Abstract
Transcript elongation by RNA polymerase is discontinuous and interrupted by pauses that play key regulatory roles. We show here that two different classes of pause signals punctuate elongation. Class I pauses, discovered in enteric bacteria, depend on interaction of a nascent RNA structure with RNA polymerase to displace the 3′ OH away from the catalytic center. Class II pauses, which may predominate in eukaryotes, cause RNA polymerase to slide backwards along DNA and RNA and to occlude the active site with nascent RNA. These pauses differ in their responses to antisense oligonucleotides, pyrophosphate, GreA, and general elongation factors NusA and NusG. In contrast, substitutions in RNA polymerase that increase or decrease the rate of RNA synthesis affect both pause classes similarly. We propose that both pause classes, as well as arrest and termination, arise from a common intermediate that itself binds NTP substrate weakly.
Keywords: transcriptional pausing, NusA, NusG
Gene expression often is regulated during RNA chain elongation. Regulation depends both on interruptions to transcription caused by pause, arrest, and termination signals encoded in the DNA and RNA and on auxiliary proteins that modify the response of RNA polymerase (RNAP) to these signals (reviewed in refs. 1 and 2). Pausing (a temporary delay in chain elongation) synchronizes transcription and translation in prokaryotes, slows RNAP to allow timely interaction of regulatory factors, and is a precursor to both arrest (complete halting without dissociation; refs. 3 and 4), and dissociation of the transcription elongation complex (TEC) at ρ-dependent and ρ-independent terminators (1).
Numerous auxiliary proteins modulate pausing in organisms from bacteria to humans. Two of these, NusA and NusG, are universally conserved among bacteria and archaebacteria (5), are typically essential to cell viability, and, respectively, inhibit or stimulate pausing by bacterial RNAP (6). NusA and NusG also modulate the termination activity of ρ protein (which dissociates paused TECs) and, together with other auxiliary proteins like λ N or Q, assemble antitermination TECs that resist pausing and termination.
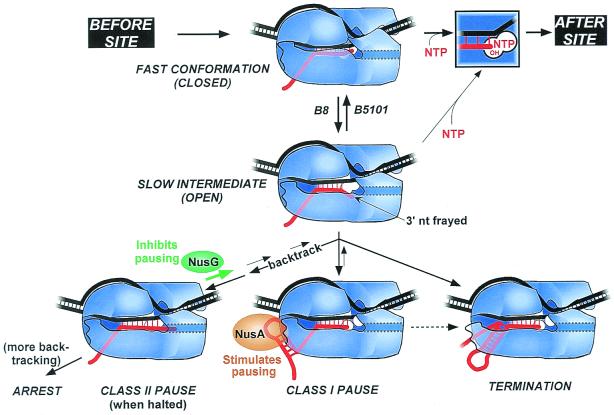
Although transcriptional pausing was first described more than two decades ago (7), no consensus pause sequence exists. Rather, different types of signals appear to inhibit alignment of the RNA 3′ OH with substrate NTP in different ways (8–12). Most if not all of these signals depend on RNAP's ability to slide back and forth along RNA and DNA chains (while preserving an ≈17-nt DNA bubble and ≈8-bp RNA⋅DNA hybrid; refs. 4, 13, 14, and references therein). These movements may produce five distinct TEC configurations (see Fig. 1): (i) backtracked (displaced RNA protruding downstream from the active site); (ii) frayed (3′ RNA nt separated from DNA); (iii) pretranslocated (3′ RNA nt blocking the NTP-binding or i+1 subsite); (vi) active (3′ RNA nt in the priming or i subsite; NTP binding site open or occupied by NTP); and (v) hypertranslocated (RNA 3′ nt pulled away from the catalytic center). During rapid elongation, RNAP fluctuates between the pretranslocated and active states.
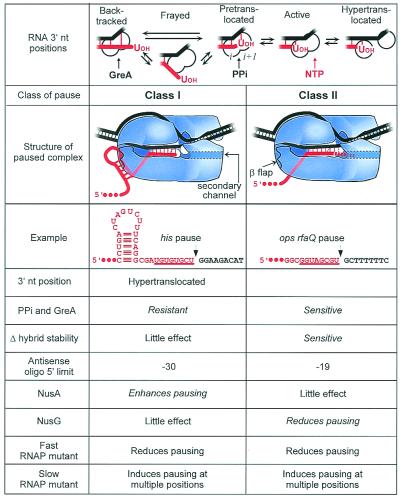
Figure 1.
Properties of two classes of pause signals. (Top) Possible positions of the RNA 3′ nt during active elongation or pausing. Structures of class I and class II paused TECs are depicted with DNA (black) entering RNAP (blue) from downstream at right and separating near the active site (white circles). RNA (red) pairs with template DNA in an eight-bp hybrid (vertical red lines), then exits under the β flap domain (37). In the examples, the RNA nt in the hybrid (in the active state) are underlined. Nucleotide addition occurs when the RNA 3′ OH and template-specified NTP simultaneously occupy the left and right halves of RNAP's bipartite active site (i and i+1, respectively). At a class I pause, interaction of the pause hairpin with the β flap domain displaces the RNA 3′ OH away from the catalytic center. At a class II pause, RNAP enters pretranslocated or backtracked conformations (dashed red line). Backtracked RNA may enter the secondary channel through which NTPs are thought to enter (gray dotted outline; see refs. 37, 46, and 47). These structures are consistent with the tabulated differences in sensitivity to pyrophosphorolysis and transcript cleavage, effect of changes in hybrid stability on pausing, 5′ limit of positions at which antisense oligos can reduce pausing, effects of NusA and NusG, and effects of fast and slow RNAP mutants.
