Abstract
Bcl-2 and related proteins are key regulators of apoptosis or programmed cell death implicated in human disease including cancer. We recently showed that cell-permeable Bcl-2 binding peptides could induce apoptosis of human myeloid leukemia in vitro and suppress its growth in severe combined immunodeficient mice. Here we report the discovery of HA14-1, a small molecule (molecular weight = 409) and nonpeptidic ligand of a Bcl-2 surface pocket, by using a computer screening strategy based on the predicted structure of Bcl-2 protein. In vitro binding studies demonstrated the interaction of HA14-1 with this Bcl-2 surface pocket that is essential for Bcl-2 biological function. HA14-1 effectively induced apoptosis of human acute myeloid leukemia (HL-60) cells overexpressing Bcl-2 protein that was associated with the decrease in mitochondrial membrane potential and activation of caspase-9 followed by caspase-3. Cytokine response modifier A, a potent inhibitor of Fas-mediated apoptosis, did not block apoptosis induced by HA14-1. Whereas HA14-1 strongly induced the death of NIH 3T3 (Apaf-1+/+) cells, it had little apoptotic effect on Apaf-1-deficient (Apaf-1−/−) mouse embryonic fibroblast cells. These data are consistent with a mechanism by which HA14-1 induces the activation of Apaf-1 and caspases, possibly by binding to Bcl-2 protein and inhibiting its function. The discovery of this cell-permeable molecule provides a chemical probe to study Bcl-2-regulated apoptotic pathways in vivo and could lead to the development of new therapeutic agents.
Bcl-2 was originally identified at the chromosomal breakpoint of t(14;18)-bearing B-cell lymphomas (1). Bcl-2 belongs to a growing family of proteins that regulate apoptosis or programmed cell death (2–4). The Bcl-2 family includes both death antagonists such as Bcl-2 and Bcl-xL and death agonists such as Bax, Bak, Bid, and Bad. These related proteins share at least one of four homologous regions termed Bcl homology (BH) domains (BH1 to BH4). As a prototypic member of this family, Bcl-2 can contribute to neoplastic cell expansion by preventing normal cell turnover caused by physiological cell death mechanisms. High levels of Bcl-2 gene expression are found in a wide variety of human cancers (5). In addition, Bcl-2 is implicated in chemoresistance as overexpression of Bcl-2 can inhibit the cell killing effect of many currently available anticancer drugs by blocking the apoptotic pathway. The expression levels of Bcl-2 proteins correlate with relative resistance to a wide spectrum of chemotherapeutic drugs and γ-irradiation. Therefore, the inhibition of the protective function of Bcl-2 protein overexpressed in tumor cells is an attractive strategy for either restoring the normal apoptotic process in these cells or making these cells more susceptible for conventional chemotherapy or radiotherapy. In this regard, cell-permeable, small molecule inhibitors of Bcl-2 may represent a new class of therapeutic agents for the treatment of cancer.
Although it is not fully understood how Bcl-2 family proteins regulate apoptotic pathways, one possible mechanism is that members of this family engage in various protein-protein interactions to form homo- and heterotypic dimers important for their biological functions (2, 3). The three-dimensional structure of Bcl-2, constructed based on the x-ray and NMR structure of the highly homologous protein Bcl-xL (6, 7), reveals a hydrophobic binding pocket that mediates protein-protein interactions involving Bcl-2 family members. This surface pocket is required for the anti-apoptotic function of Bcl-2, as studies have shown that mutations at this site abolished Bcl-2 biological function (8). Synthetic peptides that bind this surface pocket of Bcl-xL and Bcl-2 have been shown to have in vitro activity in inducing apoptosis in cell-free systems (9) and in HeLa cells (10). To develop cell-permeable peptides as in vivo regulators of Bcl-2 function, we recently synthesized Bcl-2 binding peptides containing a fatty acid as a cell-permeable moiety. Such cell-permeable Bcl-2 binding peptides were shown to induce apoptosis in vitro and have in vivo activity in suppressing human myeloid leukemia growth in severe combined immunodeficient mice (11). Taken together, these studies suggested the clinical potential of small molecule inhibitors targeting the Bcl-2 surface pocket.
