Abstract
Activation of class II gene transcription may involve alleviation of transcription repression as well as stimulation of the assembly and function of the general RNA polymerase (RNAP) II transcription machinery. Here, we investigated whether activator-reversible transcription repression by NC2 (Dr1/DRAP1) contributes to maximum induction levels in unfractionated HeLa nuclear extracts. Surprisingly, we found that depletion of NC2 does not significantly affect basal transcription, but dramatically reduces activated transcription. Immunoblot analyses revealed that the loss of activator function coincides with selective removal of the C-terminal domain (CTD)-hyperphosphorylated RNAP IIO along with NC2. Coimmunoprecipitation experiments with purified factors confirmed that NC2 interacts with RNAP IIO, but not with the unphosphorylated or hypophosphorylated RNAP IIA or CTD-less RNAP IIB forms. Finally, we demonstrate that, in contrast to previously published observations in cell-free systems reconstituted with purified factors, only the CTD-phosphorylated form of RNAP II can mediate activator function in the context of unfractionated HeLa nuclear extracts. These findings reveal an unexpected link between NC2 and transcription activation and suggest that regulation of RNAP II transcription through reversible CTD phosphorylation might be more complex than previously proposed.
Eukaryotic activators of RNA polymerase (RNAP) II-mediated transcription exert their function by stimulating the assembly and/or the function of the general transcription factor (GTF) machinery (1–3). Recent studies reveal that optimal levels of induction by activators depend on a complex array of positive- and negative-acting factors (4). Positive cofactors are thought to mediate and/or enhance signals from activation domains and include components of the GTF machinery such as TFIIA and TATA box-binding protein (TBP)-associated proteins (TAFIIs) (5–8), mediator protein-containing complexes (4, 7, 9–12), and cofactors derived from the upstream stimulatory activity (USA) (4, 12, 13). Negative-acting factors in humans include global repressors of transcription, such as negative cofactor (NC) 1 (14) and NC2 (also called Dr1/DRAP1) (15, 16), that may restrict or repress the intrinsic activity of the general RNAP II machinery in an activator-reversible manner (4, 8, 13). Indeed, NC2 was initially identified as an activity present in HeLa nuclear extracts (NEs) that, at low concentrations, selectively repressed basal transcription but not Sp1-activated transcription in a human cell-free system reconstituted with partially purified factors (15). In addition, there is evidence in yeast and mammalian cells that activation of certain class II genes involves reversal of NC2-mediated repression (17–22).
The enormous complexities involved in RNAP II transcriptional regulation constitute a major impediment for definitive biochemical studies on underlying molecular mechanisms. It has become evident that composition, factor concentration, and factor stoichiometry are critical parameters that determine both the overall activator responsiveness of cell-free systems and their dependency on various cofactor activities. Matters are further complicated by the fact that GTFs, DNA-binding regulatory factors, and their cofactors are subject to posttranslational modifications such as phosphorylation, acetylation, and poly(ADP-ribosyl)ation (23–29) and that both protein kinase and protein acetylase activities are present in the general transcription machinery and in certain cofactor complexes (1, 2, 7, 30).
A paradigm for the regulation of transcription factor activity by protein kinases is the reversible phosphorylation of the C-terminal domain (CTD) within the large RPB1 subunit of RNAP II (31). Eukaryotic cells contain two major forms of RNAP II, an unmodified or hypophosphorylated IIA form and a heavily phosphorylated IIO form (32, 33). RNA synthesis is catalyzed by RNAP IIO both in vitro and in vivo (33–35), whereas RNAP IIA appears to be preferentially assembled into preinitiation complexes (PICs) in vitro (36–38). Notably, CTD phosphorylation in vitro before PIC assembly by several CTD kinases, including casein kinase II, TFIIH, and CDK8, has been shown to inhibit RNAP II transcription activity (37, 39–41). This has led to the proposal that RNAP II cycles between the IIA and IIO forms during consecutive initiation events (31). In support of this idea, the human counterpart of a CTD phosphatase that is globally required for RNAP II transcription in yeast (42) recently has been shown to stimulate transcription in a human cell-free system reconstituted with highly purified components (43).
Compared with more defined reconstituted cell-free systems, NEs may provide more natural conditions to study factor requirements for transcription activation in vitro because they contain a complement of nuclear factors closer to the physiological situation. Here, we report results of studies designed to determine whether NC2-mediated repression of basal GTF activity contributes to optimal transcription induction in unfractionated human NEs. Surprisingly, we find that depletion of NC2 results in a dramatic reduction of activated transcription, with little or no effect on basal transcription activity. Moreover, we further demonstrate that the loss of activator function is caused to a large extent by selective removal of CTD-phosphorylated RNAP IIO along with NC2. Our data suggest that in HeLa NE, CTD-hyperphosphorylated RNAP IIO is quantitatively associated with NC2 and is required for activator function.
