Abstract
Retroviruses copy their RNA genome into a DNA molecule, but little is known of the structure of the complex mediating reverse transcription in vivo. We used confocal and electron microscopy to study the structure of human immunodeficiency virus type 1 (HIV-1) intracellular reverse transcription complexes (RTCs). Cytoplasmic extracts were prepared 3, 4, and 16 h after acute infection by Dounce homogenization in hypotonic buffer. RTCs were purified by velocity sedimentation, followed by density fractionation in linear sucrose gradients and dialysis in a large pore cellulose membrane. RTCs had a sedimentation velocity of approximately 350 S and a density of 1.34 g/ml and were active in an endogenous reverse transcription assay. Double labeling of nucleic acids and viral proteins allowed specific visualization of RTCs by confocal microscopy. Electron microscopy revealed that RTCs are large nucleoprotein structures of variable shape consisting of packed filaments ca. 6 nm thick. Integrase and Vpr are associated with discrete regions of the 6-nm filaments. The nucleic acids within the RTC are coated by small proteins distinct from nucleocapsid and are partially protected from nuclease digestion.
Human immunodeficiency virus type 1 (HIV-1), like other retroviruses, copies its RNA genome into a double-stranded DNA molecule. Reverse transcription takes places in the cytoplasm of infected cells. It starts shortly after virus infection and is likely to be synchronized with virus uncoating and trafficking through the cytosol (22). The actual process of reverse transcription is fairly well understood (for details, see reference 43 and Fig. 7). It is catalyzed by reverse transcriptase (RT), a DNA polymerase that can employ both RNA and DNA as a template and whose crystal structure has been solved (25, 27, 43). The generation of full-length viral DNA proceeds in several steps and requires degradation of RNA-DNA hybrid intermediates by the RNase H activity of RT and two template switches or strand transfer reactions that appear to be stimulated by nucleocapsid proteins (NC) (1, 34). Divalent cations and deoxyribonucleotide triphosphates (dNTPs) are necessary for template elongation that proceeds slowly compared to other DNA polymerases (43). Minus-strand DNA synthesis is primed by a partially unfolded tRNA, which is annealed to the viral genome. The positive-strand DNA is synthesized by using the minus DNA strand as a template and starts at the polypurine tract (PPT), a region in the genome that is resistant to RNase H degradation and thus acts as primer for RT (43). Interestingly, HIV-1 and other lentiviruses possess an extra PPT in the central region of the genome (8). As reverse transcription proceeds, a small region of overlap between the 5′ end and the growing 3′ end of the positive strand is formed at the central PPT, giving a short stretch of triple-stranded DNA (8).
FIG. 7.
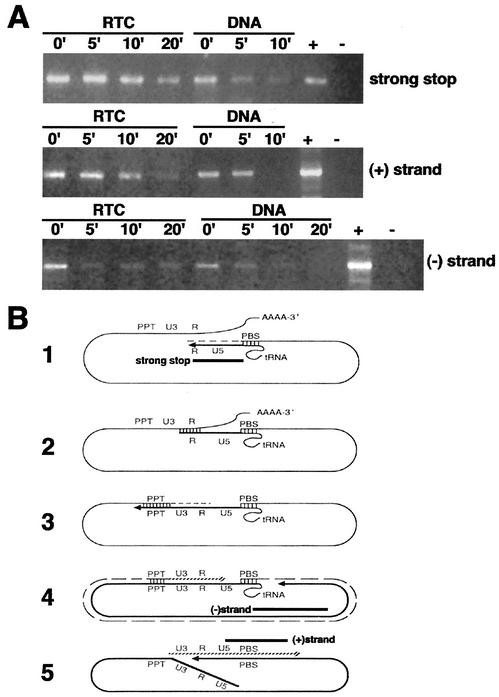
The viral DNA within the RTC is partially protected from micrococcal nuclease digestion. (A) Equilibrium density fractions containing the peak of the viral DNA (4 h postinfection) were incubated in the presence of micrococcal nuclease and 1.6 mM CaCl2. The reactions were stopped by addition of 4 mM EGTA at the indicated time points and analyzed by PCR with primers specific for negative-strand, positive-strand, and strong-stop DNAs. Naked viral DNA was prepared from the same density fractions by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation and then incubated as described above in the presence of micrococcal nuclease. −, No DNA; +, HIV-1 plasmid DNA. (B) Schematic representations of reverse transcription. The regions of the genome amplified by PCR in panel A are indicated by black bars. Diagram 1 shows the synthesis of the negative-strand strong-stop DNA starts at the primer-binding site (PBS), where a partially unfolded tRNA is bound. RNase H degrades the positive-strand RNA template so that the first strand transfer can take place. In diagram 2, a bridge is formed between the two complementary R sequences, and RT can jump on either of the RNA strands to elongate the negative-strand DNA in diagram 3. The RNA template is degraded except for the PPT. In diagram 4, the synthesis of the negative strand is completed. In diagram 5, the synthesis of the positive-strand strong-stop DNA starts at the PPT. The tRNA primer is removed, and the two strong-stop strands pair, forming a circular molecule suitable for elongation of the positive strand. At the end of the elongation process, the circular intermediate is opened into a linear double-stranded DNA molecule.
