Abstract
Reverse transcriptase PCR (RT-PCR) and real-time RT-PCR assays have been used to detect and quantify actin mRNA from yeasts and molds. Universal primers were designed based on the available fungal actin sequences, and by RT-PCR they amplified a specific 353-bp fragment from fungal species involved in food spoilage. From experiments on heat-treated cells, actin mRNA was a good indicator of cell viability: viable cells and cells in a nonculturable state were detected, while no signal was observed from dead cells. The optimized RT-PCR assay was able to detect 10 CFU of fungi ml−1 in pure culture and 103 and 102 CFU ml−1 in artificially contaminated yogurts and pasteurized fruit-derived products, respectively. Real-time RT-PCR, performed on a range of spoiled commercial food products, validated the suitability of actin mRNA detection for the quantification of naturally contaminating fungi. The specificity and sensitivity of the procedure, combined with its speed, its reliability, and the potential automation of the technique, offer several advantages to routine analysis programs that assess the presence and viability of fungi in food commodities.
Fungi are ubiquitous microorganisms that are often associated with the spoilage and biodeterioration of a large variety of foods and feedstuffs (7). Some molds can also adversely affect human and animal health, as they may produce mycotoxins that have been related to a range of pathologies, from gastroenteritis to cancer (11).
Because of the importance of fungi in food quality, quick and accurate procedures to detect and enumerate these contaminants in food commodities are essential. Traditional culture techniques for detection of food-borne fungi involve the use of selective microbiological media, followed by the isolation of pure cultures and finally the application of confirmatory tests. Although effective, these procedures are extremely labor-intensive and require several days. During the past years, a number of molecular methods based on immunological and genotypic techniques have been developed for revealing the presence of undesirable microorganisms, including fungi, in different food matrices (14, 17, 30). Among these, PCR is one of the most promising analytical tools in food microbiology and food control because of its specificity and sensitivity (27, 31). However, conventional PCR methods do not distinguish among viable, viable but nonculturable (VBNC), and dead cells. VBNC cells are defined as those cells that have lost the ability to express genes but may return to a culturable state (5). The presence of these cells limits the use of PCR for microbiological monitoring of food samples, where metabolically injured or nonviable cells are generally present after the stresses imposed during food processing. Several authors have demonstrated that DNA from cells killed by heat or other treatments serves as a template for PCR many days after cell viability has been lost (1, 13, 19).
In contrast to DNA, mRNA is turned over rapidly in viable cells; most mRNA species have half-lives measured in minutes (12). Therefore, detection of mRNA by reverse transcriptase PCR (RT-PCR), as opposed to DNA-based methods, is considered a better indicator of cell viability (25). The relationship between detection of microbial mRNA and cell viability has been investigated in a number of studies concerning bacterial pathogens and standard indicators of fecal contamination (2, 4, 5, 16, 22, 24, 26). Recently, Vaitilingom et al. (29) described a method for the detection of viable bacteria, molds, and yeasts by RT-PCR, with primers specific for an elongation factor gene, in heat-treated milk samples. To our knowledge, this is the first instance of the use of RT-PCR to detect viable fungi in foods.
The disadvantages of RT-PCR include its complexity and problems associated with its sensitivity, reproducibility, and specificity. Moreover, it suffers from the problems inherent in traditional PCR when it is used as a quantitative method (3). However, the introduction of real-time RT-PCR and the availability of modern equipment provide extended possibilities for the accurate quantification of mRNA species. Real-time RT-PCR allows the determination of the initial template concentration and, therefore, an accurate estimation of cell number. Real-time RT-PCR has several advantages over other PCR-based quantification approaches, including elimination of postamplification handling, easier automation, and processing of large numbers of samples. In addition, it has a very large dynamic range of template determination (around 6 orders of magnitude) (9). Thus, real-time RT-PCR offers an enormous potential for the quantification of a range of microorganisms of medical, alimentary, and environmental importance (8, 10, 15).
The purpose of the present study was to develop RT-PCR and real-time RT-PCR assays for the detection and quantification of yeasts and molds involved in food spoilage and contamination. Specifically, oligonucleotide primers were designed from the coding region of the actin (act) gene and were validated for the detection of fungi in pure cultures as well as in artificially and naturally contaminated food products.
MATERIALS AND METHODS
Microbial strains and growth conditions.
The strains used in this study and their sources are listed in Table 1. Yeasts were grown for 16 to 18 h at 28°C in YPD broth (2% Bacto Peptone, 1% yeast extract, and 2% glucose). Molds were cultured in malt yeast extract broth (2% malt extract, 0.1% yeast extract) for 72 h at 20 to 25°C under agitation. Solid media were prepared by the addition of agar (2%).
TABLE 1.