Materials and Methods
Promoter Constructs.
The G5Ad2ML template pTOG5ML(−51/+62) has been described (44). pEC(−111/+80), pTOHIV(−33/+80), and pTOHIV(−33/+68) were constructed by insertion of HIV-1 long-terminal repeat promoter sequences (45) into the multiple cloning site of pGEM7Zf(+/−) (Promega). pTOG5HIV(−33/+80) was obtained by insertion of a BamHI fragment containing five GAL4 binding sites derived from pYKG5E4T (46) upstream of the HIV-1 promoter in pTOHIV(−33/+68).
NEs and Proteins.
Extracts and purified proteins were stored in BC buffer (20 mM Tris⋅HCl, pH 7.9 at 4°C/20% glycerol/0.2 mM EDTA, pH 8.0/10 mM β-mercaptoethanol/0.5 mM PMSF) containing 0.1 M KCl. HeLa NEs were prepared as described (47). HeLa NE lacking NC2 was prepared by immunoaffinity chromatography on protein A-Sepharose CL4B (Amersham Pharmacia) covalently crosslinked to polyclonal anti-NC2β antibody. Immunodepletions were carried out in BC buffer containing 0.5 M KCl and 0.1% NP-40.
6His:TBP and GAL4-VP16 were expressed in Escherichia coli and purified as described (46). Bacterially expressed 6His:NC2α and 6His:NC2β were purified on Ni2+- nitrilotriacetic acid resin under denaturing conditions, mixed in equimolar amounts, and dialyzed in BC-100 to reconstitute 6His:rNC2. Next, the mixture was loaded onto Heparin Sepharose CL6B (Amersham Pharmacia) and eluted with a linear gradient from 0.1 to 0.5 M KCl in BC buffer. 6His:rNC2 complex-containing fractions were pooled, dialyzed in BC-100, loaded onto a FPLC MonoS column, and eluted with a linear gradient from 0.1 to 0.5 M KCl in BC buffer.
RNAP II from HeLa nuclear pellet was prepared as described (48, 49) with the following modification: after precipitation with (NH4)2SO4, the precipitate was dissolved in TGED buffer (50 mM Tris⋅HCl, pH 7.9 at 4°C/25% glycerol/0.5 mM EDTA/0.5 mM EGTA/2 mM DTT/0.5 mM PMSF/5 μg/ml aprotinin/2 μg/ml pepstatin/5 μg/ml leupeptin), and the (NH4)2SO4 concentration was adjusted to 1.2 M (TGED-1200). The material was loaded onto an 15-ISO (isopropyl) SOURCE (Amersham Pharmacia) column and eluted with a linear gradient from 1.2 M to 0 M (NH4)2SO4. RNAP II-containing fractions were identified by immunoblot analysis, pooled, and dialyzed in TGED-60 until the conductivity corresponded to TGED-80. The material then was loaded on a DEAE Sepharose FF (Amersham Pharmacia) column, and step-eluted with TGED-600. The resulting RNAP II preparation was about 30% pure, contained IIO, IIA, and IIB forms (see Fig. 3D), and was free of GTFs and SRB/MED components.
Figure 3.
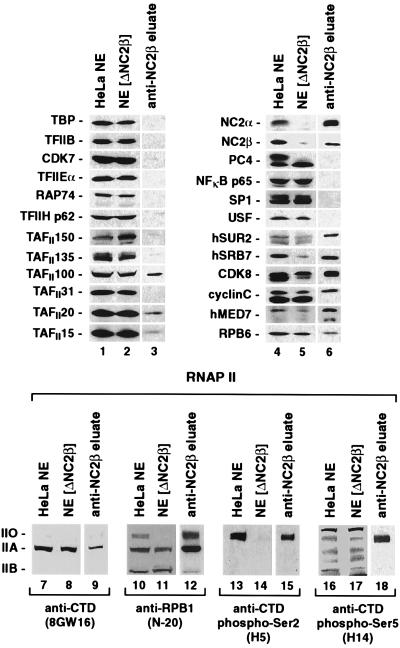
(A) RNAP II purified from HeLa nuclear pellet restores HIV-1 enhancer-dependent transcription activation in NE[ΔNC2β]. Standard transcription reactions contained 25 μg HeLa NE proteins and were analyzed as described in the legend to Fig. 1. (B) Purified recombinant 6His:tagged NC2 (6His:rNC2) after SDS/PAGE and Coomassie staining. (C) Addition of 6His:rNC2 in amounts comparable to those of NC2 present in untreated HeLa NE does not affect transcription levels in NE[ΔNC2β]. (D) 6His:rNC2 selectively interacts with the CTD-phosphorylated IIO form present in RNAP II purified from HeLa nuclear pellet. RNAP II immunoprecipitated with anti-NC2β antibodies after preincubation in the absence (lanes 5 and 6) or presence (lanes 3 and 4) of 6His:rNC2. Twenty percent of input proteins, 50% of the unbound fractions (U), and 100% of the immunoprecipitates (B) were subjected to SDS/PAGE and immunoblot analysis. The large RPB1 subunit of RNAP II was analyzed by using antibodies H5 and N-20 described in the legend to Fig. 2.