We are interested in the development and application of chemical and structural strategies for the discovery of small molecule regulators of protein biological function involved in both cell surface molecular recognition and intracellular signaling pathways (12, 13). In addition to the above described approach of using cell-permeable Bcl-2 binding peptides (11), small nonpeptidic organic compounds that interact with the surface pocket of Bcl-2 can be used as cell-permeable agents to affect Bcl-2-regulated apoptotic pathways and develop new pharmaceuticals. The computer screening approach that exploits both new computational methods and the diversity of existing compound databases for the identification of novel protein binding molecules has become a powerful tool for the discovery of nonpeptidic organic ligands (12–14). Other existing methods, such as random testing (either manually or in a high throughput fashion) of large numbers of compounds from either natural sources or synthetic combinatorial libraries, can be both costly and time-consuming. In contrast, if the three-dimensional structure of a target protein is known or can be predicted from the structure of a homologous protein, computer screening involving the virtual design of ligand molecules based on such structural information can reduce the time and cost associated with massive synthesis and assay methods. In a previous study, we used this computer screening approach to identify small nonpeptidic ligands of a CD4 surface binding pocket implicated in the oligomerization of CD4 on the T cell surface and stabilization of the super complex formed by major histocompatibility complex (MHC) class II, peptide antigen, and T cell receptor (15, 16). These CD4 inhibitors displayed potent immunoregulatory activity both in vitro and in vivo and are promising leads for the development of new therapeutics for autoimmune diseases and transplantation reactions.
In the present study, we have extended this approach to develop chemical modulators of Bcl-2-regulated apoptotic process. HA14-1, a small nonpeptidic organic ligand of the Bcl-2 surface pocket, was discovered by computer screening combined with various cell-based assays. This compound was shown to interact with soluble Bcl-2 protein and induce apoptosis of tumor cells. The structure-based discovery of this cell-permeable small molecule has important implication for the study of the in vivo apoptotic signaling pathways regulated by Bcl-2 and related proteins and development of potential anti-cancer agents.
Materials and Methods
Compound and Tumor Cells.
The identified lead compound HA14-1, ethyl 2-amino-6-bromo-4-(1-cyano-2-ethoxy-2-oxoethyl)-4H-chromene-3-carboxylate (molecular weight 409.24) was purchased from Maybridge (Cornwall, U.K.). The structure and purity of the compound was confirmed by 1H-NMR (in CDCl3) and HPLC. Because of the poor solubility of HA14-1 in PBS, the compound was first dissolved in pure DMSO (dimethylsufoxide) and then was diluted to the desired concentrations (at which the percent of DMSO was less than 0.5%) for the bioassays. It was noted that in pure DMSO HA14-1 could slowly decompose over time. Therefore, freshly prepared solution of HA14-1 was used for all experiments. For the experiments, blank 0.5% DMSO solution was used in the untreated control and showed no effect in binding and apoptosis assays.
Human myeloid leukemia (HL-60) cells were kindly provided by K. Bhalla (University of Miami School of Medicine). The cells had been transfected with pZip-Bcl-2 and overexpress Bcl-2 protein. Human lung cancer H1299 cells transfected with cytokine response modifier A (CrmA) were provided by M. S. Sheikh (State University of New York). Cells were maintained in suspension culture in RPMI 1640 medium supplemented with 10% FBS and 750 μg/ml G418 (GIBCO/BRL) and were split at 1:10 every 3–4 days.
Computer Screening Procedure.
Bcl-2 and Bcl-xL are highly homologous proteins (47.2% of sequence identity). Both contain four conserved regions denoted as BH1, BH2, BH3, and BH4 domains. Taking the NMR structure of Bcl-xL protein in the complex of Bcl-xL and BakBH3 peptide as the template (7), the three-dimensional structure of Bcl-2 protein was modeled in Homolog module of msi/insight ii 98.0 software (Biosym Technologies, San Diego). We assigned the Bcl-2 protein residues with CVFF force field potential. The Bcl-2 protein model structure was optimized with the discover program of msi/insight ii until the maximum energy derivation was less than 0.5 kcal/(mol⋅Å). The optimized model structure was used for virtual molecular database screening of a collection of 193,833 compounds from MDL/ACD 3D database (Molecular Design Limited, San Leandro, CA), taking the corresponding BakBH3 peptide binding pocket on the Bcl-2 protein as the target. The program used for screening is dock 3.5, provided by I. D. Kuntz of the University of California at San Francisco (17).