To clarify the mechanism of pausing and to examine how it is modulated by NusA and NusG, we compared the two types of pause signals that have been studied most extensively, which we call class I and class II pauses. At a class I pause, nascent RNA hairpin–RNAP interaction inhibits nucleotide addition by stabilizing the RNA 3′ OH in a frayed or hypertranslocated position (15, 16). At a class II pause, a weak RNA⋅DNA hybrid induces backtracking of RNAP to one or more states that occlude the active site with nascent RNA (9–11).
Most class I pause signals have been found in the leader regions of certain enterobacterial amino acid biosynthetic operons, where they synchronize RNAP and ribosome movement during attenuation (17). Previous studies establish that class I pause signals are multipartite. Extensive analysis of the his leader pause site reveals that this pause is largely dependent on the RNA hairpin, but also is affected by the 11-nt region between the pause hairpin and the 3′ end, the nt in the active site, and the first 14 bp of downstream DNA (15).
Previously, class II pauses have been characterized only in vitro or at arrest or termination sites, where a less stable RNA⋅DNA hybrid induces backtracking of RNAP (3, 4, 18). Backtracking can retreat RNAP by 10 or more nt, but has been observed directly only when RNAP is artificially halted by NTP deprivation at a hairpin-less pause or arrest site (4, 10, 14, 19, and references therein). We report the identification and characterization of a physiologically relevant class II pause. ops (operon polarity suppressor; ref. 20) pause sites occur in the early transcribed region of Escherichia coli operons that encode or affect synthesis of extracytoplasmic macromolecules like hemolysin. They function to permit recruitment of RfaH, an auxiliary protein that suppresses premature termination of transcription (20). We show that the his (class I) and ops (class II) signals, although sharing some features, are mechanistically distinct, differing in the paused configuration of RNAP's active site and in their responses to the elongation proteins NusA and NusG.
Materials and Methods
Sources of Oligonucleotides and Proteins.
All oligonucleotides were obtained from Operon Technologies (Alameda, CA). Wild-type and His-tagged RNAP (16), NusA (21), and GreA (22) were purified as described previously. Chromosomal nusG was cloned between NdeI and HindIII sites of pET28a (Novagen); the resulting plasmid, pIA247, encodes additional 20 amino acids (MGSSHHHHHHSSGLVPRGSH) at the nonconserved N terminus. When induced in strain BL21, NusG was soluble and constituted ≈20% of total cell protein. Cells were lysed by sonication in NTA buffer (50 mM Tris⋅HCl (pH 7.9)/300 mM NaCl/0.1 mM EDTA/0.1 mM PMSF/1 mM β-mercaptoethanol). Cleared lysate was loaded on a Ni-NTA agarose (Qiagen, Chatsworth, CA) column. After four consecutive washes with 10 vol of NTA buffer containing 0, 1, 10, and 30 mM imidazole (pH 7.9), NusG was eluted with 150 mM imidazole and dialyzed against storage buffer (10 mM Tris⋅HCl (pH 7.9)/0.1 M NaCl/50% glycerol/0.1 mM EDTA/0.1 mM DTT). His-tagged NusG was >90% pure and behaved similarly to wild-type NusG (gift from M. Gottesman, Columbia University, New York) in in vitro transcription assays.
ops Sites.
The 12-nt consensus ops signal is GGCGGTAGnnTG [identified by analysis of transcribed sequences in operons that are regulated by RfaH (20)]. Using this consensus sequence, 17 ops sites, of which 15 were followed closely by an ORF, were identified in the E. coli genome sequence (23) at positions 232196 (no ORF), 504357 (no ORF), 598983 (pheP), 605824 (yi81_2), 1035383 (hyaF), 1619170 (ydeD), 1812043 (b1730), 1880364 (b1801), 2111170 (rfbB), 2135455 (b2062), 3040562 (b2899), 3325384 (yhbY), 3435371 (smf), 3495017 (cysG), 3805820 (rfaQ), 4146046 (yijO), and 4214647 (yjaB). Several more ops sites are located on F plasmids and pathogenicity islands not in the published E. coli genome sequence (24, 25). Synthetic oligos encoding the pheP or rfaQ ops signals were cloned between SpeI and BglII sites of pCL102b (15), replacing the underlined region of the his transcribed sequence (shown from +1) ATCGAGAGGGACACGGGGAAACACCACCATCATCACCATCATCCTGACTAGTCTTTCAGGCGATGTGTGCTgGAAGACATTCAGAT with GGCGGTAGCGTgCTTTTTTC (rfaQ) or GGCGGTAGTCTGTgCGCTGT (pheP) sequences (major pause sites occur just before lowercase g). Both sites confer response to RfaH in vitro (data not shown).