Coimmunodepletion of RNAP IIO with 6His:rNC2.
RNAP II purified from HeLa nuclear pellet was incubated in the absence or presence of 6His:rNC2 in BC-300 containing 0.1% NP-40, 10 mM Na3VO4, and 10 mM NaF for 6 h at 4°C under constant agitation. Next, anti-NC2β antibody was added, and the solutions were further incubated for 2 h at 4°C. Finally, protein Sepharose A beads were added, the solutions were incubated 2 h at 4°C, and immunoprecipitates were recovered by low-speed centrifugation.
In Vitro Transcription.
Standard 60-min in vitro transcription reactions were performed as described (46). In single-round experiments, reactions were preincubated at 30°C for 60 min in the absence of NTPs to allow for PIC formation. Transcription was initiated by addition of NTPs and stopped after 1-min incubation at 30°C.
Results
Depletion of NC2 from HeLa NEs Selectively Reduces Activator-Dependent Transcription But Does Not Affect Basal Transcription Levels.
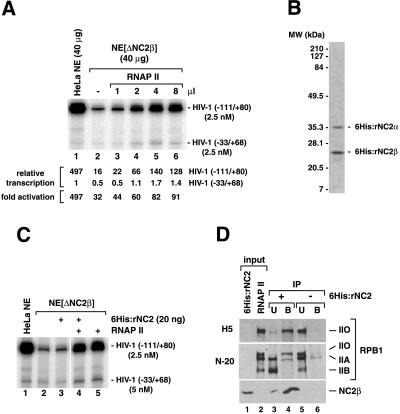
HeLa NEs lacking NC2 (NE[ΔNC2β]) were obtained by immunoaffinity chromatography on an anti-NC2β antibody column and compared with untreated NE in multiple-round and single-round transcription assays using HIV-1 long-terminal repeat promoter templates (Fig. 1). Transcription was analyzed by using plasmid templates containing the HIV-1 core promoter sequences from −33 to +80 in conjunction with either the natural HIV-1 enhancer sequences from −34 to −111 or with an artificial enhancer region comprised of five GAL4 binding sites (Fig. 1A). Fold stimulation of HIV-1 core promoter activity by transcription activators was determined by parallel primer extension analysis (using the same radiolabeled oligonucleotide) of absolute transcription levels from enhancer-containing HIV-1 promoter constructs and from an enhancer-less reference plasmid carrying HIV-1 core promoter sequences from −33 to +68. Basal transcription levels obtained from plasmid templates containing HIV-1 core promoter sequences from −33 to +80 or from −33 to +68 were indistinguishable (Fig. 1B, lanes 3 and 4).
Figure 1.
Immunodepletion of NC2 selectively reduces activated transcription in HeLa NE. Transcription reactions (20 μl) contained 25 μg NE proteins, 50 fmol of each promoter template, and 20 ng of recombinant GAL4-VP16. Transcripts were analyzed by primer extension and visualized by autoradiography and PhosphorImaging. The positions of primer extension products corresponding to correctly initiated transcripts are indicated. Relative amounts of specific transcripts and fold activations are based on PhosphorImager quantitation. (A) Schematic representation of promoter constructs used in this study. (B) Multiple-round transcription reactions were carried out for 60 min in the presence of NTPs (*). Strong enhancer-dependent HIV-1 transcription occasionally resulted in the generation of additional bands that obscured transcripts from the enhancer-less HIV-1(−33/+68) promoter template (B, lane 1). In these cases, fold activation of transcription was calculated from absolute amounts of HIV-1(−33/+68) basal transcription observed in the presence of the G5HIV(−33/+80) construct in the absence of GAL4-VP16 (B, lanes 3 and 4). (C) Single-round transcription reactions were stopped 1 min after addition of NTPs.
We found by deletion analysis that the HIV-1 enhancer region from −34 to −111, containing three binding sites for Sp1 and two binding sites for NFκB (50), is sufficient for maximal HIV-1 transcription from naked DNA templates in HeLa NEs (data not shown). As evident from Fig. 1, HIV-1 core promoter activity in HeLa NE was stimulated more than 400-fold in the presence of the minimal HIV-1 enhancer (using Sp1 and NFκB present in HeLa NE; Fig. 1B, lane 1), and more than 200-fold with GAL4-VP16 in the presence of five GAL4 binding sites (Fig. 1B, lane 5). Interestingly, absolute levels of activated transcription were 3-fold higher with Sp1/NFκB than with GAL4-VP16 in standard 60-min transcription assays, whereas GAL4-VP16 and Sp1/NFκB supported similar levels of activated transcription in single-round transcription experiments (compare Fig. 1B, lanes 1 and 5 with Fig. 1C, lanes 1 and 5). This may indicate that GAL4-VP16 and SP1/NFκB stimulate various steps of the transcription reaction differentially.