In the process of docking calculation, the interaction with the Bcl-2 surface pocket of each molecule in different orientations was scored by a shape complementarity scoring function that resembles the van der Waal attractive energy. As a result, 1,000 best-scoring molecules in the best orientation were saved. Then, the orientation of each of the 1,000 molecules in the Bcl-2 receptor was optimized automatically in sybyl 6.2 (Tripos Associates, St. Louis) environment by running a SPL program written by ourselves. Atoms of the ligand and receptor were assigned with Gästeiger-Hückel and Kollman-All charges, respectively. The binding energy between each molecule and the Bcl-2 protein was evaluated after the geometry optimization. The structures of 1,000 compounds selected from the above procedure were further examined individually and manually by both molecular modeler and medicinal chemist. Only those compounds with relatively lower binding energy, favorable shape complementarity, and/or potential of forming hydrogen bonds with the Bcl-2 protein were picked for next phase of evaluation. Fifty-three candidate molecules were chosen by the criteria, of which 28 compounds with diverse scaffold and possible drug-like properties were actually obtained for biological testing.
It should be pointed out that some organic compounds in the MDL/ACD database, such as HA14-1 identified in this study, have one or more chiral centers. For these compounds, both R and S configurations of the chiral centers were examined during the ligand docking procedure. Another issue was the multiple possible conformations of compounds, particularly those flexible molecules. For this study, because of the consideration of the available computational power, only one conformation was examined for each molecule (or each specific configuration of a chiral molecule) during the docking process. It is possible that this rigid docking procedure may miss some potential ligand candidates. Flexible docking using multiple conformations of a compound that requires more computational power can be used to improve the outcome of computer screening.
In Vitro Bcl-2 Binding Assay.
The binding affinity of organic compounds to Bcl-2 protein in vitro was determined by a competitive binding assay based on fluorescence polarization. For this assay, 5-carboxyfluorecein was coupled to the N terminus of a peptide, GQVGRQLAIIGDDINR, derived from the BH3 domain of Bak (Flu-BakBH3), which has been shown to bind to the surface pocket of the Bcl-xL protein with high-affinity (dissociation constant, KD, of ≈0.34 μM) (7). According to our molecular modeling studies and binding measurement using fluorescence polarization, the Flu-BakBH3 peptide binds the surface pocket of Bcl-2 with a similar affinity (dissociation constant, KD, of ≈0.20 μM) (S. Li, S.S., and Z.H., unpublished work). Bcl-2 used in this assay was a recombinant GST-fused soluble protein (Santa Cruz Biotechnology). Flu-BakBH3 and Bcl-2 protein were mixed in the presence or absence of organic compounds under standard buffer conditions and were incubated for 30 min. The binding of Flu-BakBH3 to Bcl-2 protein was measured on a LS-50 luminescence spectrometer equipped with polarizers using a dual path length quartz cell (500 μl) (Perkin–Elmer). The fluorophore was excited with vertical polarized light at 480 nm (excitation slit width 15 nm), and the polarization value of the emitted light was observed through vertical and horizontal polarizers at 530 nm (emission slit width 15 nm). The binding affinity of each compound for Bcl-2 protein was assessed by determining the ability of different concentrations of the compound to inhibit Flu-BakBH3 binding to Bcl-2.
Cell Viability Assay.
The dose-dependent effect of the compounds on the viability of HL-60 cells was tested by using the CellTiter 96AQ kit (Promega). In brief, the cell suspension containing 1 × 105 cells in 100 μl of medium were plated into a 96-well plate and were incubated with compounds at different concentrations. The numbers of apoptotic cells were determined by measuring the optical density on a Wallac Victor (2) counter (EG & G, Gaithersburg, MD) at 490 nm. Cell viability was also determined by Trypan blue exclusion in a hemocytometer.
DNA Fragmentation Assay.
Cells (1 × 106 cells) were incubated with compounds for the time indicated. Cells were washed once with PBS and were pelleted by centrifugation. The cell pellets were fixed in 70% ethanol for 1 h and then were resuspended in PC buffer (192 mM Na2HPO4/4 mM citric acid, pH 7.8) to extract DNA. After centrifugation for 5 min at 13,000 rpm (Eppendorf centrifuge 5417R) the supernatants were collected and incubated with 10 μg/ml RNase A for 1 h at 37°C followed by digestion with 20 μg/ml proteinase K at 50°C for 3 h. The DNA fragments were separated by electrophoresis in 2% agarose gel and were visualized by ethidium bromide staining.