Microorganisms used in this study, their origins, and RT-PCR results
| Microorganism | Straina | Isolation source | RT-PCR result with primers ACT1 and ACT2 (353-bp fragment) |
|---|---|---|---|
| Yeasts | |||
| Candida albicans | MUCL 29800T | Skin | + |
| Candida parapsilosis | MUCL 31233T | Spruce, human | + |
| Debaryomyces hansenii | MUCL 30242T | Cherry, fruit | + |
| Kluyveromyces marxianus | CBS 834 | Kefir grain | + |
| Pichia anomala | MUCL 28639T | + | |
| Pichia membranifaciens | MUCL 29895 | Wine | + |
| Rhodotorula mucilaginosa | MUCL 30403T | + | |
| Saccharomyces cerevisiae | CBS 1171T | Brewery | + |
| Zygosaccharomyces rouxii | MUCL 30254T | Concentrated grape must | + |
| Molds | |||
| Alternaria alternata | MUCL 16089 | Fish meal | + |
| Aspergillus flavus | MUCL 9068 | Melted cheese | + |
| Aspergillus niger | MUCL 13608T | + | |
| Aureobasidium pullulans | MUCL 19020 | Deteriorated army supplies | + |
| Byssochlamys fulva | MUCL 14267 | + | |
| Cladosporium herbarum | MUCL 21027 | Leaf | + |
| Galactomyces geotrichum | MUCL 15176T | Yogurt | + |
| Penicillium italicum | MUCL 15608T | Fruit, citrus species | + |
| Bacteria | |||
| Bifidobacterium breve | ATCC 15700T | Infant intestine | − |
| Enterococcus faecium | LMG 11423T | − | |
| Lactobacillus casei subsp. casei | ATCC 334 | Emmental cheese | − |
| Lactobacillus delbrueckii subsp. bulgaricus | ATCC 11842T | Bulgarian yogurt | − |
| Lactobacillus plantarum | ATCC 14917T | Pickled cabbage | − |
| Lactococcus lactis subsp. lactis | ATCC 9936T | − | |
| Streptococcus thermophilus | DSM 20617T | Pasteurized milk | − |
MUCL, BCCM/MUCL Agro-Industrial Fungi and Yeast Collection, Université Catholique de Louvain, Louvain-La-Neuve, Belgium; CBS, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands; ATCC, American Type Culture Collection, Manassas, Va.; LMG, BCCM/LMG Bacteria Collection, Laboratorium voor Microbiologie, Universiteit Gent, Ghent, Belgium; DSM; Deutsche Veterinarmedizinische Gesellschaft, Giessen, Germany.
Lactobacilli and Enterococcus faecium were grown in MRS broth (Oxoid, Milan, Italy) for 16 h at 37°C. Bifidobacterium breve was grown in MRS broth with 0.05% cysteine for 48 h at 37°C in anaerobiosis obtained with an Anaerocult A system (VWR International, Milan, Italy). Lactococcus lactis and Streptococcus thermophilus were grown in M17 medium (Oxoid) for 24 h at 37°C. Fungi were counted on YPD agar containing 50 μg of ampicillin ml−1.
Oligonucleotide primers.
The oligonucleotide primers were designed by aligning sequences of the act gene present in the major databases for yeasts and molds by using the CLUSTAL W program (28). The following fungal act gene sequences were considered for primer selection: GenBank accession numbers M64729 (Absidia glauca), AJ000335 (Botrytis cinerea), AF056976 (Cephalosporium acremonium), AF112537 (Colletotricum gloeosporioides), M22869 (Emericella nidulans), U17498 (Histoplasma capsulatum), AB003111 (Humicola grisea), M25826 (Kluyveromyces lactis), U78026 (Neurospora crassa), AF056975 (Penicillium chrisogenum), L21183 (Pneumocystis carinii), V01288 (Saccharomyces cerevisiae), V01290 (Saccharomyces pastorianus), Y00447 (Schizosaccharomyces pombe), and X07463 (Thermomyces lanuginosus).
Potential primers for reverse transcription of the specific mRNA species and amplification of the cDNA were subsequently analyzed with the OLIGO (version 3.4; National Biosciences Inc., Plymouth, Minn.) and MACAW (version 2.0.5; National Center for Biotechnology Information, Bethesda, Md.) programs. The following primers were chosen: ACT1 (5′-CTGGGAYGAYATGGARAAGAT-3′) and ACT2 (5′-GYTCRGCCAGGATCTTCAT-3′).
Genomic DNA extraction.
Genomic DNA was isolated from fungi by the procedure of Querol et al. (23), modified as follows: cells were resuspended in 500 μl of lysis buffer (25 mg of Rhizoctonia solani lytic enzyme ml−1, 1 M sorbitol, 0.1 M EDTA [pH 7.5]) and incubated for 2 h at 45°C. Genomic DNA was extracted from 1 ml of bacterial cultures in late-exponential-growth phase as described by Marmur (18).
Total-RNA extraction from pure cultures and food samples.
Cells from late-exponential-phase cultures (2 ml) were harvested by centrifugation (at 5,500 × g for 5 min) and resuspended in LETS buffer (200 mM LiCl, 20 mM EDTA, 20 mM Tris-HCl [pH 8.0], 0.4% sodium dodecyl sulfate). Then 300 μl of acid phenol (pH 4.3; Sigma-Aldrich, Milan, Italy)-chloroform-isoamyl alcohol (25:24:1), 1 μl of diethyl pyrocarbonate (DEPC; Sigma-Aldrich), and about 60 mg of acid-washed glass beads were added. The preparations were treated by alternating 1-min cycles of vortexing with incubation on ice for about 5 min. Extracts were then centrifuged at 15,000 × g for 10 min at 4°C. The supernatant was extracted again with an equal volume of chloroform-isoamyl alcohol (24:1). These steps were repeated until a clear interface between aqueous and organic layers was obtained after centrifugation. Total RNA was precipitated with 2 volumes of ice-cold 100% ethanol and 0.1 volume of 3 M potassium acetate and was left at −80°C for 1 h before the nucleic acids were pelleted at 15,000 × g for 15 min at 4°C. The pellet was washed with 70% ethanol and resuspended in 30 μl of sterile DEPC-treated water. The sample was stored at −80°C until use.