Surprisingly, depletion of NC2 from HeLa NE had little or no effect on HIV-1 core promoter activity but selectively reduced activated transcription levels (Fig. 1 B and C), with Sp1/NFκB-activated transcription being affected to a somewhat greater extent (≈20-fold, Fig. 1B, lanes 1 and 2) than GAL4-VP16-activated transcription (3- to 5-fold, Fig. 1B, lanes 5 and 6). Activated transcription levels in NE[ΔNC2β] were reduced to a similar extent in single- and multiple-round transcription assays (compare Fig. 1 B and C), indicating that NC2 depletion affected activator function mainly at the level of productive PIC formation. Taken together, these observations suggested that in unfractionated HeLa NEs NC2 is not required for efficient repression of basal transcription but is associated with an activity required for high levels of transcription activation.
NC2 Depletion from HeLa NE Coincides with Selective Removal of the CTD-Hyperphosphorylated RNAP IIO Form.
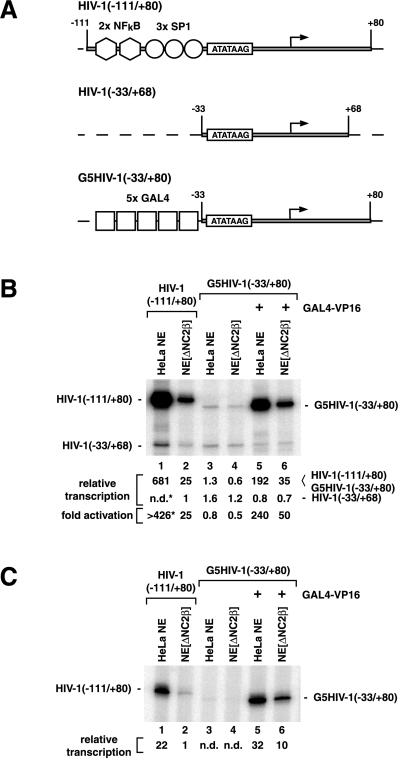
To investigate the loss of activator function in NE[ΔNC2β], we examined the protein levels of GTFs, activators, and known coactivator activities in untreated HeLa NE, NE[ΔNC2β], and glycine eluates of the anti-NC2β antibody column by immunoblot analysis. As evident in Fig. 2, NC2α was efficiently depleted along with NC2β, suggesting that NC2α (DRAP1) is quantitatively associated with NC2β (Dr1) in HeLa NEs (lanes 4–6). Sp1, NFκB, USF (upstream stimulatory factor), and the GTFs TBP, TFIIB, -IIE, -IIF, and -IIH were not retained on the anti-NC2β antibody column (lanes 1–6). These results suggested that NC2 depletion from NE did not significantly affect the protein levels of GTFs or transcription activators that mediate HIV-1 enhancer function.
Figure 2.
Immunoblot analysis of untreated HeLa NE and NC2β-depleted HeLa NE (NE[ΔNC2β]). Equal amounts of NE protein (30 μg) and 10 μl of anti-NC2β column eluate were subjected to 4–16% gradient SDS/PAGE, transferred to nitrocellulose, and analyzed with antibodies to the proteins indicated. Various antibodies were used to test for the presence of the large RPB1 subunit of RNAP II. mAb 8WG16 (Babco, Richmond, CA) is specific for the CTD and detects mainly the unphosphorylated, CTD-containing RNAP IIA form (lanes 7–9). The polyclonal antibody N-20 (Santa Cruz Biotechnology) recognizes an epitope located at the N terminus of RPB1 and therefore detects CTD-containing (IIA, IIO) and CTD-less (IIB) RPB1 (lanes 10–12). mAbs H5 and H14 (Babco) detect only the phosphorylated IIO form of the CTD and are specific for phosphoserine 2 (H5; lanes 13–15) and phosphoserin 5 (H14; lanes 16–18).