In Vitro Transcription Reactions.
Transcription templates were prepared by PCR amplification from pCL102b (his pause signal), pIA237 (rfaQ pause signal), and pIA251 (pheP pause signal). Transcription complexes halted at position A29 by UTP deprivation were formed in 40 nM in transcription buffer (20 mM Tris⋅HCl/20 mM NaCl/14 mM MgCl2/14 mM β-mercaptoethanol/0.1 mM Na2EDTA) with 32P-derived from [α-32P]CTP (NEN; 3000 Ci/mmol) as described previously (24) and kept on ice before use. Transcription was restarted by addition of GTP to 10 μM, ATP, CTP, and UTP to 150 μM each, and heparin to 100 μg/ml. Oligos, if present, were added to 500-fold molar excess relative to the TEC. Samples were removed at times shown in the figures, and, after a final 5-min incubation with 250 μM each, NTP (chase) were processed, separated by electrophoresis, and quantified as described previously (24). Pause half-life (the time during which half of the complexes reenter the elongation pathway) and pause efficiency (fraction of transcribing RNAP molecules that pause) were determined by nonlinear regression analysis (24).
Results
ops Is a Class II Pause Signal.
Previous characterization of ops led us to suspect it is a class II pause signal that plays a regulatory role in vivo. RNAP pauses in vivo at an ops site about 160 nt into the hly operon (25). Because no inverted repeat elements occur near this site, it is unlikely to function via a pause hairpin and might represent a class II pause. We further defined ops and assessed its biological significance by searching the E. coli genome sequence for the ops consensus (see Materials and Methods). We identified 15 candidate ops sites that were followed by an ORF; of these ORFs, 12 are known or predicted to encode or affect synthesis of an extracytoplasmic macromolecule. Two representative ops elements (from upstream of the pheP and rfaQ genes) were cloned downstream from the T7 A1 promoter. Both sites induced pausing in vitro by purified E. coli RNAP (e.g., Fig. 2, Right).
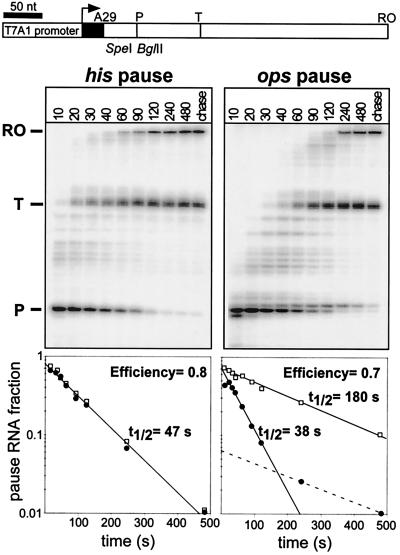
Figure 2.
Pausing at the his and ops pause sites. (A) Preformed [α-32P]CMP-labeled A29 complexes were incubated with 10 μM GTP, 150 μM ATP, CTP, and UTP on the his (Left) or ops (Right) template. Samples were taken at the times indicated in seconds above each lane. Prominent transcripts (and lengths for his and ops templates, respectively) are P, pause RNA transcript (71 and 62 nt); T, terminated transcript (150 and 138 nt); and RO, run-off RNA transcript (381 and 369 nt). The fractions of the pause RNA (closed circles) were plotted against time; the pause half-life and efficiency were determined as described previously (24). Fractions of pause RNAs from preformed complexes (open squares; gels not shown) were determined after paused TECs were halted by NTP deprivation for ≈5 min.
RNAP Pauses at the ops and his Pause Sites in Vitro.
To compare the response of RNAP to class I (his) and class II (ops) pause signals, we performed side-by-side in vitro transcription assays on templates that differed only in the vicinity of the pause sites (see Materials and Methods). The class I his pause (Fig. 2, Left) and the class II pheP pause (Fig. 2 Right) were comparable in efficiency (80% and 70%, respectively) and half-life (47 s and 38 s, respectively; see Materials and Methods). Three adjacent pauses are apparent on the pheP template; the major pause is at position U64, and two shorter pauses are at U62 and C66 (the half-life and efficiency were calculated for the major site only). RNAP recognized the rfaQ (ops, class II) pause site with similar efficiency (75%), but with a shorter half-life (23 s; data not shown).
Escape from the his pause site fit a single exponential, suggesting that his paused complexes were homogenous (24). Consistent with this interpretation, incubation of the his paused complex for as long as 48 h before addition of NTPs did not change its half-life (Fig. 2). In contrast, escape from the ops pause did not fit a single exponential. The fast and slow components of the pause RNA decay curve suggest that at least two distinct paused conformations formed at this site (Fig. 2). Whereas the fast escaping conformation constituted the majority during elongation, the slow component became predominant upon halting of RNAP at the ops site. RNAP “walked” to this site by step-wise addition of subsets of NTPs escaped with a half-life of 180 s upon addition of all four NTPs (Fig. 2). This behavior is similar to that of the “temporarily arrested complexes” (10) that are prone to backtracking when halted by NTP deprivation.