Despite our detailed knowledge of reverse transcription, little is known about the intracellular viral complex that mediates the process in vivo. A better understanding of its structure and function may help to elucidate fundamental aspects of retrovirus replication and reveal potential targets for antiviral agents. Pioneering studies on the yeast Ty retrotransposable elements indicated that reverse transcription of the Ty genome occurs within virus-like particles of ca. 60-nm diameter containing RT, integrase (IN), and capsid (CA) proteins (15, 21). Remarkably, p30Gag CA proteins were also found in murine leukemia virus (MLV) preintegration complexes (PICs) (4, 19), and some mutations in CA were found to block MLV infection at early steps of reverse transcription (38, 39, 42). These data led to the hypothesis that retroviral reverse transcription takes place in a shell-like structure permeable to dNTPs that is derived from the extracellular viral core (4). Such a shell would provide an optimal environment by preventing the loss of factors essential for reverse transcription and protection from cellular nucleases.
Recent biochemical and electron microscopy (EM) data on HIV-1 have, at least in part, challenged such view. First, incubation of purified HIV-1 virions with dNTPs induces reverse transcription in vitro in the so-called endogenous reverse transcription assay (49, 50). EM analysis of HIV-1 virions subjected to this assay showed profound alterations in virion morphology and disruption of the conical core even in the absence of detergents (51). Similarly, the native ribonucleoprotein complex of avian myeloblastosis virus that is active in an endogenous reverse transcription assay does not have a core-like structure (9). Second, if cyclophilin A is not incorporated in HIV-1 virions, initiation of reverse transcription is affected in vivo, presumably due to an effect on the disassembly of the viral core after entry (5). Third, HIV-1 intracellular reverse transcription complexes (RTCs) isolated from acutely infected cells in the absence of detergents contain very little p24Gag CA proteins (19, 26, 31). More recently, scanning EM data on acutely infected cells indicate that reverse transcription takes place in a large and elongated structure that bears no resemblance to a core (29). Taken together, these data suggest that reverse transcription of HIV-1 genome is coupled with uncoating of the viral core.
In an attempt to elucidate the structure of the HIV-1 RTC and the conformation of the nucleic acids within this complex, we have purified functional intracellular RTCs from acutely infected cells and examined them by confocal microscopy and EM. Our results indicate that HIV-1 RTCs are large nucleoprotein complexes of variable shape consisting of packed filaments 6 nm wide that contain the nucleic acids. The results also suggest that small cellular proteins may be part of the complexes.
MATERIALS AND METHODS
Cell culture and recombinant virus production.
HeLa and 293T cells were grown in Dulbecco modified Eagle medium (Gibco Laboratories, Paisley, United Kingdom) supplemented with 10% fetal calf serum (Helena Bioscience, Newcastle, United Kingdom) and 2 mM glutamine at 37°C in a humidified atmosphere containing 5% CO2. HIV-1 vectors were prepared by transient transfection in 293T cells, treated with DNase I and purified through a 25 to 45% sucrose cushion as described previously (19) by using vector plasmid pHR′SINcPPT, pCMVΔR8.2 (expressing viral core proteins) or pCMVΔR8.9 (expressing viral core proteins but no Vpr or accessory proteins), and pMD.G (expressing vesicular stomatitis virus glycoprotein [VSV-G]) (13, 32, 52).
Purification of RTCs.
Hypotonic cytoplasmic extracts were prepared from infected and uninfected cells as previously described (18, 19). Cytoplasmic extracts were loaded onto a 5 to 20% continuous sucrose gradient in 50 mM sodium phosphate buffer (pH 7.4) containing 1 mM dithiothreitol (DTT), 20 μg of aprotinin/ml, and 20 μg of leupeptin/ml, and centrifuged at 23,000 rpm for 1 h at 4°C in a Sorvall AH-650 rotor. Fractions (0.4 ml each) were collected by puncturing the bottom of the tube, the density was measured by weighing 100 μl of each fraction, and the position of the viral DNA was monitored by PCR with primers specific for the strong stop or positive-strand DNA as previously described (15). The S value was calculated according to the method of McEwen (30). Calibration of the system was performed by independently running 32S-labeled poliovirus, intact MLV, and naked viral DNA through identical sucrose gradients. Fractions of the sedimentation velocity gradient containing the peak of the viral DNA were diluted in a 20% sucrose solution in 50 mM sodium phosphate buffer (pH 7.4) containing 1 mM DTT, 20 μg of aprotinin/ml, and 20 μg of leupeptin/ml and then mixed with a cold 70% sucrose solution in D2O to obtain a continuous 20 to 70% gradient by using a gradient maker (Biocomp, Fredericton, N.B., Canada). Gradients were centrifuged at 35,000 rpm at 4°C for 20 h in a Sorvall AH-650 rotor and divided into 12 fractions by puncturing the bottom of the tube. The density was calculated by weighing 100 μl of each fraction, and the position of the viral DNA was monitored by PCR as before. The fraction containing the peak of the viral DNA was diluted 1:1 in phosphate-buffered saline (PBS), loaded onto a 1-ml cellulose Float-A-Lyzer (300,000-Da cutoff; Spectrum laboratories, Cheshire, United Kingdom), and dialyzed against large volumes of PBS at 4°C.