Total RNA was also extracted from food specimens that either were naturally contaminated with fungi (Table 2) or, alternatively, were artificially inoculated with different concentrations of S. cerevisiae and Byssochlamys fulva. Microbial cells were recovered from yogurt samples by the procedure of Drake et al. (6). RNA was extracted as described above. For pasteurized food products, each sample was initially diluted 1:3 with sterile 0.9% NaCl and centrifuged at 5,500 × g for 5 min. Total RNA was isolated by the procedure described above, except for the addition of an equal volume of 2× CTAB (N-acetyl-N,N,N-trimethylammonium bromide) buffer (2% [wt/vol] CTAB [VWR International], 1% [wt/vol] polyvinyl pyrrolidone [VWR International], 100 mM Tris-HCl [pH 8.0], 20 mM EDTA [pH 8.0], 1.4 mM NaCl, and DEPC-purified water) to the supernatant before extraction with chloroform-isoamyl alcohol. An aliquot of RNA was examined by gel electrophoresis.
TABLE 2.
Quantification of yeasts and molds contaminating commercial yogurts and pasteurized food product specimens by plate count and real-time RT-PCR
| Food category | Sample no. | Log10 CFU ml−1 | CT (mean ± SD) |
|---|---|---|---|
| Yogurt | |||
| Plain | 1 | 6.6 | 16.58 ± 0.149 |
| 2 | 4.6 | 20.45 ± 0.326 | |
| 3 | 6.9 | 16.16 ± 0.036 | |
| With fruit | 1 | 6.9 | 17.82 ± 0.454 |
| 2 | 6.0 | 19.37 ± 0.075 | |
| 3 | 4.8 | 20.35 ± 0.202 | |
| 4 | 3.8 | 21.86 ± 0.079 | |
| With cereal | 1 | 7.0 | 16.23 ± 0.025 |
| Milk | 1 | 6.2 | 18.11 ± 0.030 |
| Cheese mousse with fruit | 1 | 5.9 | 17.06 ± 0.829 |
| 2 | 6.9 | 15.74 ± 0.056 | |
| Fruit juice | |||
| Orange | 1 | 5.0 | 19.41 ± 0.703 |
| 2 | 4.8 | 20.29 ± 0.674 | |
| 3 | 3.0 | 23.27 ± 3.251 | |
| Tangerine | 1 | 5.0 | 19.29 ± 0.269 |
| 2 | 4.0 | 21.29 ± 0.278 | |
| Pineapple | 1 | 4.0 | 21.11 ± 0.130 |
| Peach | 1 | 6.9 | 17.89 ± 0.454 |
| Apricot | 1 | 5.7 | 19.02 ± 0.259 |
| Pear | 1 | 3.0 | 23.74 ± 0.362 |
| Mixed fruit | 1 | 3.0 | 23.28 ± 0.367 |
| Fruit preserves | 1 | 5.0 | 20.22 ± 0.139 |
| 2 | 7.0 | 16.21 ± 0.253 | |
| 3 | 3.8 | 21.50 ± 0.556 | |
| 4 | 5.0 | 19.68 ± 1.047 | |
| 5 | 4.5 | 20.89 ± 0.295 | |
| 6 | 3.6 | 22.54 ± 0.234 |
Contaminating genomic DNA was removed from total RNA by incubation with 10 U of RNase-free DNase I (Boehringer Mannheim, Mannheim, Germany) in a 20-μl reaction mixture containing 10 μl of RNA, 4 U of RNase inhibitor (RNaseOUT; Invitrogen, S. Giuliano Milanese, Italy), DNase assay buffer (100 mM sodium acetate-0.5 mM MgSO4), and DEPC-treated water. The reaction mixture was incubated first for 30 min at 37°C and then for 5 min at 60°C. A PCR was performed to check for any contaminating DNA as described below. When necessary, the DNase treatment was repeated. RNA concentrations were determined spectrophotometrically.
RT-PCR.