Antisense Oligos Distinguish the Two Classes of Pause Sites.
Antisense oligos inhibit hairpin-mediated pauses (class I) and backtracking (which is associated with class II pausing) by distinct mechanisms (3, 14, 16). Antisense oligos inhibit hairpin-dependent pausing by blocking hairpin formation. We recently demonstrated that antisense oligos dramatically reduce the his pause half-life (up to 20-fold; ref. 16 and Fig. 3A Left), even when oligos disrupt the 5′ stem of the hairpin at a distance of 30 nt from the 3′ end of the RNA. However, antisense oligos prevent backtracking by pairing to nascent RNA immediately adjacent to RNAP (3, 14).
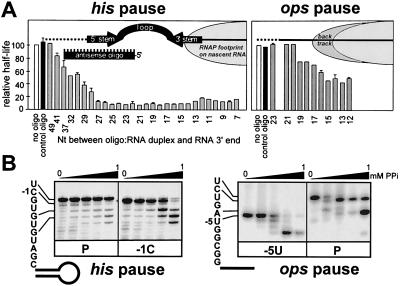
Figure 3.
Oligos and pyrophosphate affect the his and ops pause complexes differently. (A) The pause half-lives are plotted (as a fraction of a “no-oligo” control) by the 3′-most nucleotide of the nascent RNA that remains outside the RNA:oligo duplex (equivalent to the 5′-terminal base of the 22-nt oligonucleotides). Each value is an average of at least two independent measurements. (B) Immobilized TECs were halted along the templates encoding the his (Left) or ops (Right) pause signals at the positions indicated below each panel and treated with increasing concentrations of PPi (0, 0.001, 0.01, 0.1, and 1 mM) as described previously (16).
We tested which antisense oligos could inhibit pausing at the ops site. As expected for a backtracking mechanism, oligos reduced pausing only when they could pair 19 nt or closer from the RNA 3′ end (Fig. 3A Right). Surprisingly, the maximal reduction in pausing was only 2-fold. We interpret this result to mean that RNAP backtracks only a few bp at the ops site when NTPs are present, because much larger effects of antisense oligos on arrest by E. coli or human RNAPII in complexes backtracked by six or more nt were reported previously (3, 14). It is unlikely that the observed differences in oligo effects reflect altered annealing kinetics because the his and ops transcripts were identical up to position 51 and the same set of oligos was used for both templates.
RNAP Is Prone to Backtracking When Halted at the ops but Not the his Pause Site.
GreA and PPi induce cleavage of nascent RNA in backtracked and pretranslocated conformations, respectively, but not in the active conformation (Fig. 1). Because his paused complexes are resistant to GreA-stimulated transcript cleavage (22) and pyrophosphorolysis (ref. 16 and Fig. 3B Left) even though a 3′-terminal UMP usually is sensitive to pyrophosphorolysis (26), we suggested that his pause RNA is frayed or hypertranslocated rather than pretranslocated or backtracked (15, 16). Note that the 3′-terminal UMP of the his pause RNA is much more resistant to PPi than the 3′ terminal CMP of the −1 RNA. In contrast, RNAP halted at the rfaQ ops site was exceptionally sensitive to both PPi (the ops pause RNA was cleaved to position −1 even at 1 μM PPi, whereas the −5U complex remained relatively resistant to cleavage; Fig. 3B Right), and GreA-induced cleavage (data not shown), indicating that it is in pretranslocated and backtracked conformations.
Backtracking of RNAP appears to be facilitated by a weak RNA⋅DNA hybrid in the active conformation as well as by formation of a relatively stronger hybrid upon backtracking (9, 10). For instance, arrest can be prevented by incorporation of hybrid-stabilizing analogs into the RNA chain (11). Thus, if the energetics of the RNA⋅DNA hybrid (4) are important for escape from a class II pause, we would expect base analogs to have large effects, whereas base analogs might have little if any effect on class I pausing. To test this prediction, we incorporated analogs into the ops and his RNA⋅DNA hybrid (underlined in Fig. 1) at multiple positions (to enhance observed effects) and away from the 3′end of the RNA (to avoid changes in the vicinity of the active site). Consistent with other evidence indicating a backtracked pause, incorporation of IMP at −7 and −8 positions of the ops RNA (Fig. 1) increased the pause half-life 3-fold (data not shown). In contrast, incorporation of hybrid-stabilizing 5-iodo-UMP at −7, −5, and −3 or hybrid-destabilizing IMP at −4 and −6 positions (Fig. 1) did not affect the rate of escape from the his pause (data not shown). We conclude that hybrid stability plays a role in escape from a class II, but not a class I pause. Because RNAP appears to assume multiple conformations at the ops pause (Fig. 2), sensitivity to PPi, GreA, and IMP is most easily explained by formation of both pretranslocated and backtracked conformations at the class II ops pause.
The his and ops Pause Sites Respond Differently to NusA and NusG.