PCR.
PCR was performed in a final volume of 50 μl containing 1× PCR buffer, a 100 μM concentration of each dNTP, 2.5 mM MgCl2, 5 U of Taq polymerase (Perkin-Elmer), and 30 pmol of each primer. Primer sequences were as follows: strong-stop forward primer, 5′-GGCTAACTAGGGAACCCACTG-3′; strong-stop reverse complementary primer, 5′-CTGCTAGAGATTTTCCACACTGAC-3′; positive-strand forward primer, 5′-AGGGCTAATTCACTCCCAACGAAG-3′; positive-strand reverse complementary primer, 5′-GCCGTGCGCGCTTCAGCAAGC-3′; negative-strand forward primer, 5′-AAGGACCGCGCGACCTGGTGC-3′ (corresponding to the 3′ end of the phosphoribosyltransferase gene present in the viral vector); and negative-strand reverse complementary primer, 5′-CTGCTAGAGATTTTCCACACTGAC-3′. Next, 5 μl of the equilibrium density fractions or 1.5 μl of the sedimentation velocity fractions were used as a template for the PCR. Cycle parameters were as follows: 94°C for 3 min the first cycle; 94°C for 1 min, 55°C for 30 s, and 68°C for 1 min for 27 to 35 cycles; followed by one final extension cycle at 68°C for 10 min. PCR product were resolved on a 1% agarose plus 2% NuSieve gel and visualized by using ethidium bromide. Quantitative PCR was performed in duplicate in the same conditions described above by using twofold serial dilutions of the template and pHR′SINcPPT HIV plasmid. After 30 cycles, PCR bands were quantified by using Kodak Digital Science 1D image analysis software, and the linear region of the reaction was then selected to calculate the RTC copy number, assuming that 1 fg of a 7-kb plasmid DNA corresponds to 1.3 × 102 molecules.
Reverse transcription assay.
Endogenous reverse transcription reactions were carried out in 30 μl of buffer (20 mM Tris-HCl [pH 8.1], 15 mM NaCl, 6 mM MgCl2, 1 mM DTT, 2 mM concentrations of each dNTP). Then, 5 μl from the density equilibrium fractions were added to the buffer and incubated for 4 to 6 h at 37°C. The products of reverse transcription were detected by PCR with 5 μl of the endogenous reaction as a template with primers specific for the positive-strand DNA. The dNTP concentrations were adjusted in individual PCRs to maintain a final concentration of 100 μM for each dNTP throughout all samples. Serial dilutions of the relevant HIV plasmid were amplified in parallel. After 27 to 30 cycles of PCR, bands were resolved on 1% agarose plus 2% NuSieve gels and quantified by the Kodak Digital Science 1D image analysis software.
Immunochemistry and confocal microscopy.
Rabbit antiserum to HIV-1 Vpr (residues 1 to 46) from Jeffrey Kopp was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institutes of Health; rabbit antisera R5 and R4 to HIV-1 IN peptide 1 and peptide 2, respectively (from S. Ranjbar), were obtained through the Centralised Facility for AIDS Reagents, Potters Bar, United Kingdom. Aliquots of the density fractions (10 μl) were diluted in 2 volumes of PBS containing 8 μg of Polybrene/ml and incubated onto tissue culture plastic slides for 1 h at 37°C in a humidified atmosphere. Samples were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature and incubated with anti-Vpr (1:250) or anti-IN (1:300) antibodies for 1 h at room temperature, followed by incubation with TRITC (tetramethyl rhodamine isothiocyanate)-conjugated anti-rabbit secondary antibody (1:300; Dako, Ely, United Kingdom) and YOYO-1 (Molecular Probes, Eugene, Oreg.), diluted 1:10,000 in PBS, for 1 h at room temperature. Samples were mounted with Vectashield mounting medium and analyzed on a Bio-Rad MRC 1024 confocal microscope equipped with a krypton-argon laser. Images were acquired sequentially and merged later by using Lasersharp confocal assistant software (Bio-Rad).