Mitochondrial Membrane Potential Assay.
To determine changes in the mitochondrial membrane potential, 1 × 106 cells were incubated with 3,3′-dihexyloxacarbocynine iodide [DiOC6(3), 40 nM] at 37°C for 15 min followed by flow cytometry analysis.
Immunoblotting.
Total cell lysate (50 μg) was resolved on 12% SDS/PAGE and was transferred to a nitrocellulose membrane. Membranes were blocked in Tris-buffered saline (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl) with 5% dry milk and 0.1% Tween-20 at 4°C overnight. Antibodies to detect the cleavage of caspase-3 (PharMingen, catalog no. 65906E; diluted to 1:1,000), caspase-9 (PharMingen, catalog no. 68086E; diluted to 1:1,000), and poly(ADP-ribose)-polymerase (PARP) (Boehringer Mannheim, catalog no. 1835238; diluted to 1:2,000) were used for 1 h incubation at room temperature. Immune complexes were detected with horseradish perioxidase conjugated goat anti-rabbit IgG antibody (Oncogene Research Products) and were visualized by using the ECL system (Amersham Pharmacia).
Results
Identification of HA14-1 by Computer Screening.
The surface pocket formed by domains BH1 to BH3 in the model structure of Bcl-2 protein was used as the target site for computer screening to search for potential inhibitory molecules. In particular, we focused on potential ligands that interact with the site surrounded by a number of important residues, including F104, R107, Y108, D111, F112, A113, Q118, L119, T132, V133, E136, L137, G145, R146, I147, V148, A149, F153, and R207. Of 28 compounds that were finally selected and obtained for actual biological testing, HA14-1 was found to possess the desired biological activity in binding to the Bcl-2 surface pocket and inducing apoptosis of HL-60 cells (Fig. 1). It should be noted that HA14-1 has two chiral centers located at the C4 position and the carbon atom attached with the cyano group, respectively. The compound used for biological experiments was a mixture of diastereomers as assessed by HPLC and NMR. The absolute configurations of the biologically active diastereomer need to be determined in further experiments.
Figure 1.
(A) Structure of HA14-1 (ethyl 2-amino-6-bromo-4-(1-cyano-2-ethoxy-2-oxoethyl)-4H-chromene-3-carboxylate), a nonpeptidic organic compound that binds Bcl-2 protein. Note that HA14-1 has two chiral centers (marked by asterisks) located at the C4 position and the carbon atom attached with the cyano group, respectively. (B) Structural model for the complex of HA14-1 with the Bcl-2 surface pocket as predicted by computer docking calculation.
Binding Affinity of HA14-1 to Bcl-2 Protein.
The binding of the compounds to Bcl-2 protein was evaluated in vitro by a competitive fluorescence polarization assay. For this assay, the N terminus of a Bak BH3 peptide, which has high affinity binding to the Bcl-2 surface pocket, was labeled with 5-carboxyfluorecein (Flu-BakBH3). The binding affinity of the compounds to the Bcl-2 surface pocket was determined by their dose-dependent competition with the binding of Flu-BakBH3. Among the compounds tested, HA14-1 was found to bind the designated pocket on Bcl-2 with the IC50 of ≈9 μM in competing with the Bcl-2 binding of Flu-BakBH3 (Fig. 2). The interaction of HA14-1 with the Bcl-2 surface pocket appeared to be specific for the chemical structure of the compound as a series of synthetic analogs derived from HA14-1 containing various modifications were found to have widely different Bcl-2 binding activities (N. Yu, D.L., S.S., and Z.H., unpublished work).
Figure 2.
Binding of HA14-1 to Bcl-2 protein in vitro as measured by a competitive fluorescence polarization assay. The data points represent the mean of three independent experiments.
HA14-1 Induces Apoptosis of HL-60 Cells.