Recognition of pause and termination sites by E. coli RNAP is affected by both NusA and NusG, whose effects on transcription in vitro are opposite. NusA inhibits nucleotide addition at some sites, such as the his and trp pause sites (17, 27), and induces pausing at sites not evident in its absence (2, 28). In contrast, NusG accelerates transcription both in vivo and in vitro (29–31). Both NusA and NusG interact with core RNAP, but their effects on transcription in vitro are noncompetitive (6). These proteins could bind to different sites on the same RNAP molecule and exert their effects independently, perhaps even at the same template positions. Alternatively, NusA and NusG could recognize distinct conformers of the TEC that form at different template positions in response to specific nucleic acid sequences or structures.
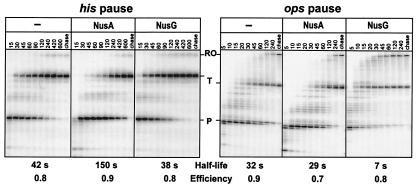
We tested whether NusA and NusG change RNAP's behavior as it transcribes through the his and ops sites under identical in vitro conditions. NusA slowed escape from the his pause site by a factor of three, but had almost no effect on the escape from the ops site. In contrast, NusG accelerated escape from the major ops pause site almost 5-fold with little effect at the ops C66 site or at the his pause site (Fig. 4). Because NusG accelerates transcription through other regions of the template only moderately (compare rates of arrival at the terminator in Fig. 4 Left), we conclude that it preferentially stimulates escape from the ops pause site. Recognition of the his pause site was moderately increased by NusA (from 80% to 90% efficiency) but was unaffected by NusG. Both NusA and NusG reduced pause efficiency at the ops site from 90% to 70% and 80%, respectively. These observations support the conclusion of Burns et al. (6) that NusA and NusG affect transcript elongation noncompetitively, and are consistent with previously demonstrated effects of NusA at hairpin-dependent pause sites (17) and NusG on pausing at a hairpin-less pause site in the λ PL transcript (30). However, NusG was previously reported to reduce pausing at the hairpin-dependent trp pause site (29). We repeated these experiments and observed little effect of NusG (reduction of half-life from 43 to 40 s; data not shown); the earlier report may have overestimated the effect of NusG because of the more rapid arrival of complexes at the trp pause site in its presence.
Figure 4.
Elongation factors NusA and NusG preferentially target his and ops pause complexes, respectively. [α-32P]CTP-labeled A29 complexes (40 nM) were preformed on his or ops pause templates. Transcription was allowed to resume in the absence of additional factors (Left) or in the presence of 50 nM NusA (Center) or NusG (Right). The pause half-life and efficiency (see Materials and Methods) are indicated below each panel.
Elongation Through Both Pause Sites Is Affected by Mutant RNAPs.
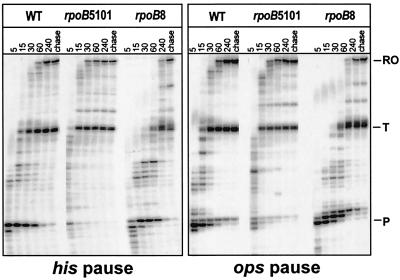
Recognition of regulatory signals also is affected by amino acid substitutions in RNAP that can either decrease or enhance both pausing and termination. One explanation is that these substitutions favor conformational states that hold the RNA 3′ end more or less tightly in the active site, leading to rapid or slow nucleotide addition, respectively (32). We decided to test fast and slow mutant RNAPs for pausing at the his and ops sites by using RpoB5101 (fast; TI563, PS560 in RNAP's β subunit) and RpoB8 (slow; βQP513). These mutant enzymes were identified by genetic screens for altered termination phenotypes and exhibit dramatic effects on elongation in vitro (33–36). However, the substitutions cannot affect catalysis directly because they are located in the rifampicin-binding site, more than 20 Å from the active site (37). RpoB5101 mutant RNAP transcribes through both his (Fig. 5 Left) and ops (Right) sites with a significant reduction in both half-life and efficiency; the short half-lives of those complexes prevent accurate measurements of the efficiency of pause site recognition. RpoB8 mutant RNAP pauses at several positions in the vicinity of both sites, presumably because of backtracking (36); however, the efficiency and pause half-life of the his or ops TEC remain relatively unchanged. Thus, fast and slow mutant RNAPs affect both classes of pause sites similarly.
Figure 5.
Fast and slow RNAP mutants recognize his and ops pause sites similarly. [α 32P]-CTP-labeled A29 complexes were formed on his (Left) or ops (Right) pause templates with wild-type (WT), RpoB5101 and RpoB8 RNAPs and then incubated with NTPs (see Materials and Methods).
Discussion
Our results lead to three principal conclusions. First, RNAP recognizes two classes of pause signals that lead either to RNA 3′ OH displacement from the active site (the RNA is frayed or hypertranslocated; class I) or to threading of the RNA past the catalytic center (backtracking; class II). Second, the effects of NusA and NusG are pause class-specific: NusA enhances pausing at class I sites, whereas NusG inhibits pausing at class II sites. Third, pausing at both classes of sites is affected similarly by substitutions in RNAP that increase or decrease the rate of RNA synthesis. These conclusions lead us to propose that (i) pause sites and the proteins that regulate them are functionally specialized, and (ii) the fundamental mechanisms by which RNAP recognizes pause, arrest, and termination sites share a common slow intermediate.