Nuclease treatments.
Micrococcal nuclease treatments were performed in 25 μl of buffer (10 mM Tris-HCl [pH 7.4], 160 mM KCl, 5 mM MgCl2) containing 1 mM DTT, 20 μg of aprotinin/ml, 20 μg of leupeptin hemisulfate/ml, 15 U of S7 micrococcal nuclease (Boehringer Mannheim), and 1.6 mM CaCl2. Then, 5 μl from the equilibrium density fractions containing the peak of the retroviral DNA was added to the nuclease mixture, and samples were incubated on ice. The reaction was stopped by addition of 4 mM EGTA and kept on ice. Then, 5-μl aliquots from the same reaction were analyzed by semiquantitative PCR with primers specific for the strong-stop, negative- and positive-strand DNA, respectively. The nuclease sensitivity of the intact RTCs was compared to that of equal amounts of protein-stripped RTCs.
EM.
Suspensions of RTCs (2 to 4 μl) were adsorbed onto carbon-coated 400 mesh grids (glow-discharged before use) for 20 to 30 min at room temperature. Grids were washed briefly on a large drop of ultrapure distilled water and transferred onto a drop of negative stain for ca. 20 s before we drained the excess stain onto filter paper. We used routinely 4% sodium silicotungstate (STA) at pH 7.6 and pH 8.2 or 2% uranyl sulfate. For positive staining of nucleic acids, grids were submerged into 1% ethanolic uranyl acetate (UA) diluted 1:5 in acetone for 5 min. Next, the grids were washed in absolute alcohol, followed by distilled water and dried from absolute alcohol. This procedure reduced substantially the number of dark precipitates sometimes observed in a UA-stained specimen. To relax the complexes, 20 μl of the suspension was mixed 1:1 with 0.2 M glycine buffer (pH 10) containing 2 M NaCl and 10 mM EDTA (GNE) and incubated at 4°C for 3 to 24 h. For gold immunolabeling, grids with adsorbed RTC were submerged into TBG conditioning buffer (0.2 M Tris-buffered saline [pH 8.2], 0.1% bovine serum albumin, 10% fish gelatin) for 5 min and then incubated with the specific antibody for 60 min at room temperature, followed by three washes in TBG for 5 min each. Next, the grids were incubated with secondary 5-nm gold-conjugated antibodies for 45 min, followed by three washes in TBG and transfer into Tris-buffered saline. After a brief wash in distilled water grids were negatively stained as described above. Nonimmune rabbit sera were used as controls for polyclonal antibodies or a dilute TBG for monoclonal antibodies. To visualize the nucleic acids, suspensions of RTCs were incubated at 37°C for 1 to 2 h in PBS containing 5 mM EDTA and 20 μg of proteinase K/ml. Samples were precipitated in 2.5 volumes of absolute ethanol in the presence of 0.3 M sodium acetate and then resuspended in 20 μl of PBS. Specimens were examined by using a Philips CM 20 electron microscope or a JEOL 1010 microscope operated at 80 kV.
RESULTS
Purification of functional HIV-1 RTCs.
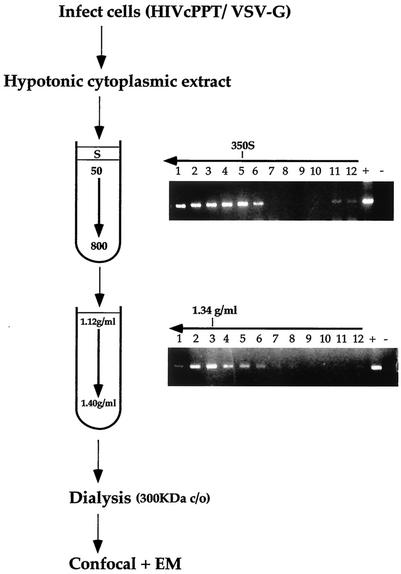
Intracellular HIV-1 complexes were purified by using a modified protocol shown previously to yield functional HIV-1 and MLV RTCs (Fig. 1) (18, 19). Cells were acutely infected at a high multiplicity of infection (≥10) with an HIV-1-based vector pseudotyped with the VSV-G envelope. Virus pseudotyped with VSV-G envelope is highly infectious, and intracellular RTCs can be readily extracted in mild conditions, in contrast to those formed by wild-type HIV-1, which can be extracted only in high-ionic-strength buffers (6). Although wild-type and VSV-G-pseudotyped HIV-1 use a different entry route, the resulting RTCs are generally believed to be similar (19, 29). Infection was synchronized by preincubating the cells at 4°C to allow for virus binding to the receptor. Cells were treated with trypsin, and cytosolic extracts were prepared by Dounce homogenization in hypotonic buffer at 3, 4, and 16 h postinfection. Extracts were subjected to centrifugation at 50,000 × g for 1 h in a 5 to 20% linear sucrose gradient. PCR on individual fractions detected the peak of the viral DNA sedimenting at approximately 350 S. Contamination with plasmid DNA was excluded by amplification of a region present in the plasmid but absent in the reverse-transcribed viral DNA. The 350 S fraction containing the viral DNA peak was subjected to density fractionation in a 20 to 70% linear sucrose gradient. The peak of viral DNA was then recovered in fractions with a density of ca. 1.34 g/ml, a finding consistent with previous reports (19, 23), and was dialyzed by using a 300-kDa pore cellulose membrane. Uninfected cells were processed in exactly the same way in parallel and used as negative controls in all experiments.