RT-PCRs were conducted in either separate (two-step) or single (one-step) reactions by using a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer [PE] Applied Biosystems, Foster City, Calif.). For two-step RT-PCR, each reaction mixture (10 μl) contained 2 μl of first-strand buffer (Invitrogen), 0.3 mM deoxynucleoside triphosphates (dNTP), 5 ng of the random hexamer pd(N)6 (Amersham Pharmacia) μl−1, 20 U of RNaseOUT, 10 mM dithiothreitol, 1 μM reverse primer ACT2, 100 U of SuperScript II RNase H reverse transcriptase (Invitrogen), and 3 to 5 μl of RNA. The reaction mixture was incubated first at 42°C for 50 min and then at 70°C for 15 min. An aliquot (2.5 μl) of the resulting cDNA was amplified by PCR using a 25-μl mixture that contained 1× PCR buffer (Invitrogen), 1.5 mM MgCl2, 300 μM dNTP, 1.5 μM each primer (ACT1 and ACT2), and 1.25 U of Platinum Taq polymerase (Invitrogen). After an initial incubation at 95°C for 2 min, 30 cycles of the following temperature conditions were used: 95°C for 30 s, 52°C for 1 min, and 72°C for 45 s. A final extension at 72°C for 5 min was performed. One-step RT-PCR was carried out in a final volume of 25 μl containing 1× PCR buffer, 0.3 mM dNTP, 1.5 μM each primer (ACT1 and ACT2), 5 ng of random hexamer μl−1, 10 U of RNaseOUT, 50 U of SuperScript II RNase H reverse transcriptase, and 3 μl of RNA template. After incubation at 42°C for 50 min, 1.25 U of Platinum Taq polymerase and 1.5 mM MgCl2 were added and the reaction was processed as described above.
To increase the sensitivity of RT-PCR, a second amplification was performed in a 25-μl reaction mixture containing 2.5 μl of the amplification product under the same PCR conditions. Several controls were routinely run: a negative control with no template, a positive control with DNA from S. cerevisiae, and a DNase control with DNase-treated RNA. PCR products (5 μl) were analyzed by electrophoresis in agarose gels stained with ethidium bromide.
To confirm the identity of the amplification products, amplicons of the expected sizes were gel purified and cloned into pGEM-T (Promega Corp., Madison, Wis.). The insert of selected recombinant clones was sequenced at the Centro Genoma Vegetale-ENEA CR Casaccia (Rome, Italy) by using T7 and SP6 primers.
Heat inactivation of cultured S. cerevisiae.
An exponentially growing S. cerevisiae CBS 1171T culture was diluted in YPD broth to yield two suspensions containing 108 and 105 CFU ml−1, respectively (as determined by the standard plate count method). Heat treatment involved incubation of the broth-containing tubes (5 ml) at 60°C in a water bath for 20 min. Samples of each cell suspension were removed during treatment (at 1-min intervals), at the end of treatment, and during subsequent incubation at 25°C for 24 h (after 30 min and after 1 to 3, 5, 7, 10, 12, 14, 16, 18, 20, 22, and 24 h). Each sample was analyzed for cell viability by a standard plate count and for the presence of act DNA and mRNA as described above, in addition to fluorescence.
For the latter method, dual staining of the samples with propidium iodide (PI) and fluorescein diacetate (FDA) (Sigma-Aldrich) was performed as described by Ormerod (21). Briefly, samples were centrifuged (at 5,500 × g for 2 min), and the cell pellets of S. cerevisiae were resuspended in phosphate-buffered saline at about 106 CFU ml−1. Fluorogenic substrates were added to the cell suspensions at final concentrations of 10 μg ml−1 for FDA and 2.3 μg ml−1 for PI. The suspensions were then incubated in the dark at room temperature for 10 min. Stained samples were observed with a Leica epifluorescence microscope (LEITZ DM RB) equipped with a 50-W mercury arc lamp and with a green/red filter block (excitation wavelengths of 450 to 490 and 530 to 575 nm and emission wavelengths of 520 and 635 to 640 nm for green and red fluorescence, respectively). Uptake of PI (orange/red fluorescence) indicates dead cells, while accumulation of FDA (bright green fluorescence) indicates viable cells.
Real-time RT-PCR.
To quantify cDNA generated by reverse transcription from target RNA, real-time PCR with SYBR Green I was performed by using SYBR Green PCR master mix in a GeneAmp 5700 sequence detection system (both from PE Applied Biosystems). The 25-μl reaction mixture contained 1× SYBR Green PCR master mix (PE Applied Biosystems), 1.5 μM each primer, and 2.5 μl of the template (reverse transcription reaction product). The amplification program was carried out as described above. MicroAmp Optical 96-well reaction plates with optical caps were used. Three replicates of each sample and the various controls were processed. Fluorescence was measured at the end of the annealing-extension phase of each cycle. A threshold value for the fluorescence of all samples was set manually. The reaction cycle at which the PCR product exceeded this fluorescence threshold was identified as the threshold cycle (CT).
RESULTS
Primer design and specificity of PCR.
Primers ACT1 and ACT2 were designed by comparison of the act gene sequences available in the current databases for yeasts and molds. These candidate primers were selected on the basis of lowest degeneracy and amplify a 353-bp fragment from fungi.
To evaluate the effectiveness and specificities of this primer pair, conventional PCR was performed using purified DNA from a panel of seven bacterial strains and five yeast cultures (Candida albicans, Kluyveromyces marxianus, Pichia anomala, Pichia membranifaciens, and S. cerevisiae) frequently associated with foods (Table 1). A DNA fragment of the expected size was found with all the target fungi tested (as an example, see Fig. 1, lane 8), while no specific amplicon was produced from any of the bacteria tested. Moreover, preliminary experiments demonstrated that these primers were also suitable for RT-PCR.
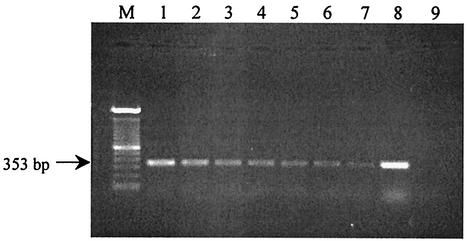
FIG. 1.