Pause Sites and Regulatory Proteins May Be Functionally Specialized.
The existence of at least two pausing mechanisms raises the possibility that different pause classes have distinct biological functions. For example, one or another of these paused states could be a preferred target for ρ-dependent termination or for recruitment of antitermination factors such as RfaH. Alternatively, a backtracked or hairpin-stabilized pause could resist ρ action. Future studies may reveal additional types of pause signals. For instance, a nascent RNA hairpin conceivably could stabilize a backtracked pause if it were positioned farther from the RNA 3′ end (e.g., see ref. 38).
The differential effects of NusA and NusG establish that regulatory proteins can have greater effects on certain classes of pauses. NusA enhancement of pausing probably depends on the nascent RNA hairpin component of class I pause sites because NusA stabilizes hairpin interaction with RNAP. NusA crosslinks to the his pause hairpin (our unpublished results), protects the trp pause hairpin from RNase digestion (27), and enhances pausing to greater or lesser extent when hairpin sequences are changed (39). If transcript release were inhibited by hairpin-RNAP interaction, stabilization of this interaction by NusA would explain why NusA inhibits ρdependent termination even though it slows the rate of transcription (ref. 6 and references therein).
NusG, in contrast, accelerated elongation at the hairpin-dependent pause sites only modestly but had a large effect at the class II sites. It seems probable that NusG acts by inhibiting backtracking of RNAP through contacts to the enzyme itself (40), to nucleic acid (41), or to both. If transcript release were inhibited by backtracking, inhibition of backtracking by NusG would explain why NusG enhances ρ-dependent termination even though it stimulates the rate of transcription (ref. 6 and references therein).
Different target specificity of NusA and NusG also may explain why elongation factor requirements vary among organisms. In E. coli, where at least two classes of pause sites exist, both nusA and nusG are essential in wild-type cells. The primary role of NusA in E. coli, however, appears to be in ρ-dependent termination because nusA becomes dispensable once ρ function is compromised (42). In contrast, Bacillus subtilis nusG is dispensable, whereas nusA cannot be deleted even when ρ is inhibited (5). Perhaps hairpin-dependent rather than ρ-dependent termination dominates in Bacillus, and NusA plays an essential role at some hairpin-dependent terminators. The situation differs in eukaryotes, where RNAPII, at least, appears to pause via the class II pathway preferentially. Pausing by human RNAPII is induced at U-tracks and accompanied by backtracking (2, 38). RNA hairpins, when important, appear to act by inhibiting backtracking (38, 43). Consistently, no NusA homologue has been identified to date in a eukaryote, but both yeast and humans express NusG homologues (44). A complete understanding of the interplay between pausing and elongation factor action will first require fully elucidating the mechanisms of pausing and then determining how factors modulate these mechanisms through interactions with RNAP, RNA, and DNA.
Pausing, Arrest, and Termination May Share a Common Slow Intermediate.
We suggest that all pause, arrest, and termination signals share the common feature of initially triggering formation of a slow intermediate in which the RNA 3′ OH is frayed away from the DNA template and catalytic center (Fig. 6). In the slow intermediate, NTP binding would be weak and nucleotide addition would be slow because the 3′ OH and NTP must both coordinately bond to the catalytic Mg2+ ion before reaction (45). The delay in transcription caused by formation of slow intermediate would allow time for rearrangement into paused, arrested, or termination conformations.
Figure 6.
Slow intermediate mechanism. RNAP is depicted as in Fig. 1. At most template positions, fast translocation from the pretranslocated state to the active state allows tight NTP binding (assisted by a properly positioned 3′OH) and rapid transcription (top horizontal pathway)]. When RNAP encounters a pause, arrest, or termination site, it isomerizes to a slow intermediate in which the RNA 3′ end frays away from the DNA. A slight conformational opening of RNAP may precede and accelerate this change or may accompany it. Further rearrangement of the slow intermediate produces the different classes of paused, arrested, or terminating complexes. Escape of the slow intermediate back to the elongation pathway occurs by weak NTP binding and recapture of the 3′ OH in the active site. Amino acid substitutions in RNAP favor or disfavor the slow intermediate, whereas elongation factors NusA and NusG stabilize hairpin-RNAP interaction or inhibit backtracking, respectively, at later steps in the pathway. Whether termination sometimes involves hairpin-RNAP interaction (dotted line) and whether it occurs via hairpin-induced bubble collapse or RNA pull-out (18, 36) remains to be determined.
It seems likely that formation of the slow intermediate is facilitated or accompanied by a modest opening of RNAP's crab-claw-like shape (46), even though such a conformational change is not an obligatory feature of a slow-intermediate mechanism. The crystal structures of bacterial RNAP (37) and yeast RNAPII (47) suggest that RNAP can adopt more closed or open conformations around the channels that bind RNA and DNA. Further, observations of single RNAP molecules during transcription (48) suggest that a slow state can persist for multiple rounds of nucleotide addition, which would require an altered protein conformation. In the fast (closed) state, tight interaction of RNAP with the RNA⋅DNA hybrid would restrict the 3′ OH to a position that optimizes NTP-binding and rapid nucleotide addition (15). In the slow (open) state, weaker RNAP contacts to the RNA⋅DNA hybrid could facilitate fraying of the 3′ OH away from the active site.