Next, we examined the protein levels of known coactivators. Although minute amounts of TAFII100 and TAFII20/15 were detected in the anti-NC2β column eluate, the NE levels of these TAFIIs were essentially unaffected (Fig. 2, lanes 1–3). In contrast to the control NE, NE[ΔNC2β] was found to be devoid of the inactive phosphorylated form Fig. 2 (upper band) of PC4 (13) but contained an increased level of the active unphosphorylated form Fig. 2 (lower band) of PC4 (lanes 4–6). Because PC4 was undetectable in the anti-NC2β column eluate, we attribute the loss of phosphorylated PC4 in NE[ΔNC2β] to PC4 dephosphorylation during the experimental procedures rather than to codepletion with NC2. Interestingly, NC2 depletion also coincided with a significant reduction of hSRB7 protein levels and a moderate reduction of hSUR2, hMED7, CDK8, and cyclin C protein levels; and corresponding amounts of these factors were detected in the anti-NC2β antibody eluate (Fig. 2, lanes 4–6). These factors were previously identified as components of mammalian coactivator complexes that are related to SRB/MED protein-containing coactivator complexes in yeast and that can mediate activator functions in vitro (4, 7, 9–11, 51). Finally, and most relevant for this study (see below), we found that NE[ΔNC2β] was essentially devoid of CTD-hyperphosphorylated RNAP IIO, but contained RNAP IIA and RNAP IIB in amounts comparable to those found in untreated NE (Fig. 2, lanes 7–18). Importantly, corresponding amounts of RNAP IIO were detected in the anti-NC2β antibody column eluate. Taken together, our analysis revealed that NC2 depletion coincided with a partial depletion of SRB/MED-protein complex components and a selective and quantitative depletion of RNAP IIO.
The CTD-Phosphorylated Form of RNAP II Is Required for Activator Function in Unfractionated HeLa NE.
The results of the immunoblot analysis pointed to a deficiency in SRB/MED protein-containing coactivator complexes and/or RNAP IIO as possible cause for the limited activator responsiveness in NE[ΔNC2β]. Indeed, earlier immunoprecipitation studies already had indicated coactivator functions of hSRB7 and/or associated factors in HeLa NEs (46). However, we found that addition of RNAP II purified from HeLa nuclear pellet was sufficient to selectively stimulate activated, but not basal, HIV-1 transcription levels in NE[ΔNC2β] (Fig. 3A). Immunoblot analysis revealed that this RNAP II preparation contained a mixture of RNAP IIO, IIA, and IIB forms (Fig. 3D) and was free of GTFs and SRB/MED protein-containing coactivator complex components that were partially depleted in NE[ΔNC2β] (data not shown). Given that NE[ΔNC2β] was selectively depleted of the RNAP IIO form (Fig. 2), this implied that transcription activation in NE[ΔNC2β] was stimulated by RNAP IIO present in our RNAP II preparation.
Addition of highly purified recombinant NC2 (6His:rNC2, Fig. 3B) in amounts comparable to those present in untreated HeLa NE (0.5 ng NC2/μg NE protein; data not shown) to NE[ΔNC2β] had little or no effect on basal or activated transcription levels, either in the absence or presence of added purified RNAP II (Fig. 3C, lanes 2–5). However, equal amounts of 6His:rNC2 strongly repressed transcription in a system reconstituted with highly purified GTFs and RNAP II (data not shown). Efficient repression in NEs required 10-fold higher 6His:rNC2 amounts (= 5 ng NC2/μg NE protein) as compared with the reconstituted system and affected basal and activated transcription to the same extent, consistent with previous observations (ref. 52, data not shown). Taken together, these findings suggested that NC2 is not required to support high levels of transcription activation in HeLa NE.
This conclusion raised the question whether depletion of RNAP IIO along with NC2 may have been caused fortuitously, for example by cross-reactivity of the anti-NC2β antibody toward an RNAP IIO-associated factor distinct from NC2. To address this issue, purified RNAP II containing forms IIO, IIA, and IIB was preincubated in the presence and absence of 6His:rNC2, and subsequently immunoprecipitated with anti-NC2β antibody (Fig. 3D). Immunoblot analysis of the bound and unbound fractions revealed efficient and selective depletion of RNAP IIO that absolutely depended on preincubation with 6His:rNC2. Although minute amounts of RNAP IIA were detectable in the anti-NC2β immunoprecipitate, similar amounts of RNAP IIA also were retained in the absence of 6His:rNC2 (Fig. 3D, lane 4 versus lane 6) and indicate weak nonspecific interactions with the antibody or the protein A beads. We therefore conclude that NC2 interacts selectively with CTD-hyperphosphorylated RNAP IIO and, further, that RNAP IIO in HeLa NE is quantitatively associated with NC2.