FIG. 1.
Scheme of the method used to purify intracellular HIV-1 RTCs. HeLa cells were infected at a high multiplicity of infection with an HIV-1-based vector pseudotyped with VSV-G. After 2 h of incubation at 4°C, cells were transferred at 37°C for 3, 4, or 16 h, and cytoplasmic extracts were prepared by Dounce homogenization in hypotonic buffer. Extracts were loaded onto a 5 to 20% linear sucrose gradient and subjected to velocity sedimentation. The position of the viral DNA in the gradient was detected by PCR with primers specific for the strong-stop DNA. The peak of the viral DNA was consistently found sedimenting at approximately 350 S. Fractions 4 and 5 were subjected to density fractionation into a 20 to 70% linear sucrose gradient, and the position of the viral DNA was detected as before by PCR. The fractions having a density of ca. 1.34 g/ml contained the peak of the viral DNA and were dialyzed in a large pore cellulose membrane (300,000-Da cutoff). Dialyzed samples were used for confocal and EM analyses. Arrows indicate the direction of the gradients.
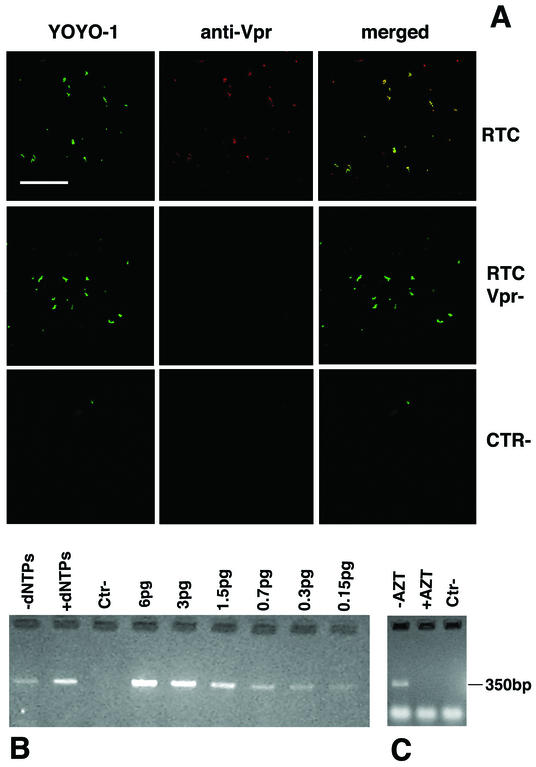
To test whether purified RTCs were functional, aliquots from the 1.34-g/ml density fractions were subjected to an endogenous reverse transcription assay in the absence of detergents. This assay examines the integrity of the RT machinery and its ability to synthesize full-length viral DNA. Samples were incubated for several hours in the presence or absence of exogenous dNTPs and analyzed by semiquantitative PCR with primers specific for the positive-strand DNA, a late reverse transcription product (Fig. 2B). The addition of exogenous dNTPs consistently stimulated the synthesis of near full-length viral DNA in RTCs isolated 3 and 4 h postinfection. RTCs isolated 16 h postinfection had a reduced ability to reverse transcribe (not shown). No viral DNA was found in samples of uninfected cells processed in the same way (Fig. 2B). To exclude any contamination with plasmid DNA, cells were infected in the presence of 200 μM zidovudine (AZT), and RTCs were extracted as described above. The amount of strong-stop DNA was substantially reduced in AZT-treated samples but was still enough to locate RTCs in the gradients during purification. Purified RTCs from AZT-treated cells were subjected to the endogenous RT reaction in the presence of dNTPs. The positive-strand DNA was not detected in these controls (Fig. 2C), suggesting that the purified complexes were bona fide RTCs.
FIG. 2.
