the conserved residues are near the site. This difference appears to be due to the insertion of two residues (Leu50 and Ala51) in the loop of QORTt relative to QOREc, as is evident from the superposition of the two structures. In addition, QOREc has characteristic polar or charged residues Tyr46, Asn240, and Arg263, but in QORTt, they are replaced by the nonpolar residues Leu43, Ala219, and Gly241. These structural differences in the substrate-binding sites of QORTt and QOREc may provide information about the physiological substrate of QORTt, which has yet to be characterized.
FIG. 8.
Characteristic residues around the substrate-binding sites of QORTt (a) and QOREc (b). The residues in magenta are conserved in both QORTt and QOREc. Cyan and yellow indicate characteristic residues in QORTt and QOREc, respectively. NADPH is shown in green; the active C-4 atom is shown in dark green.
Acknowledgments
We thank Kazuko Sumiguchi-Agari of RIKEN Harima Institute for the preparation of QORTt labeled with SeMet and Ryoji Masui for help with the CD measurements. We also thank Hisanobu Sakai and Masahide Kawamoto of the Japan Synchrotron Radiation Research Institute (JASRI) for aid with data collection using the synchrotron radiation of BL41XU, SPring-8. Synchrotron radiation experiments were performed with the approval of JASRI (2002A0313-NL1-np).
This work was supported in part by a grant-in-aid for Scientific Research on Priority Areas (Biological Machinary) to K.F. (no. 11169223) and by the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Brünger, A. T. 1992. The Rfree value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. [DOI] [PubMed] [Google Scholar]
- 2.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Cedergren-Zeppezauer, E. 1983. Crystal-structure determination of reduced nicotinamide adenine dinucleotide complex with horse liver alcohol dehydrogenase maintained in its apo conformation by zinc-bound imidazole. Biochemistry 22:5761-5772. [DOI] [PubMed] [Google Scholar]
- 4.Colonna-Cesari, F., D. Perahia, M. Karplus, H. Eklund, C.-I. Brändén, and O. Tapia. 1986. Interdomain motion in liver alcohol dehydrogenase. Structural and energetic analysis of the hinge bending mode. J. Biol. Chem. 261:15273-15280. [PubMed] [Google Scholar]
- 5.Duhaiman, A. S., N. Rabbani, A. A. AlJafari, and A. S. Alhomida. 1995. Purification and characterization of ζ-crystallin from the camel lens. Biochem. Biophys. Res. Commun. 215:632-640. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, K. J., J. D. Barton, J. Rossjohn, J. M. Thorn, G. L. Taylor, and D. L. Ollis. 1996. Structural and sequence comparisons of quinone oxidoreductase, ζ-crystallin, and glucose dehydrogenase. Arch. Biochem. Biophys. 328:173-183. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, K. J., J. M. Thorn, J. A. Daniher, N. E. Dixon, and D. L. Ollis. 1994. Crystallization and preliminary X-ray diffraction studies on a soluble Escherichia coli quinone oxidoreductase. J. Mol. Biol. 240:501-503. [DOI] [PubMed] [Google Scholar]
- 8.Eklund, H., B. Nordström, E. Zeppezauer, G. Söderlund, I. Ohlsson, T. Boiwe, B.-O. Söderberg, O. Tapla, C.-I. Brändén, and Å. Åkeson. 1976. Three-dimensional structure of horse liver alcohol dehydrogenase at 2.4 Å resolution. J. Mol. Biol. 102:27-59. [DOI] [PubMed] [Google Scholar]
- 9.Eklund, H., J.-P. Samama, L. Wallén, C.-I. Brändén, Å. Åkeson, and T. A. Jones. 1981. Structure of a triclinic ternary complex of horse liver alcohol dehydrogenase at 2.9 Å resolution. J. Mol. Biol. 146:561-587. [DOI] [PubMed] [Google Scholar]
- 10.Ernster, L., R. W. Estabrook, P. Hochstein, and S. Orrenius. 1987. Editors of DT diaphorase; a quinone reductase with special functions in cell metabolism and detoxification. Chem. Scripta 27A:1-207. [Google Scholar]
- 11.Gonzalez, P., P. V. Rao, and J. S. Zigler. 1993. Molecular cloning and sequencing of zeta-crystallin/quinone reductase cDNA from human liver. Biochem. Biophys. Res. Commun. 191:902-907. [DOI] [PubMed] [Google Scholar]
- 12.Harris, M., and T. A. Jones. 2001. Molray—a web interface between O and the POV-Ray ray tracer. Acta Crystallogr. D 57:1201-1203. [DOI] [PubMed] [Google Scholar]
- 13.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 14.Komori, H., R. Masui, S. Kuramitsu, S. Yokoyama, T. Shibata, Y. Inoue, and K. Miki. 2001. Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism. Proc. Natl. Acad. Sci. USA 98:13560-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of proteins. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 16.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 6:283-291. [Google Scholar]