Agarose gel electrophoresis of RT-PCR products amplified from serial dilutions of S. cerevisiae RNA extracts. Lane M, 100-bp DNA ladder; lanes 1 through 7, amplification from 25 ng, 15 ng, 5 ng, 250 pg, 150 pg, 50 pg, and 25 pg of S. cerevisiae RNA in the RT-PCR mixture, respectively; lane 8, amplification from 20 ng of S. cerevisiae DNA; lane 9, negative control.
Optimization of RNA extraction and specific RT-PCR assay.
For extraction of total RNA, effective means of cell disruption, separation of nucleic acids from proteins, and purification of RNA from contaminating genomic DNA are required. Several methods of extracting total RNA from yeasts and molds were tested; the most reproducible and efficient results were obtained when cells were disrupted by a mechanical procedure, by using glass beads and incorporating acid phenol (pH 4.3) in the extraction mixture. To remove traces of DNA, different conditions of treatment with DNase I were applied. Use of 10 U of DNase I was found to reliably remove contaminating DNA and prevent false-positive RT-PCR results, whereas an adequate buffer composition was of primary importance to ensure RNA integrity.
First-strand cDNA synthesis was primed by using specific primers, random hexamers, and/or oligo(dT) primers, either alone or in combination. In terms of reaction efficiency and specificity, use of the gene-specific primer ACT2 in combination with random hexamers gave the most satisfactory results. Under our conditions, the SuperScript II reverse transcriptase significantly increased the yield of RT-PCR products over that obtained with Moloney murine leukemia virus reverse transcriptase. PCR conditions were also optimized by using 1.5 mM MgCl2 and 1.5 μM each primer at an annealing temperature of 52°C. No significant differences in amplification efficiencies between one- and two-step RT-PCRs were observed. The one-tube method was the quickest and required fewer manipulations.
The RT-PCR assay carried out on DNase-treated RNA from the panel of fungal microorganisms listed in Table 1 consistently produced the expected 353-bp amplification product (data not shown). The identities of the RT-PCR products from some fungal strains were confirmed by DNA sequence analysis. As expected, no amplification product of the appropriate size was detected from any of the bacteria listed in Table 1.
Assessment of RT-PCR detection limits.
The sensitivity and detection limits of the RT-PCR assay were tested on dilutions of total RNA (from 1,500 ng μl−1 to 25 pg μl−1) extracted from S. cerevisiae and on S. cerevisiae cells (107 to 10 CFU ml−1). The intensity of the stained 353-bp amplification product detected on the agarose gel decreased with the concentration of RNA (the range from 25 ng to 25 pg is shown in Fig. 1) or cells (data not shown). It was possible to detect the amplification product with either 25 pg of total RNA μl−1 or 10 CFU of S. cerevisiae ml−1.
Effects of heat treatment on cell viability and mRNA detection.
The suitability of act mRNA, in comparison with act DNA, as an indicator of fungal cell viability in an RT-PCR assay was evaluated by using S. cerevisiae as a reference species. The stability of both molecules was assessed on the basis of the ability to support amplification on samples taken at various times during and after heat treatment at 60°C for 20 min. Cell viability was simultaneously evaluated by standard plate count and fluorescence. Two initial concentrations of S. cerevisiae cells, namely, 105 (sample A) and 108 (sample B) CFU ml−1, were examined. A progressive linear reduction of S. cerevisiae population counts in YPD plates (about 1 logarithmic unit every min of treatment) was observed, until no colonies were detected after 5 and 8 min for samples A and B, respectively. For sample A, no RT-PCR amplification of act mRNA from S. cerevisiae was detected after 20 min of heat treatment, while the signal from DNA was still observed even after 24 h of incubation at 25°C. For sample B, both mRNA and DNA signals were detectable after 24 h of incubation, even if no growth was observed in either plates or broth. In order to corroborate these results, fluorescence microscopy was performed using FDA and PI dyes. This analysis revealed that, in contrast with the cells in sample A, a number of cells in sample B maintained their metabolic activity throughout the treatment and subsequent incubation periods.
RT-PCR analysis of artificially contaminated food samples.
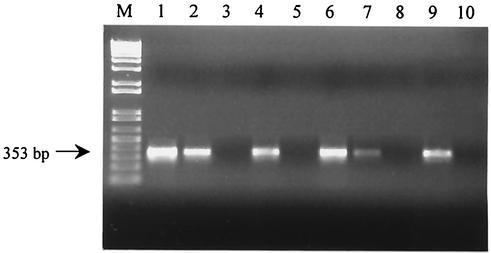
The ability to detect viable fungal cells in pasteurized food products by RT-PCR amplification of act mRNA was evaluated. Analyses were conducted on three different food matrices (yogurts, fruit juices, and fruit preserves), which were artificially contaminated with S. cerevisiae and B. fulva, selected as model species for yeasts and molds, respectively, at 105 and 107 CFU g−1. RT-PCR showed successful amplification in all the contaminated samples, while no act mRNA or DNA was detected in uninoculated foods. The detection limits of RT-PCR were also assessed in these food systems. Serial dilutions of S. cerevisiae ranging from 106 to 1 CFU g−1 were added to each selected food product. The act amplicon was detected down to concentrations of 104 CFU g−1 in yogurt and 103 CFU g−1 in fruit juice and fruit preserves. Furthermore, after a second PCR amplification, an act signal was detected at levels corresponding to 103 CFU g−1 in yogurt and 102 CFU g−1 in fruit juice and fruit preserves. Selected examples of RT-PCR on contaminated foods are shown in Fig. 2.