The ability of HA14-1 in binding to the surface pocket of Bcl-2 critical for its anti-apoptotic function raises the possibility that HA14-1 may antagonize Bcl-2 function and trigger apoptosis in cells. To examine the effect of HA14-1 on cell viability, HL-60 cells were treated with various concentrations of HA14-1 for 4 h. HA14-1 induced the death of HL-60 cells in a dose-dependent manner (Fig. 3). At 50 μM, HA14-1 caused the lost of viability in more than 90% of the cells.
Figure 3.
Effect of HA14-1 on the viability of HL-60 cells. The cells were incubated with HA14-1 at different concentrations for 4 h. The cell viability was determined by using MTS method in 96-well plate.
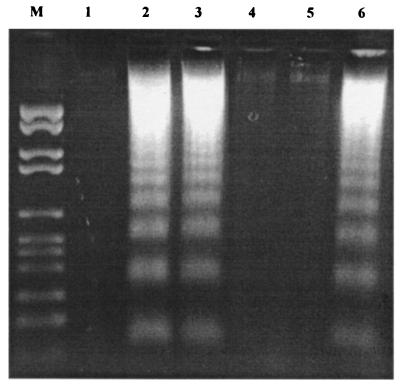
The effect of the compounds was further examined by DNA fragmentation, a characteristic marker of apoptotic cells. HL-60 cells treated with 50 μM HA14-1 by 3 h displayed the characteristic pattern of DNA fragmentation (Fig. 4A). The DNA fragmentation induced by HA14-1 was completely blocked by the preincubation of the cells with a broad-spectrum caspase inhibitor, zVAD-fmk.
Figure 4.
Apoptosis induced by HA14-1 in HL-60 cells as indicated by DNA ladders (A) and cleavage of caspase-9, caspase-3 and PARP (B). DNA ladders were visualized on 2% of agarose gel after the cells were treated with 50 μM HA14-1 compound at the designated time. M, DNA marker. Lanes: 1, control; 2, 1 h of HA14-1 treatment; 3, 2 h of HA14-1 treatment; 4, 3 h of HA14-1 treatment; 5, 4 h of HA14-1 treatment; 6, preincubation with zVAD-fmk for 2 h before the treatment with HA14-1 for 4 h. The cleavage of caspase-3, -9, and PARP was shown by Western blot analysis using polyclonal rabbit antibodies.
HA14-1 Induces Apoptosis via Caspase Activation.
To investigate the downstream events of Bcl-2-regulated apoptotic signaling in cells treated with HA14-1, we performed Western blot analysis to examine caspase activation. The cleavage of caspase-9 started by 1 h of treatment of HA14-1 and became more evident at 4 h (Fig. 4B). The cleavage of caspase-3 was also observed, but started only by 3 h under the same experimental condition, indicating that the cleavage of caspase-3 occurred after that of caspase-9. With the activation of caspase-9 and caspase-3, the cleavage of PARP was also observed. These data are consistent with a mechanism by which HA14-1 antagonizes Bcl-2 function, thereby initiating the activation of caspase-9 followed by caspase-3 and, eventually, cleavage of their cellular substrates, such as PARP.
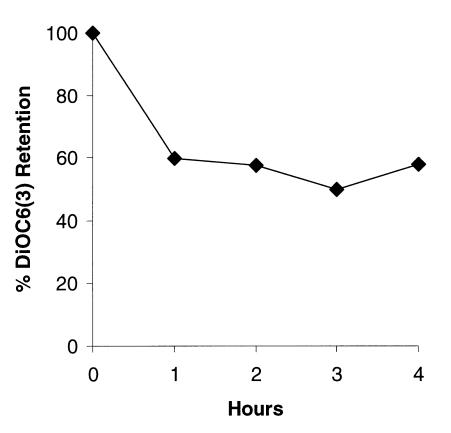
HA14-1 Causes the Decrease in Mitochondria Membrane Potential.