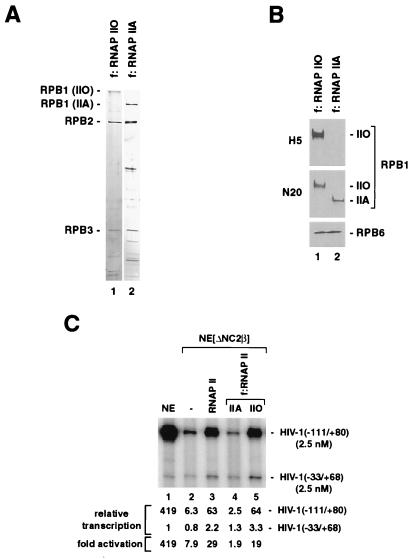
Although partial depletion of SRB/MED protein complex components may have contributed to the loss of activator responsiveness in NE[ΔNC2β], our results strongly suggested that high levels of transcription activation in unfractionated human NEs requires (i) RNAP IIO or (ii) an RNAP IIO-associated coactivator activity present in RNAP II purified from HeLa nuclear pellet. To distinguish these possibilities, hypophosphorylated/unmodified RNAP IIA and CTD-phosphorylated RNAP IIO were purified to near homogeneity (Fig. 4 A and B), from a cell line expressing a FLAG epitope-tagged RNAP II subunit, by an immunoaffinity procedure similar to that described previously (53) and then tested for their ability to restore activator function in NE[ΔNC2β]. As shown in Fig. 4C, selective stimulation of activator-dependent HIV-1 transcription in NE[ΔNC2β] was observed either with conventionally purified RNAP II containing RNAP IIO (Fig. 3D, lane 2) or with immunoaffinity-purified RNAP IIO, but not with immunoaffinity-purified RNAP IIA. These data demonstrate that RNAP II requires CTD phosphorylation to efficiently mediate activator functions in NE and suggest that regulation of RNAP II activity via reversible CTD phosphorylation might be more complex than previously proposed.
Figure 4.
(A) Analysis of immunoaffinity-purified RNAP IIO and RNAP IIA by SDS/PAGE and silver staining. (B) Comparative immunoblot analysis of immunoaffinity-purified RNAP IIO and RNAP IIA. The large RPB1 subunit of RNAP II was analyzed by using antibodies H5 (Babco) and N-20 (Santa Cruz Biotechnologies) described in the legend of Fig. 2. (C) High levels of transcription activation in human NEs requires CTD-phosphorylated RNAP IIO. NE[ΔNC2β] lacking RNAP IIO (Fig. 2) was supplemented with equal amounts (RPB6 content) of RNAP II purified from HeLa nuclear pellet (lane 3) containing IIO, IIA, and IIB forms (Fig. 3D), immunoaffinity-purified RNAP IIA (f:RNAP IIA, lane 4), and immunoaffinity-purified RNAP IIO (f:RNAP IIO, lane 5).
Discussion
NC2 Depletion Does Not Affect Basal Transcription Levels in Unfractionated HeLa NEs.
Previous observations suggested that high levels of transcription induction by transcription activators may involve, at least in part, alleviation from NC2 repression (7, 8, 13, 17–19). However, depletion of NC2 from HeLa NE had no appreciable effect on basal transcription levels (Fig. 1). Although this result is inconsistent with a major role of NC2 in restricting basal transcription in unfractionated human NEs, it does not exclude important physiological functions of NC2 as a negative regulator of transcription through the previously proposed mechanism involving direct interactions with TBP (TFIID) (52, 54). First, NC2 activity in NEs might be redundant with, and obscured by, other negative-acting factors (4, 13). Second, certain coactivator activities that can mediate activator function in cell-free systems also can repress (basal) transcription. These include USA components such as PC2, PC3 (topoisomerase I), PC4 and HMG-1 (13), human SRB/MED complex components (41, 55), and TAFIIs (6, 46). Consequently, it is anticipated that composition and factor stoichiometry will determine the extent to which individual NCs (i.e., NC2) affect transcription in cell-free systems.
Selective Association of NC2 with CTD-Phosphorylated RNAP IIO.
Immunodepletion of NC2 from NEs resulted in selective and quantitative depletion of CTD-hyperphosphorylated RNAP IIO (Fig. 2). Moreover, recombinant NC2 associated selectively with the IIO form, but not with the IIA or IIB forms, in RNAP II purified from HeLa nuclear pellet (Fig. 3D). We were unable to demonstrate any NC2 coactivator functions in our in vitro transcription assays. However, some human mediator components were coimmunoprecipitated with NC2 and RNAP IIO (Fig. 2) and, in yeast, NC2 is directly linked to the mediator/RNAP II holoenzyme through the identification of NC2 as a suppressor of mutations in SRB4 and SRB6 (21). Moreover, a previous study had indicated dual functions of NC2 in yeast, both as a repressor at most genes and as a positive cofactor required for the activation of certain genes (22). In light of these observations it is tempting to speculate that a subpopulation of cellular NC2, possibly associated with mediator components, might function as a positive-acting cofactor in conjunction with RNAP IIO to activate transcription at certain genes. Further studies in more defined cell-free systems will be needed to clarify the functional relevance of the observed NC2 interactions with RNAP IIO.
CTD-Hyperphosphorylated RNAP IIO Is Required for Activated Transcription in HeLa NEs.