Analysis of purified HIV-1 RTCs by confocal microscopy and endogenous reverse transcription assay. (A) Purified RTCs isolated 4 h postinfection were adsorbed onto plastic tissue culture dishes, fixed, and labeled with the nucleic acid dye YOYO-1 and an anti-Vpr polyclonal antibody. Images were acquired sequentially and merged by using the Confocal Assistant software. Mutant RTCs lacking Vpr (RTC Vpr−), samples from uninfected cells (CTR−), and nonimmune rabbit sera were used as controls. Scale bar, 15 μm. (B) Endogenous RT assay on the equilibrium density fractions containing the peak of the viral DNA (4 h postinfection). Samples were incubated for 6 h at 37°C in the presence or absence exogenous dNTPs and then subjected to PCR with primers specific for the positive-strand DNA (expected band size is 350 bp). Serial dilutions of the HIV-1 vector plasmid were amplified in parallel. Ctr−, uninfected cells. (C) RTCs were extracted from cells infected in the presence (+AZT) or absence (−AZT) of 200 μM AZT, purified, and subjected to an endogenous RT assay in the presence of dNTPs. The expected band size for the positive-strand DNA is 350 bp. Lower-molecular-weight bands are PCR artifacts. Ctr−, uninfected cells.
Analysis of purified RTCs by confocal microscopy.
To further verify the purity and identity of the complexes, we adapted a method used previously to visualize single viral particles in tissue culture supernatants by fluorescence microscopy (37, 45). Dialyzed fractions were adsorbed on to plastic slides, fixed, and labeled with an antiserum to Vpr, a protein that remains associated with the RTC (19, 23), and YOYO-1, a sensitive nucleic acids dye that allows visualization of single DNA and RNA molecules. Confocal microscopy detected colocalization of Vpr and nucleic acids in more than 90% of fluorescent particles (Fig. 2A). Similar results were obtained with an anti-IN rabbit polyclonal antibody (results not shown but see Fig. 3). No colocalized particles were seen in samples containing mutant RTCs lacking Vpr when rabbit nonimmune serum was used or in samples purified from uninfected cells (Fig. 2A). YOYO-1-labeled particles were seen only occasionally in uninfected controls (Fig. 2A). These experiments showed that purified RTCs could be visualized specifically by confocal microscopy.
FIG. 3.
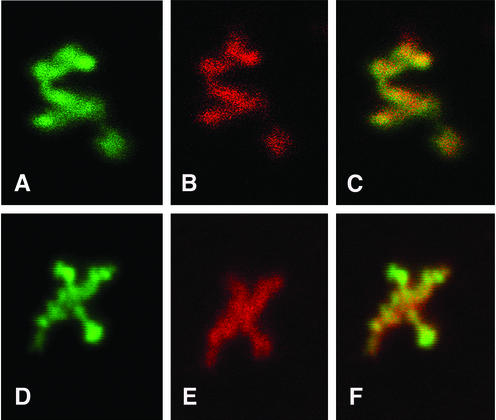
Analyses of purified RTC by confocal microscopy at ×6,300 magnification. Purified RTCs (4 h postinfection) were adsorbed onto plastic tissue culture dishes, fixed, and doubly labeled with YOYO-1 (A and D) and anti-Vpr (B) or anti-IN (E) antibodies. Images were acquired sequentially at ×6,300 magnification by using an optical zoom and merged by using Lasersharp Confocal Assistant software (C and F).
To examine the structure of intracellular HIV-1 RTCs, purified complexes were doubly labeled with YOYO-1 and the anti-Vpr or anti-IN antibodies and examined by confocal microscopy at higher magnification by using an optical zoom (Fig. 3). Images were acquired sequentially and merged to detect specific patterns of colocalization. Nucleic acids in the complex appeared partially condensed. Both IN and Vpr colocalized with the nucleic acids, but Vpr produced a weaker signal than IN (Fig. 3). Although the complexes had, overall, a similar size, no specific shape could be identified.
Analysis of purified RTCs by EM.
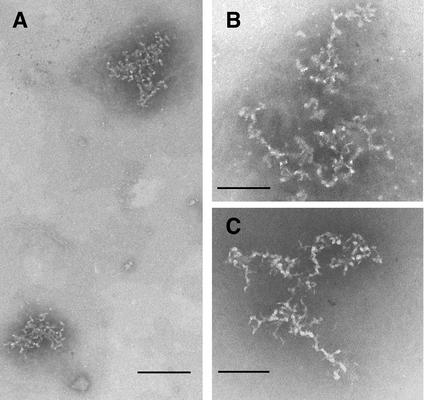
Confocal microscopy indicated that dialyzed fractions contained purified RTCs and an endogenous RT assay showed that purified complexes were functional, suggesting that their structure was not grossly perturbed. Thus, transmission EM was used to investigate RTCs in more detail. Samples negatively stained with STA at pH 7.6 or 8.2 showed the presence of relatively large structures consisting of a mesh-like network of flexible filaments (Fig. 4). Higher-magnification images showed complexes consistent with the pattern observed by confocal microscopy; these were mainly composed of packed filaments that presumably contained the nucleic acids (Fig. 4B). Measurements of the width of the filaments in the complexes indicated that the average was 6.1 nm (standard deviation = 0.88, n = 293). Negative staining of the samples with uranyl sulfate produced similar images (not shown). Similar structures were found in samples prepared 3, 4, and 16 h postinfection. However, the number of complexes found 16 h postinfection was too low to perform a detailed comparative analysis with RTCs isolated at earlier time points. Most complexes were a few hundred nanometers across, which would be consistent with the size of a partially condensed DNA molecule of ca. 5 kb (the size of the retroviral vector used in the present study) (41). Larger accumulations of the typical structures were sometimes observed; these accumulations presumably resulted from the aggregation of several complexes during the purification procedure.