Recent studies have shown that overexpression of Bcl-2 prevents the loss of potential across the mitochondrial membrane (18, 19). The decrease in mitochondria membrane potential can result in the release of mitochondrial protein cytochrome c into cytosol, where it forms a complex with Apaf-1, thus activating caspase-9 in the caspase cascade that finally leads to apoptosis (20, 21). Overexpression of Bcl-2 protein in HL-60 cells inhibits this chain reaction by blocking the release of cytochrome c from mitochondria (22, 23). Thus, the early loss of mitochondria membrane potential is another hallmark for apoptosis. We studied the changes in mitochondria membrane potential as measured by potential sensitive dye DiOC6 (3) in HL-60 cells. The significant decrease in mitochondria membrane potential as indicated by the decline of DiOC6 (3) retention was observed within 1 h in cells treated with 50 μM HA14-1 (Fig. 5). This seemed to correlate with the activation of caspase-9, which also occurred at a similar time point (Fig. 4B). It was also consistent with the observation of DNA fragmentation, which started approximately 2 h after the change in mitochondria membrane potential (Fig. 4A).
Figure 5.
Decrease in mitochondrial membrane potential caused by HA14-1. HL-60 cells were cultured in the presence of 50 μM HA14-1 for different time points. Changes in mitochondrial membrane potential after HA14-1 treatment were examined by using DiOC6(3) fluorescence dye.
Apoptosis Induced by HA14-1 Is Distinct from that by Fas.
To further study the mechanism of HA14-1 action, we tested the effect of HA14-1 on human lung cancer H1299 cells transfected with cytokine response modifier A (CrmA), a 38-kDa cowpox virus-encoded serpin protein known to inhibit apoptosis induced by the death receptor Fas and tumor necrosis factor receptor type 1 (24–26). CrmA was not able to inhibit apoptosis triggered by HA14-1 whereas it blocked apoptosis induced by anti-human CD95 (Fas/APO-1) antibody (Fig. 6). This supported the notion that HA14-1 exerts its biological effect through the Bcl-2-regulated apoptotic pathway, which is distinct from that mediated by Fas.
Figure 6.
Apoptosis of lung cancer H1299 cells induced by HA14-1 cannot be blocked by CrmA. The effects of HA14-1 and Fas antibody as a positive control were tested in human lung cancer H1299 cells transfected with either plasmid containing neomycin resistance gene (in lanes 1–3) or the same vector containing CrmA gene (in lanes 4–6). Cells were treated with Fas antibody or HA14-1 for 24 h and were analyzed for DNA ladders on 2% agarose gel. Lanes: 1 and 4, untreated control; 2 and 5, treatment with Fas antibody; 3 and 6, treatment with 50 μM HA14-1.
Apoptosis Induced by HA14-1 Depends on Apaf-1.
Apaf-1 is the protease-activating factor that interacts with procaspase-9 and cytochrome c and induces the activation of caspase-9 (20, 27, 28). Bcl-2 inhibits this Apaf-1 pathway by preventing the release of cytochrome c from the mitochondria. Unlike Bcl-2-regulated apoptotic signaling pathway that requires Apaf-1, Fas-mediated apoptosis in mouse cells is independent of Apaf-1 (29, 30). Using Apaf-1-deficient mouse embryonic fibroblast (Apaf-1−/−) (28), we examined whether Apaf-1 is involved in the apoptosis induced by HA14-1. As determined by cell count using Trypan blue staining, it was found that HA14-1 had little apoptotic effect on Apaf-1−/− cells (<5% cell death, similar to that of untreated cells) (Fig. 7). In contrast, HA14-1 induced >90% cell death in NIH 3T3 cells, which are Apaf-1+/+. The death of Apaf-1+/+ cells induced by HA14-1 requires caspase activation because preincubation of the cells with zVAD-fmk for 2 h completely blocked this effect. These data are consistent with a mechanism by which HA14-1 induces Apaf-1 activation, most likely by inhibiting Bcl-2 function and promoting cytochrome c release from the mitochondria.
Figure 7.
Effect of HA14-1 on Apaf-1−/− and Apaf-1+/+ cells. Apaf-1-deficient mouse embryonic fibroblast (Apaf-1−/−) and NIH 3T3 (Apaf-1+/+) cells were treated with 50 μM HA14-1 for 24 h. Percent cell death was determined by using Trypan blue staining. To examine the role of caspases in the death of Apaf-1+/+ cells induced by HA14-1, cells were preincubated with zVAD-fmk for 2 h before the treatment of the compound.