FIG. 2.
Agarose gel electrophoresis of RT-PCR products amplified from RNAs extracted from food products inoculated with serial dilutions of S. cerevisiae. Lane M, 1-kb-plus DNA ladder; lanes 1 to 3, RT-PCR amplification from contaminated yogurt samples (105, 104, and 103 CFU ml−1, respectively); lanes 4 and 5, second PCR on RT-PCR products from contaminated yogurt samples (103 and 102 CFU ml−1); lanes 6 to 8, RT-PCR amplification from samples of contaminated fruit preserves (104, 103, and 102 CFU ml−1); lanes 9 and 10, second PCR on RT-PCR products from samples of contaminated fruit preserves (102 and 10 CFU ml−1).
Real-time RT-PCR analysis of food samples.
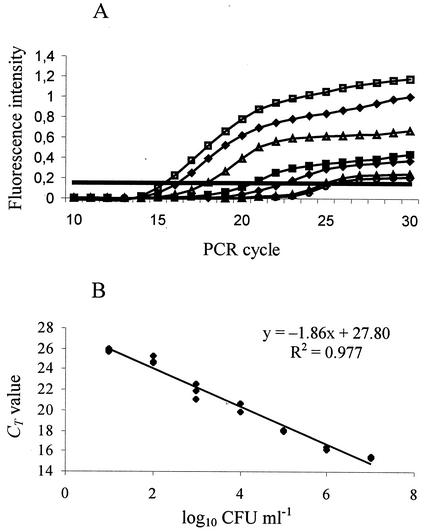
In order to obtain a more precise quantification of fungal contaminants in pasteurized food products, a real-time RT-PCR assay was optimized. In a first step, a mixed culture of selected yeasts and molds (Debaryomyces hansenii, S. cerevisiae, Zygosaccharomyces rouxii, B. fulva, Galactomyces geotrichum, and Penicillium italicum) was diluted from 107 to 10 CFU ml−1. Total RNA was isolated in triplicate from each dilution and subjected to real-time RT-PCR. The threshold value for this analysis was chosen to be 0.18, as it is included within the range of logarithmic increase of fluorescence (Fig. 3A), avoiding the signal from the negative controls. The data were plotted on a graph relating CT values to the log input CFU (Fig. 3B). A linear response was observed over more than 6 orders of magnitude; the slope of the curve was −1.86, and the square regression coefficient after the linear regression was 0.977.
FIG. 3.
Real-time RT-PCR assays of serial dilutions of mixed cultures of yeasts and molds. (A) Increase in fluorescence intensity with the number of PCR cycles. Fungal contaminants were present at 7 (□), 6 (♦), 5 (Δ), 4 (▪), 3 (⋄), 2 (▴), and 1 (○) log10 CFU ml−1. The straight horizontal line indicates the threshold value. (B) CT values plotted against the log10 CFU ml−1 derived from the plate count; three repetitions for each dilution are indicated.
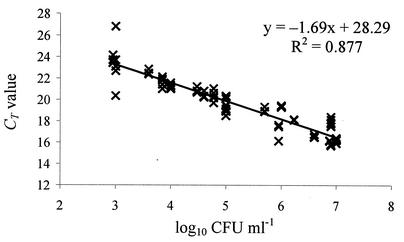
In a subsequent step, the optimized real-time RT-PCR assay was run on 27 food samples (Table 2). These products were collected from local retailers, had different origins and compositions, and showed some evidence of spoilage, probably associated with yeast and mold activity. Standard plate counts revealed the presence of fungal contaminants at levels ranging from 3 to 7 log10 CFU ml−1 in these samples (Table 2). A mixed heterogeneous population of yeasts and/or molds was generally observed in all these products. RNA was isolated in triplicate from each product and subjected to real-time RT-PCR. The results obtained are shown in Table 2. Agarose gel electrophoresis of real-time RT-PCR mixtures consistently showed amplicons of the expected size (353 bp) for all samples, while nonspecific RT-PCR products were absent (data not shown). A linear regression was performed on CT values plotted against CFU per milliliter (Fig. 4). The experimental curve obtained showed a slope of −1.69 and a regression coefficient of 0.877.
FIG. 4.
Quantification of yeasts and molds by real-time RT-PCR in spoiled commercial food products (see Table 2 for details). The CT values are plotted against the log10 CFU per milliliter derived from the plate count; three repetitions for each specimen are reported.
DISCUSSION
The need for more-rapid and -sensitive methods for the detection of food-borne fungi is increasing, particularly because these contaminants lead to considerable economic losses and may produce toxic metabolites as well. In the present study, RT-PCR and real-time RT-PCR assays were used to detect and quantify yeasts and molds associated with deterioration of yogurts and pasteurized food products. For these purposes, a fragment of the act gene was chosen as a target for the specific detection of fungal cells because this gene is essential for cell growth, is expressed throughout the cell cycle, and encodes one of the most abundant and best-conserved eukaryotic proteins. The act gene is also absent in prokaryotic cells. Recently, the gene encoding dermatophyte actin has been successfully used as a marker for rapidly assessing fungal viability in therapeutic efficacy testing of antimycotics using RT-nested PCR (20).