- 17.Li, R., M. A. Bianchet, P. Talalay, and L. M. Amzel. 1995. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. USA 92:8846-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilley, P. E., N. P. J. Stamford, S. G. Vasudevan, and N. E. Dicon. 1993. The 92-min region of the Escherichia coli chromosome: location of the ubiA and alr genes. Gene 129:9-16. [DOI] [PubMed] [Google Scholar]
- 19.Mano, J., Y. Torii, S. Hayashi, K. Takimoto, K. Matsui, K. Nakamura, D. Inzé, E. Babiychuk, S. Kushnir, and K. Asada. 2002. The NADPH:quinone oxidoreductase P1-ζ-crystallin in Arabidopsis catalyzes the a,b-hydrogenation of 2-alkenals: detoxication of the lipid peroxide-derived reactive aldehydes. Plant Cell Physiol. 43:1445-1455. [DOI] [PubMed] [Google Scholar]
- 20.Mano, J., E. Babiychuk, E. Belles-Boix, J. Hiratake, A. Kimura, D. Inzé, S. Kushnir, and K. Asada. 2000. A novel NADPH:diamide oxidoreductase activity in Arabidopsis thaliana P1 ζ-crystallin. Eur. J. Biochem. 267:3661-3671. [DOI] [PubMed] [Google Scholar]
- 21.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, A., R. Kato, R. Masui, A. Yamagishi, T. Oshima, and S. Kuramitsu. 1996. An aspartate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus HB8. J. Biochem. 119:135-144. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowsky, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 24.Persson, B., J. S. Zigler, and H. Jornvall. 1994. A super-family of medium-chain dehydrogenases/reductases (MDR). Eur. J. Biochem. 226:15-22. [DOI] [PubMed] [Google Scholar]
- 25.Rao, P. V., C. M. Krishna, and J. S. Zigler. 1992. Identification and characterization of the enzymatic activity of zeta-crystallin from guinea pig lens. J. Biol. Chem. 267:96-102. [PubMed] [Google Scholar]
- 26.Rao, P. V., and J. S. Zigler. 1991. ζ-Crystallin from guinea pig lens is capable of functioning catalytically as an oxidoreductase. Arch. Biochem. Biophys. 167:1221-1228. [DOI] [PubMed] [Google Scholar]
- 27.Rao, P. V., and J. S. Zigler. 1992. Purification and characterization of zeta-crystallin/quinone reductase from guinea pig liver. Biochim. Biophys. Acta 1117:315-320. [PubMed] [Google Scholar]
- 28.Rodokanaki, A., R. K. Holmes, and T. Borrás. 1989. Zeta-crystallin, a novel protein from guinea pig lens is related to alcohol dehydrogenases. Gene 78:215-224. [DOI] [PubMed] [Google Scholar]
- 29.Shimomura, Y., K. Sumiguchi-Agari, R. Masui, S. Kuramitsu, and K. Fukuyama. 2002. Overproduction, crystallization and preliminary X-ray diffraction analysis of a quinone oxidoreductase from Thermus thermophilus HB8. Acta Crystallogr. D 58:1365-1367. [DOI] [PubMed] [Google Scholar]
- 30.Sugishima, M., H. Sakamoto, Y. Kakuta, Y. Omata, S. Hayashi, M. Noguchi, and K. Fukuyama. 2002. Crystal structure of rat apo-heme oxygenase-1 (HO-1): mechanism of heme binding in HO-1 inferred from structural comparison of the apo and heme complex forms. Biochemistry 41:7293-7300. [DOI] [PubMed] [Google Scholar]
- 31.Thorn, J. M., J. D. Barton, N. E. Dixon, D. L. Ollis, and K. J. Edwards. 1995. Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH. J. Mol. Biol. 249:785-799. [DOI] [PubMed] [Google Scholar]
- 32.Tumminia, S. J., P. V. Rao, J. S. Zigler, and P. Russell. 1993. Xenobiotic induction of quinone oxidoreductase activity in lens epithelial cells. Biochim. Biophys. Acta 1203:251-259. [DOI] [PubMed] [Google Scholar]
- 33.Wada, T., T. Yamazaki, S. Kuramitsu, and Y. Kyogoku. 1999. Cloning of the RNA polymerase α subunit gene from Thermus thermophilus HB8 and characterization of the protein. J. Biochem. 125:143-150. [DOI] [PubMed] [Google Scholar]
- 34.Walden, H., G. S. Bell, R. J. Russell, B. Siebers, R. Hensel, and G. L. Taylor. 2001. Tiny TIM: a small, tetrameric, hyperthermostable triosephosphate isomerase. J. Mol. Biol. 306:745-757. [DOI] [PubMed] [Google Scholar]
- 35.Wittung-Stafshede, P. 2002. Role of cofactors in protein folding. Acc. Chem. Res. 35:201-208. [DOI] [PubMed] [Google Scholar]
- 36.Yagi, T. 1991. Bacterial NADH-quinone oxidoreductases. J. Bioenerg. Biomembr. 23:211-225. [DOI] [PubMed] [Google Scholar]
- 37.Yokoyama. S., H. Hirota, T. Kigawa, T. Yabuki, M. Shirouzu, T. Terada, Y. Ito, Y. Matsuo, Y. Kuroda, Y. Nishimura, Y. Kyogoku, K. Miki, R. Masui, and S. Kuramitsu. 2000. Structural genomics projects in Japan. Nat. Struct. Biol. 7(Suppl.):943-945. [DOI] [PubMed] [Google Scholar]