Discussion
We report here the discovery of HA14-1, a small molecule that binds to a surface functional pocket of Bcl-2 protein and induces apoptosis of tumor cells. The in vitro activity of HA14-1 in inducing apoptosis of tumor cells was characterized by using the HL-60 cell line. HL-60 cells were used to test the effect of the compounds because Bcl-2 protein overexpressed in these cells has been shown to block the apoptosis-inducing effects of various anti-cancer drugs (23). For example, in this study, HL-60 cells did not undergo apoptosis as measured by DNA fragmentation with the treatment of 50 μM etoposide, a widely used anti-cancer drug, during a 4-h incubation period (data not shown). In contrast, 50 μM HA14-1 induced extensive apoptosis in these cells within 3 h of treatment, suggesting that HA14-1 could effectively overcome the resistance of HL-60 cells, presumably through binding to Bcl-2 protein and blocking its anti-apoptotic function. Several lines of evidence are consistent with this mechanism of action of HA14-1: (i) HA14-1 binds to the surface functional pocket of recombinant soluble Bcl-2 protein in vitro; (ii) induction of apoptosis by HA14-1 involves the activation of caspase-9 followed by caspase-3 and decrease in mitochondria membrane potential; (iii) HA14-1 induces apoptosis through the pathway distinct from the Fas/tumor necrosis factor receptor pathway; and (iv) the apoptotic effect of HA14-1 depends on Apaf-1. It should be emphasized that more experiments are necessary to further elucidate the detailed mechanism of HA14-1 action and its specificity for Bcl-2-regulated apoptotic pathways. In addition, it remains to be determined whether HA14-1 can also target other Bcl-2 related anti-apoptotic proteins such as Bcl-xL. These issues should be addressed in future studies.
To our knowledge, HA14-1 is the first reported Bcl-2 binding organic compound identified by using the de novo computer-aided design strategy based on the predicted structure of Bcl-2 protein. Together with our recent design of synthetic cell-permeable Bcl-2 binding peptides (11), they demonstrate two different approaches for the chemical modulation of protein biological function involved in intracellular signaling pathways. These synthetic cell-permeable agents offer powerful chemical tools for understanding the biology of their macromolecular targets inside the cell. The mechanism by which Bcl-2 and related proteins regulate apoptosis is still not fully defined. Various models have been proposed, including the formation of heterodimer of Bcl-2 and Bax (31), or ion channel formation (32–34) to prevent the release of cytochrome c and apoptosis-inducing factor from the mitochondria. The discovery of Bcl-2 binding small molecules such as HA14-1 shown here should be useful for further elucidation of Bcl-2 function and mechanism. Furthermore, HA14-1 represents a promising lead for the development of more potent and specific agents targeting Bcl-2-regulated apoptosis implicated in human disease, particularly cancer. Other approaches to reducing Bcl-2 protein levels by using antisense oligonucleotides (35) or single-chain antibodies (36) have previously been shown to enhance tumor cell chemosensitivity. The present study demonstrates an attractive alternative strategy of using protein structure-based computer screening to identify small molecule ligands of Bcl-2 that may hold the promise as novel cell-permeable and nonimmunogeneic therapeutic agents.
Acknowledgments
We thank Drs. K. Bhalla and M. S. Sheikh for providing HL-60 and H1299 cells, respectively, Dr. Philip Tsichlis for critical reading of the manuscript, and Drs. S. Li, N. Yu, J. Luo, X. Zeng, Z. Luo, and other members of the Huang lab for helpful discussion. This work was supported by grants (to Z.H.) from the American Cancer Society and the Sidney Kimmel Foundation for Cancer Research.
Abbreviations
- BH domain
Bcl homology domain
- CrmA
cytokine response modifier A
- PARP
poly(ADP-ribose)-polymerase
References
- 1.Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce C M. Science. 1985;229:1390–1393. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- 2.Adams J, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 3.Chao D, Korsmeyer S. Annu Rev Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 4.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 5.Reed J C. Curr Opin Oncol. 1999;1:68–75. doi: 10.1097/00001622-199901000-00014. [DOI] [PubMed] [Google Scholar]
- 6.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Chang B S, Thompson C B, Wong S L, et al. Nature (London) 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 7.Sattler M, Liang H, Nettesheim D, Meadows R P, Harlan J E, Eberstadt M, Yoon H S, Shuker S B, Chang B S, Minn A J, et al. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 8.Yin X M, Oltvai Z N, Korsmeyer S J. Nature (London) 1994;369:321–323. doi: 10.1038/369321a0. [DOI] [PubMed] [Google Scholar]
- 9.Cosulich S, Worrall V, Hedge P, Green S, Clarke P. Curr Biol. 1997;7:913–920. doi: 10.1016/s0960-9822(06)00410-6. [DOI] [PubMed] [Google Scholar]
- 10.Holinger E P, Chittenden T, Lutz R J. J Biol Chem. 1999;274:13298–13304. doi: 10.1074/jbc.274.19.13298. [DOI] [PubMed] [Google Scholar]
- 11.Wang J L, Zhang Z J, Choksi S, Shan S, Lu Z, Croce C M, Alnemri E S, Korngold R, Huang Z. Cancer Res. 2000;60:1498–1502. [PubMed] [Google Scholar]
- 12.Huang, Z. (2000) Pharmacol. Ther., in press.