FIG. 4.
Visualization of purified RTCs (3 and 4 h postinfection) by negative stain EM. Samples were adsorbed onto carbon-coated grids and negatively stained with 4% STA (pH 8.2). (A) Two complexes in different orientations are visible after negative staining at ×88,000 magnification. Scale bar, 140 nm. (B) Higher-magnification image of a typical complex as detected at ×180,000 magnification. Scale bar, 85 nm. (C) Samples (20 μl) were mixed 1:1 with GNE buffer (pH 10) for 3 h, washed, and negatively stained with STA (pH 8.2). Note the indication of continuity of the filaments after relaxation of the complexes. Scale bar, 85 nm.
IN binds to the 6-nm filaments.
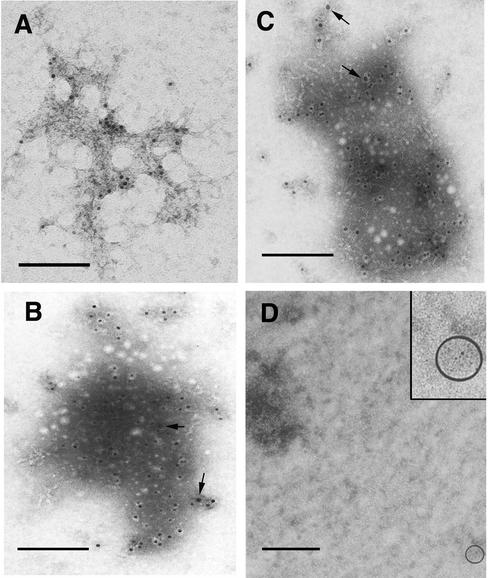
To confirm that the nucleoprotein complexes seen by EM were HIV-1 RTCs, samples were gold immunolabeled with the same anti-Vpr and anti-IN antibodies used for the confocal microscopy studies. The complexes were indeed labeled by both specific antisera but not by nonimmune rabbit serum (Fig. 5). Some gold particles were seen at a distance from the complexes, suggesting that some Vpr and IN had dissociated. The frequency of gold particles in RTC-containing samples was always higher than in control grids. More gold particles were observed on grids labeled with the anti-IN antibody, and small clusters of gold particles were found in these grids presumably reflecting IN oligomerization (Fig. 5B and C) (35). Gold labeling experiments showed that IN and Vpr were not distributed evenly throughout the 6-nm filaments in the complexes. This discrete distribution of IN can be appreciated more clearly in Fig. 5C and may reflect a forming intasome (47). Complexes were only occasionally immunolabeled by a monoclonal antibody against nucleocapsid protein (NC p7gag), and most of the gold particles were scattered throughout the grid. The same antibody specifically labeled HIV-1 virions treated with detergents to expose the cores and virus particles in thin sections of infected cells (data not shown). Several monoclonal antibodies to RT were tested, but none showed convincing immunolabeling of the complexes, presumably because too few RT molecules remained associated with the nucleic acids. This result is consistent with the finding that most RT proteins dissociate upon disassembly of the viral core (19, 20).
FIG. 5.
Gold immunolabeling of purified RTCs (4 h postinfection) with anti-Vpr polyclonal antibody (A), with anti-IN antibody (B and C) or nonimmune rabbit serum (D), followed by incubation with 5-nm colloidal gold-conjugated secondary antibody. Samples were stained with ethanolic UA in panel A or STA (pH 7.6) in panels B, C, and D. The arrows in panels B and C point to small clusters of gold particles. The round white dots in panels B and C are sublimation artifacts of the STA staining. The circled area in panel D contains a gold particle and is magnified in the inset. Scale bar, 100 nm.
Small proteins coat the nucleic acids in the complexes.