Although we cannot exclude with certainty the alternative hypothesis that various paused, arrested, or terminating complexes arise directly by independent rearrangements of a rapidly elongating TEC, we favor the slow-intermediate mechanism for several reasons. First, fluctuation of RNAP between fast and slow states is required to explain the misincorporation kinetics of E. coli RNAP (49) and the elongation kinetics of both wheat germ RNAPII (50) and yeast RNAPIII (51). Whether RNAP switches conformations randomly or forms the slow conformation at pauses and the fast conformation when pauses are absent remains to be determined.
Second, previously reported crosslinking results support the existence of the slow intermediate. Immediately after RNAP is halted at a potential arrest site, it remains competent for transcript elongation. However, the 3′-terminal base crosslinks to β residues 1097–1107 (52), which are >20 Å from the reactive 3′ nt position predicted by aligning the catalytic Mg2+ ions in the T7 replication complex (53) and the bacterial RNAP (37). Upon formation of the arrested complex, the 3′ base crosslink shifts to β′ residues 932-1035 within RNAP's secondary channel (Fig. 6). The initial movement of the 3′ nt away from the catalytic Mg2+ ion before arrest appears to correspond to formation of the slow intermediate (Fig. 6), and to fraying of the RNA 3′ end away from the template DNA (15).
Third, the existence of a common slow intermediate provides a reasonable explanation for the sharing of some features among pause, arrest, and termination signals. The 3′-terminal nucleotide at these sites is usually U or C, followed by an incoming GTP or ATP (26). This combination of pyrimidine and purine bases probably slows nucleotide addition and favors the loss of the 3′ OH from the active site. All these signals also induce backtracking when RNAP is halted either just before the site or at the site when nascent hairpins are absent or their formation is inhibited (10, 18, 19, 54). For intrinsic terminators, which encode both an RNA hairpin that is characteristic of a class I pause and a U-rich 3′-proximal region that alone creates a class II pause (18, 36), backtracking is paradoxical because reverse threading of RNA should inhibit hairpin formation. However, the coexistence of these two features can be explained if 3′-proximal sequences that can cause backtracking first trigger formation of the slow intermediate from which either hairpin formation or backtracking are possible.
The Slow-Intermediate Mechanism Can Explain Pausing, Arrest, Termination, and Antitermination.
Once the slow intermediate forms, pause, arrest, or termination signals direct further rearrangement of the TEC to conformations that differ in the position of the 3′ nt (Fig. 6). At a class I pause site, RNA hairpin interaction with RNAP's β flap domain displaces the RNA from the active site (15, 16). This interaction also may stabilize the open conformation of RNAP (46) and consequently either hypertranslocation (depicted in Figs. 1 and 6) or fraying of the RNA 3′ OH. At a class II pause site, RNAP probably populates both pretranslocated and one or more reversibly backtracked states based on its biphasic pausing kinetics (Fig. 2) and its sensitivities to PPi and GreA (Fig. 3). Although the non-hairpin components of a class I pause signal (e.g., his) contribute to formation of and slow escape from the paused state (21), they do not create a class II pause when the pause hairpin is disrupted by antisense oligos (18). Rather, special features of a class II site (e.g., ops) must be required for efficient pausing; whether these features extend beyond the 12-nt ops consensus, making class II pause signals also multipartite, remains to be determined.
At an arrest site, further backtracking traps RNAP, possibly because the protruding transcript binds to RNAP's secondary channel (47). At an intrinsic terminator, hairpin formation either opens the transcript exit channel (Fig. 6) or pulls the transcript out although the exit channel (18, 36, 46).
The slow-intermediate mechanism offers an attractive explanation for antitermination. Antiterminator proteins (like RfaH, λN, and λQ) could make RNAP resistant to pausing, arrest, and termination by keeping the enzyme from flexing open into the slow intermediate. This idea is especially attractive because different antitermination proteins could stabilize the closed conformation through different interactions with RNAP. Fast and slow RNAP mutants (32–36) could be explained by opposite effects on the same interconversion between fast and slow RNAP conformations. Thus, the slow-intermediate mechanism can explain the known behaviors of RNAP during transcript elongation and the activities of proteins that regulate it. We now must determine the locations of the paused 3′ OH more precisely and whether the postulated open and closed conformations of RNAP can be observed directly.
Acknowledgments
We thank members of the Landick laboratory, Tamas Gaal, and Cathleen Chan for suggesting revisions during preparation of the manuscript. This work was supported by National Institutes of Health Grant GM38660.