- 13.Huang Z, Li S, Korngold R. Biopolymers. 1998;43:367–382. doi: 10.1002/(SICI)1097-0282(1997)43:5<367::AID-BIP3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.Kuntz I D. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Gao J, Satoh T, Friedman T, Edling A, Koch U, Choksi S, Han X, Korngold R, Huang Z. Proc Natl Acad Sci USA. 1997;94:73–78. doi: 10.1073/pnas.94.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Satoh T, Korngold R, Huang Z. Immunol Today. 1998;19:455–462. doi: 10.1016/s0167-5699(98)01325-5. [DOI] [PubMed] [Google Scholar]
- 17.Meng E C, Shoichet B K, Kuntz I D. J Comp Chem. 1992;13:505–524. [Google Scholar]
- 18.Marchetti P, Castedo M, Susin S A, Zamzami N, Hirsch T, Macho A, Haeffner A, Hirsch F, Geuskens M, Kroemer G. J Exp Med. 1996;184:1155–1160. doi: 10.1084/jem.184.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marzo I, Brenner C, Zamzami N, Susin S A, Beutner G, Brdiczka D, Remy R, Xie Z H, Reed J C, Kroemer G. J Exp Med. 1998;187:1261–1271. doi: 10.1084/jem.187.8.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 22.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A, Cai J, Peng T-I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 24.Los M, Van de Craen M, Penning L, Schenk H, Westendorp M, Baeuerle P, Droge W, Krammer P, Fiers W, Schulze-Osthoff K. Nature (London) 1995;375:81–83. doi: 10.1038/375081a0. [DOI] [PubMed] [Google Scholar]
- 25.Enari M, Hug H, Nagata S. Nature (London) 1995;375:78–81. doi: 10.1038/375078a0. [DOI] [PubMed] [Google Scholar]
- 26.Tewari M, Quan L T, O'Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 27.Zou H, Li Y, Liu X, Wang X. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 28.Saleh A, Srinivasula S M, Acharya S, Fishel R, Alnemri E S. J Biol Chem. 1999;274:17941–17945. doi: 10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida H, Kong Y Y, Yoshida R, Elia A J, Hakem A, Hakem R, Penninger J M, Mak T W. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- 30.Cecconi F, Alvarez-Bolado G, Meyer B I, Roth K A, Gruss P. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- 31.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 32.Minn A J, Velez P, Schendel S L, Liang H, Muchmore S W, Fesik S W, Fill M, Thompson C B. Nature (London) 1997;385:353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 33.Schendel S L, Xie Z, Montal M O, Matsuyama S, Montal M, Reed J C. Proc Natl Acad Sci USA. 1997;94:5113–5118. doi: 10.1073/pnas.94.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesinger P H, Gross A, Yin X M, Yamamoto K, Saito M, Waksman G, Korsmeyer S J. Proc Natl Acad Sci USA. 1997;94:11357–11362. doi: 10.1073/pnas.94.21.11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen B, Schlagbauer-Wadl H, Brown B, Bryan R, van Elsas A, Muller M, Wolff K, Eichler H, Pehamberger H. Nat Med. 1998;4:232–234. doi: 10.1038/nm0298-232. [DOI] [PubMed] [Google Scholar]
- 36.Piche A, Grim J, Rancourt C, Gomez-Navarro J, Reed J C, Curiel D T. Cancer Res. 1998;58:2134–2140. [PubMed] [Google Scholar]