Earlier studies had demonstrated an important role of reversible CTD phosphorylation in the regulation of RNAP II transcription activity. RNAP II actively engaged in transcription is highly phosphorylated both in vitro and in vivo (33–35). In cell-free systems, purified hypophosphorylated RNAP IIA can be efficiently assembled into functional PICs (36–38) and is heavily phosphorylated by the CTD kinase activity of TFIIH (31, 56) during the transition from transcription initiation to elongation. The fact that CTD phosphorylation before PIC formation can inhibit the transcription activity of purified RNAP IIA (37, 39–41) suggested that RNAP II cycles between the IIA and IIO forms, and that RNAP IIA is the form required for transcription initiation complex formation (31). In contrast, our data suggest that high levels of activated transcription in unfractionated HeLa NEs are not supported by the RNAP IIA and IIB forms, but require the presence of RNAP IIO (Fig. 4). How can we reconcile our observations with the results of earlier studies?
First, it should be pointed out that, although in vitro phosphorylation of purified RNAP IIA by CDK8, TFIIH, or casein kinase II before PIC assembly inhibits subsequent RNAP II function, natural RNAP IIO purified from eukaryotic cells is active in both promoter-independent and promoter-dependent transcription assays in vitro (36, 53, 57). Consequently, CTD hyperphosporylation per se is not necessarily inhibitory for RNAP II transcription activity. Furthermore, given that the RNAP II CTD in mammals contains 52 tandem repeats of the consensus heptamer YSPTSPS and that five of the seven residues within this sequence are potential targets for numerous protein kinases (31, 58), excessive phosphorylation of purified RNAP IIA through a particular CTD kinase in vitro may be of little physiological relevance. Regulation of RNAP II function through CTD kinases and phosphatases in vivo may involve instead very specific and perhaps quite subtle qualitative changes in the CTD phosphorylation pattern that might not be readily detectable in SDS/PAGE analyses (58).
Second, although RNAP IIO purified from eukaryotic cells appeared to be less active than purified RNAP IIA in earlier studies using partially purified factors, recent studies in cell-free systems reconstituted with highly purified factors found no difference between purified RNAP IIO and RNAP IIA in either basal (43, 53) or activated transcription (53). A lower activity of purified RNAP IIO as compared with RNAP IIA was observed only when efficient PIC formation was compromised by changing the salt conditions of the transcription reaction significantly (43). Thus, the relative in vitro transcription activities of RNAP IIO and IIA forms appear to depend on the particular cell-free system and the reaction conditions. Interestingly, for both RNAP IIA and RNAP IIO forms, we noted significantly higher basal transcription activity in a minimal system reconstituted with highly purified factors than in depleted NEs (data not shown). This finding may indicate the presence of negatively acting activities in NEs that repress RNAP II function independently of the CTD phosphorylation status. Because activator function requires RNAP IIO in NEs but appears to be independent of CTD phosphorylation in a highly purified reconstituted transcription system (53), NEs also may contain positive-acting cofactors specific for RNAP IIO.
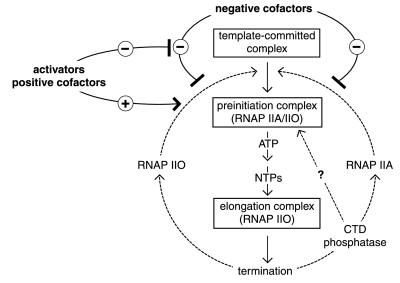
In conclusion, our observations may be explained by the working model outlined in Fig. 5. We propose that the function of RNAP II in NEs is repressed by negative factors independent of the phosphorylation status of the CTD. Activators may selectively reverse repression of RNAP IIO function by negative factors and, at the same time, selectively stimulate functional PIC assembly by RNAP IIO, presumably through specific cofactors that have yet to be identified. It is important to note that the observed requirement for RNAP IIO to mediate high levels of activator-dependent transcription in NE and the commonly accepted model of RNAP II cycling between IIO and IIA forms during consecutive initiation events are not mutually exclusive. For example, and consistent with the current dogma, the RNAP IIO that is active in NEs may be dephosphorylated before or during PIC formation (before initiation); in this case the preferential function of RNAP IIO over RNAP IIA may reflect the existence of an obligatory RNAP II recycling pathway that is only accessible to the RNAP IIO form. Reconstitution of RNAP IIO-dependent transcription activation in biochemically well-defined cell-free systems will be required to decipher the detailed molecular mechanisms of transcription regulation via CTD phosphorylation and to identify the factors involved.
Figure 5.
A working model explaining the requirement of CTD-phosphorylated RNAP IIO for high levels of transcription activation in unfractionated HeLa NEs. Transcription by CTD-hyperphosphorylated RNAP IIO and unphosphorylated RNAP IIA may be equally repressed by negative (co)factors that are present in unfractionated HeLa NEs but that are absent in systems reconstituted with purified components. Transcription activators may selectively stimulate RNAP IIO activity either directly and/or indirectly by reversing the effect of negative factors (see Discussion).