The fact that the act gene is highly conserved allowed us to select suitable regions for designing a set of primers that allowed for its specific and efficient amplification from a wide variety of fungi associated with spoilage of pasteurized products. One-step RT-PCR is faster and has a higher sensitivity than the two-step reaction, suggesting its possible application in the detection of yeast and mold species in the food industry.
Correlations between act mRNA and DNA detection and cell viability were made on heat-treated cultures of S. cerevisiae. The amplification of DNA in all samples clearly demonstrated that dead cells could be detected by PCR. To prevent the detection of dead microorganisms, a propagation step prior to the PCR analysis has been proposed (16); however, even if this approach increases the sensitivity of the assay, its use is restricted to culturable cells. Moreover, culture enrichment of many filamentous fungi is time-consuming and could also cause a selection of fungal species whose growth rates and/or cell numbers are higher than those of other microorganisms present in the sample.
In samples with a lower initial number of cells (105 CFU ml−1), act mRNA was undetectable after heat treatment, while it persisted in samples with 108 CFU ml−1. This observation was supported by FDA-PI fluorescence analysis, which showed that the latter cultures contained a mixture of dead and damaged cells. The physiological condition of these cells can be compared with the VBNC state, a survival mechanism described for numerous bacteria. When in this state, bacteria can no longer grow on conventional culture media but have metabolic activity, maintain pathogenicity, and, in some cases, may return to active growth when optimal conditions are restored (5). Recently, Del Mar Lleò et al. (5) demonstrated the expression of a gene during the VBNC state in an Enterococcus faecalis population by RT-PCR and used it as a marker for cell viability. The RT-PCR assay developed in this study detected the presence of act mRNA in nonculturable but potentially viable cells of S. cerevisiae under conditions similar to the VBNC state. Thus, detection of act mRNA could be correlated with viability, and this gene can be considered a suitable indicator of metabolic activities of the cell.
Our RT-PCR analysis also indicated that it was possible to detect the act gene of S. cerevisiae at concentrations as low as 10 CFU ml−1 in broth cultures, while the sensitivity was reduced in real food matrices (103 CFU g−1 in yogurt and 102 CFU g−1 in fruit juice and fruit preserves after a second PCR amplification). In fact, problems in applying an RT-PCR assay to foods can arise from various factors such as the presence of substrates chelating magnesium ions necessary for PCR, degradation of nucleic acids and/or primers by RNases or DNases, and direct inhibition of DNA polymerase and reverse transcriptase. The degree of inhibition is greatly dependent on the type of food. In this study, yogurts, fruit juices, and fruit preserves were chosen as models because they have different compositions and complexities. Furthermore, due to their low pH, high water content, and high content of carbohydrates and organic acids, these foods can frequently be spoiled by fungal contaminants. The removal of food components such as proteins, fats, and polysaccharides can positively affect the yield and quality of extracted nucleic acids. Therefore, the level of contaminating substances was reduced by the introduction of specific additional steps during the processes of extraction from the different food samples. The lower sensitivity of the assay for yogurt versus fruit juice and preserves was probably due to a greater loss of cells (and the consequent minor RNA yield) deriving from the more complex extraction procedure required for this kind of food. Nonetheless, these results demonstrated the suitability of RT-PCR for detecting fungal contaminants in food matrices; it could be successfully adopted for use with other foods.
Real-time RT-PCR allows for the detection and quantification of fungal contaminants in about 10 h. Using RNA extracts from dilutions of mixed cultures of yeasts and molds, preliminary experiments demonstrated a good linear correlation between the total cell number and the fluorescence signals. The linear regression of the data obtained from food samples showed a regression coefficient value (R2 = 0.877) lower than the previous value from pure cultures (R2 = 0.977). However, this correlation can be considered acceptable because two additional variables were introduced, namely, the food matrix effect and the presence of unknown fungal populations. Indeed, the chemical and genetic complexity of the naturally contaminated food samples, often associated with a large amount of bacteria, might affect either the quality or the yield of total RNA, or both.
In conclusion, we have developed a sensitive, high-throughput real-time RT-PCR assay for detection and quantification of viable naturally occurring yeasts and molds in a range of food commodities based on amplification of act mRNA. This procedure could offer several advantages in routine analysis for assessing food quality in either industrial or quality control settings. The assay can easily be extended to other food items and to a variety of food-monitoring initiatives, for instance, in the traceability of fungal contaminants as well as in verification of the Hazard Analysis and Critical Control Point (HACCP) system.