Abbreviations
- RNAP
RNA polymerase
- TEC
transcription elongation complex
- ops
operon polarity suppressor
Note Added in Proof
While this paper was in final production, Pasman and von Hippel also proposed that NusG acts principally to inhibit backtracking by RNAP (55).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Richardson J, Greenblatt J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F, et al., editors. Washington, DC: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 2.Uptain S, Kane C, Chamberlin M. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 3.Komissarova N, Kashlev M. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mote J J, Reines D. J Biol Chem. 1998;273:16843–16852. doi: 10.1074/jbc.273.27.16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingham C, Dennis J, Furneaux P. Mol Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- 6.Burns C, Richardson L, Richardson J. J Mol Biol. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- 7.Maizels N M. Proc Natl Acad Sci USA. 1973;70:3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin J R, Chamberlin M J. J Mol Biol. 1987;196:61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- 9.Guajardo R, Sousa R. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 10.Komissarova N, Kashlev M. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 11.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert W. In: RNA Polymerase. Losick R, Chamberlin M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1977. pp. 193–205. [Google Scholar]
- 13.Korzheva N, Mustaev A, Nudler E, Nikiforov V, Goldfarb A. Cold Spring Harbor Symp Quant Biol. 1998;63:337–345. doi: 10.1101/sqb.1998.63.337. [DOI] [PubMed] [Google Scholar]
- 14.Reeder T, Hawley D. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 15.Chan C, Wang D, Landick R. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 16.Artsimovitch I, Landick R. Genes Dev. 1998;12:3110–3122. doi: 10.1101/gad.12.19.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landick R, Turnbough C, Jr, Yanofsky C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F, et al., editors. Washington, DC: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 18.Gusarov I, Nudler E. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 19.Nudler E, Kashlev M, Nikiforov V, Goldfarb A. Cell. 1995;81:351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- 20.Bailey M, Hughes C, Koronakis V. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt M, Chamberlin M. Biochemistry. 1984;23:197–203. doi: 10.1021/bi00297a004. [DOI] [PubMed] [Google Scholar]
- 22.Feng G, Lee D N, Wang D, Chan C L, Landick R. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 23.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 24.Landick R, Wang D, Chan C. Methods Enzymol. 1996;274:334–352. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 25.Leeds J, Welch R. J Bacteriol. 1997;179:3519–3527. doi: 10.1128/jb.179.11.3519-3527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aivazashvili V A, Bibilashvili R S, Vartikyan R M, Kutateladze T A. Mol Biol. 1981;15:653–667. (English trans.). [PubMed] [Google Scholar]
- 27.Landick R, Yanofsky C. J Mol Biol. 1987;196:363–377. doi: 10.1016/0022-2836(87)90697-8. [DOI] [PubMed] [Google Scholar]
- 28.Theissen G, Pardon B, Wagner R. Anal Biochem. 1990;189:254–261. doi: 10.1016/0003-2697(90)90117-r. [DOI] [PubMed] [Google Scholar]
- 29.Burova E, Hung S C, Sagitov V, Stitt B L, Gottesman M E. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung S, Gottesman M. J Mol Biol. 1995;247:428–442. doi: 10.1006/jmbi.1994.0151. [DOI] [PubMed] [Google Scholar]
- 31.Zellars M, Squires C. Mol Microbiol. 1999;32:1296–1304. doi: 10.1046/j.1365-2958.1999.01442.x. [DOI] [PubMed] [Google Scholar]
- 32.Landick R. Science. 1999;284:598–599. doi: 10.1126/science.284.5414.598. [DOI] [PubMed] [Google Scholar]
- 33.Landick R, Stewart J, Lee D. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 34.Jin D J, Gross C A. J Biol Chem. 1991;266:14478–14485. [PubMed] [Google Scholar]
- 35.Pan T, Artsimovitch I, Fang X, Landick R, Sosnick T. Proc Natl Acad Sci USA. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yarnell W S, Roberts J W. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 37.Zhang G, Campbell E A, Minakhin L, C, R, Severinov K, Darst S A. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 38.Palangat M, Meier T, Keene R, Landick R. Mol Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- 39.Chan C L, Landick R. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Mason S W, Greenblatt J. Genes Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- 41.Liao D, Lurz R, Dobrinski B, Dennis P P. J Bacteriol. 1996;178:4089–4098. doi: 10.1128/jb.178.14.4089-4098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng C, Friedman D. Proc Natl Acad Sci USA. 1994;91:7543–7547. doi: 10.1073/pnas.91.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dedrick R L, Kane C M, Chamberlin M J. J Biol Chem. 1987;262:9098–9108. [PubMed] [Google Scholar]
- 44.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, et al. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steitz T, Smerdon S, Jager J, Joyce C. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 46.Mooney R A, Landick R. Cell. 1999;98:687–690. doi: 10.1016/s0092-8674(00)81483-x. [DOI] [PubMed] [Google Scholar]
- 47.Fu J, Gnatt A L, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 48.Davenport R, Wuite G, Landick R, Bustamante C. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 49.Erie D A, Hajiseyedjavadi O, Young M C, von Hippel P H. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 50.de Mercoyrol L, Soulie J, Job C, Job D, Dussert C, Palmari J, Rasigni M, Rasigni G. Biochem J. 1990;269:651–658. doi: 10.1042/bj2690651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuzaki H, Kassavetis G A, Geiduschek E P. J Mol Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 52.Markovtsov V, Mustaev A, Goldfarb A. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature (London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 54.Wang D, Meier T, Chan C, Feng G, Lee D, Landick R. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 55.Pasman Z, von Hippel P H. Biochemistry. 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]