Acknowledgments
We thank Michael Meisterernst for 6His:NC2α and 6His:NC2β bacterial expression vectors, Arnold Berk for the anti-hSUR2 antibody, Jörg Kaufmann for anti-TAFII150 antibody, and Yi Wei Jiang for anti-hMED7 antibody. We thank Jenny Douglas, Sohail Malik, Ernest Martinez, and Camilo Parada for critical reading of the manuscript. This work was supported by Marie Curie Research Institute core funding to T.O. and by National Institutes of Health Grants CA42567 and AI37327 to R.G.R.
Abbreviations
- GTF
general transcription factor
- RNAP
RNA polymerase
- NC
negative cofactor
- CTD
C-terminal domain
- TBP
TATA box-binding protein
- USA
upstream stimulatory activity
- PIC
preinitiation complex
- NE
nuclear extract
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140202297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140202297
References
- 1.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 3.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 4.Roeder R G. Cold Spring Harbor Symp Quant Biol. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Verrijzer C P, Tjian R. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 7.Hampsey M. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee T I, Young R A. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 9.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 10.Myers V E, Young R A. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 11.Hampsey M, Reinberg D. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 12.Malik, S. & Roeder, R. G. (2000) Trends Biochem. Sci., in press. [DOI] [PubMed]
- 13.Kaiser K, Meisterernst M. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 14.Meisterernst M, Roy A L, Lieu H M, Roeder R G. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 15.Meisterernst M, Roeder R G. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 16.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 17.Kraus V B, Inostroza J A, Yeung K, Reinberg D, Nevins J R. Proc Natl Acad Sci USA. 1994;91:6279–6282. doi: 10.1073/pnas.91.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 19.Caswell R, Bryant L, Sinclair J. J Virol. 1996;70:4028–4037. doi: 10.1128/jvi.70.6.4028-4037.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Na J G, Hampsey M, Reinberg D. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prelich G. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 24.Long J J, Leresche A, Kriwacki R W, Gottesfeld J M. Mol Cell Biol. 1998;18:1467–1476. doi: 10.1128/mcb.18.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akoulitchev S, Reinberg D. Genes Dev. 1998;12:3541–3550. doi: 10.1101/gad.12.22.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 27.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 28.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Nature (London) 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 29.Oei S L, Griesenbeck J, Schweiger M, Ziegler M. J Biol Chem. 1998;273:31644–31647. doi: 10.1074/jbc.273.48.31644. [DOI] [PubMed] [Google Scholar]
- 30.Struhl K, Moqtaderi Z. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 31.Dahmus M E. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 32.Dahmus M E. J Biol Chem. 1981;256:3332–3339. [PubMed] [Google Scholar]
- 33.Cadena D L, Dahmus M E. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- 34.Weeks J R, Hardin S E, Shen J, Lee J M, Greenleaf A L. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien T, Hardin S, Greenleaf A, Lis J T. Nature (London) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 36.Laybourn P J, Dahmus M E. J Biol Chem. 1990;265:13165–13173. [PubMed] [Google Scholar]
- 37.Chesnut J D, Stephens J H, Dahmus M E. J Biol Chem. 1992;267:10500–10506. [PubMed] [Google Scholar]
- 38.Lu H, Flores O, Weinmann R, Reinberg D. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serizawa H, Conaway J W, Conaway R C. Nature (London) 1993;363:371–374. doi: 10.1038/363371a0. [DOI] [PubMed] [Google Scholar]
- 40.Hengartner C J, Myer V E, Liao S-M, Wilson C J, Koh S S, Young R A. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 42.Kobor M S, Archambault J, Lester W, Holstege F C P, Gileadi O, Jansma D B, Jennings E G, Kouyoumdjian F, Davidson A R, Young R A, Greenblatt J. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 43.Cho H, Kim T-K, Mancebo H, Lane W S, Flores O, Reinberg D. Genes Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oelgeschläger T, Chiang C-M, Roeder R G. Nature (London) 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 45.Rosen C A, Sodroski J G, Haseltine W A. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 46.Oelgeschläger T, Tao Y, Kang Y K, Roeder R G. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 47.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodo H G, 3rd, Blatti S P. Biochemistry. 1977;16:2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- 49.Reinberg D, Roeder R G. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 50.Jones K A, Peterlin B M. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 51.Boyer T G, Martin M E, Lees E, Ricciardi R P, Berk A J. Nature (London) 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 52.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 53.Kershnar E, Wu S-Y, Chiang C-M. J Biol Chem. 1998;273:34444–34453. doi: 10.1074/jbc.273.51.34444. [DOI] [PubMed] [Google Scholar]
- 54.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 55.Gu W, Malik S, Ito M, Yuan C-X, Fondell J D, Zhang X, Martinez E, Qin J, Roeder R G. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 56.Svejstrup J Q, Vichi P, Egly J-M. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 57.Kim W-Y, Dahmus M E. J Biol Chem. 1989;264:3169–3176. [PubMed] [Google Scholar]
- 58.Dahmus M E. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]