REFERENCES
- 1.Allmann, M., C. Hofelein, E. Koppel, J. Luthy, R. Meyer, C. Niederhauser, B. Wegmuller, and U. Candrian. 1995. Polymerase chain reaction (PCR) for detection of pathogenic microorganisms in bacteriological monitoring of dairy products. Res. Microbiol. 146:85-97. [DOI] [PubMed] [Google Scholar]
- 2.Bej, A. K., W. Y. Ng, S. Morgan, D. D. Jones, and M. H. Mahbubani. 1996. Detection of viable Vibrio cholerae by reverse-transcriptase polymerase chain reaction (RT-PCR). Mol. Biotechnol. 5:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 4.Cook, M., and W. H. Lynch. 1999. A sensitive reverse transcriptase PCR assay to detect viable cells of the fish pathogen Renibacterium salmoninarum in Atlantic salmon (Salmo salar L.). Appl. Environ. Microbiol. 65:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Mar Lleò, M., S. Pierobon, M. C. Tafi, C. Signoretto, and P. Canepari. 2000. mRNA detection by reverse transcription-PCR for monitoring viability over time in an Enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl. Environ. Microbiol. 66:4564-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake, M. A., C. L. Small, K. D. Spence, and B. G. Swanson. 1996. Rapid detection and identification of Lactobacillus spp. in dairy products by using the polymerase chain reaction. J. Food Prot. 59:1031-1036. [DOI] [PubMed] [Google Scholar]
- 7.Filtenborg, O., J. C. Frisvad, and U. Thrane. 1996. Moulds in food spoilage. Int. J. Food Microbiol. 33:85-102. [DOI] [PubMed] [Google Scholar]
- 8.Grutzing, V., S. C. Nold, J. Zhou, and J. M. Tiedje. 2001. Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR. Appl. Environ. Microbiol. 67:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 10.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussein, H. S., and J. M. Brasel. 2001. Toxicity, metabolism and impact of mycotoxins on human and animals. Toxicology 167:101-134. [DOI] [PubMed] [Google Scholar]
- 12.Ingle, C. A., and S. R. Kushner. 1996. Development of an in vitro mRNA decay system for Escherichia coli: poly(A) polymerase I is necessary to trigger degradation. Proc. Natl. Acad. Sci. USA 93:12926-12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kappe, R., N. Okeke, C. Fauser, M. Mainwald, and H. G. Sonntag. 1998. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J. Med. Microbiol. 47:811-820. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, H., M. Morita, Y. Yabuta, K. Kuzushima, K. Kato, S. Kojima, T. Matsuyama, and T. Morishima. 1999. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 37:132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein, P. G., and V. K. Juneja. 1997. Sensitive detection of viable Listeria monocytogenes by reverse-transcriptase PCR. Appl. Environ. Microbiol. 63:4441-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, S., R. R. Marquardt, and D. Abramanson. 2000. Immunochemical detection of molds: a review. J. Food Prot. 63:281-291. [DOI] [PubMed] [Google Scholar]
- 18.Marmur, L. J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 19.Masters, C. I., J. A. Shallcross, and B. M. Mackey. 1994. Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J. Appl. Bacteriol. 77:73-79. [DOI] [PubMed] [Google Scholar]
- 20.Okeke, C. N., R. Tsuboi, M. Kawai, M. Hiruma, and H. Ogawa. 2001. Isolation of an intron-containing partial sequence of the gene encoding dermatophyte actin (ACT) and detection of a fragment of the transcript by reverse transcription-nested PCR as a means of assessing the viability of dermatophytes in skin scales. J. Clin. Microbiol. 39:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ormerod, M. G. 1994. Flow cytometry: a practical approach, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 22.Patel, B. K., D. K. Banerjee, and P. D. Butcher. 1993. Determination of Mycobacterium leprae viability by polymerase chain reaction amplification of 71-kDa heat-shock protein mRNA. J. Infect. Dis. 168:799-800. [DOI] [PubMed] [Google Scholar]
- 23.Querol, A., E. Barrio, T. Huerta, and D. Ramón. 1992. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 58:2948-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sails, A. D., F. J. Bolton, A. J. Fox, D. R. A. Wareing, and D. L. A. Greenway. 1998. A reverse transcriptase polymerase chain reaction assay for the detection of thermophilic Campylobacter spp. Mol. Cell. Probes 12:317-322. [DOI] [PubMed] [Google Scholar]
- 25.Sheridan, G. E. C., C. I. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheridan, G. E. C., E. A. Szabo, and B. M. Mackey. 1999. Effect of post-treatment holding conditions on detection of tufA mRNA in ethanol-treated Escherichia coli: implications for RT-PCR-based indirect viability tests. Lett. Appl. Microbiol. 29:375-379. [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan, B., and P. Feng. 1994. Rapid detection of food-borne pathogenic bacteria. Annu. Rev. Microbiol. 48:401-426. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaitilingom, M., F. Gendre, and P. Brignon. 1998. Direct detection of viable bacteria, molds and yeasts by reverse transcriptase PCR in contaminated milk samples after heat treatment. Appl. Environ. Microbiol. 64:1157-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Vossen, J. M., and H. Hofstra. 1996. DNA based typing, identification and detection systems for food spoilage microorganisms: development and implementation. Int. J. Food Microbiol. 33:35-49. [DOI] [PubMed] [Google Scholar]
- 31.Zur, G., E. M. Hallerman, R. Sharf, and Y. Kashi. 1999. Development of a polymerase chain reaction-based assay for the detection of Alternaria fungal contamination in food products. J. Food Prot. 62:1191-1197. [DOI] [PubMed] [Google Scholar]