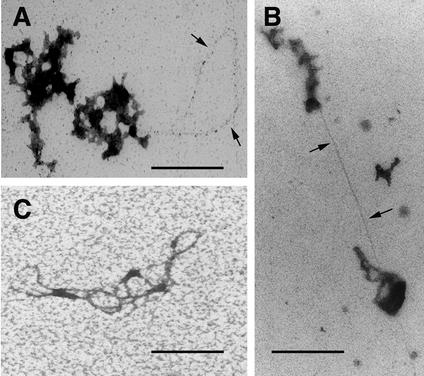
In an attempt to relax the complexes, samples were incubated in glycine buffer at pH 10 in the presence of 1 M NaCl for 3 h, followed by negative staining with STA at pH 8.2. Some relaxation was observed, and the images suggested a continuity of the filaments in the complexes (Fig. 4C). Interestingly, despite the harsh conditions, this treatment did not strip all proteins from the filaments (Fig. 4C). Thus, to visualize the nucleic acids more clearly, suspensions of RTCs were incubated with proteinase K for 1 or 2 h, ethanol precipitated, and stained with ethanolic UA after exposure to ethidium bromide. After this treatment, nucleic acids were revealed as thin filaments up to 0.8 μm long protruding from the complexes, which, in some cases, were almost completely digested (Fig. 6A and B). Complete uncoating of the nucleic acids was achieved by incubating samples with guanidine thiocyanate, a strong denaturing agent for proteins (Fig. 6C) (3).
FIG. 6.
Visualization of nucleic acids in purified RTCs. Samples (3 and 4 h postinfection) were treated with proteinase K for 1 h (A) or 2 h (B), ethanol precipitated, adsorbed onto carbon-coated 400-mesh grids, washed in distilled water, and stained with 1% ethanolic UA diluted 1:5 in acetone for 5 min. (C) Bundle of nucleic acid filaments from a preparation deproteinized with guanidine thiocyanate. The arrows point to nucleic acids. Scale bar, 150 nm.
Overall, the complexes appeared similar to the 11-nm “thin” fibers, which typically appear in histone H1-depleted chromatin (48). Digestion of thin chromatin fibers with DNase I gives the typical beads-on-a-string filaments composed of DNA regularly packed into nucleosomes separated by 15- to 20-nm intervals (44). To test whether the 6-nm filaments had a similar organization, samples were incubated in the presence of DNase I, the reaction was stopped by the addition of EDTA, and the RTCs were examined by EM. Although a control naked DNA plasmid of about 10 kb was reduced to 150- to 200-bp fragments as detected by agarose gel electrophoresis, RTC samples never showed the presence of beads-on-a-string filaments (not shown). This indicated that nucleic acids in the complexes are unlikely to be organized into nucleosomes and that proteins coat the nucleic acids.
DNA in the complexes is partially protected from nuclease digestion.
EM data indicated that the nucleic acids in the RTCs are associated with proteins, which presumably protect the complex from attack by cellular nucleases. To examine this possibility, samples containing RTCs purified 3 and 4 h postinfection were treated with micrococcal nuclease, and the reaction was stopped at the time points indicated in Fig. 7 by the addition of EGTA. Nuclease-treated samples were then analyzed by PCR with primers specific for the strong stop DNA and for the elongated negative- and positive-strand DNAs. Control samples were treated with proteinase K, phenol-chloroform extracted, and ethanol precipitated. DNA in control samples was digested faster than DNA in RTC samples. In RTC samples, the strong-stop DNA and the positive-strand DNA were partially resistant to nuclease attack. The negative-strand DNA was more sensitive to nuclease digestion, although digestion was never complete, even after 20 min of incubation (Fig. 7). These results are consistent with the EM data and indicate that the nucleic acids in the RTC are, at least in part, protected from micrococcal nuclease digestion.
DISCUSSION
We have investigated the structure of intracellular HIV-1 RTCs purified after acute infection. RTCs were extracted under mild conditions and in the absence of detergents in an attempt to preserve their structure. An endogenous reverse transcription assay showed that RTCs were able to reverse transcribe, suggesting that no major alteration of their structure occurred during purification. YOYO-1 labeling of nucleic acids and antibody labeling of Vpr and IN allowed specific identification and visualization of RTCs by confocal microscopy. At high magnification, confocal microscopy images showed that purified RTCs consisted mainly of condensed clusters of nucleic acids of variable shape and that IN and Vpr were clearly part of the complexes at an early stage, findings in agreement with biochemical data obtained previously (7, 16, 19, 26, 31). The confocal microscopy technique is straightforward and sensitive enough to study nucleic acids at a single molecule level but has some limitations with respect to the detailed structural analysis of RTCs. First, intercalating molecules such as YOYO-1 tend to stretch nucleic acids and increase their length artificially (41), and second, the size of the molecule observed is proportional to the intensity of emitted light rather than to the actual molecule dimensions. EM analyses complemented the confocal microscopy results, adding greater definition and detail. EM of the purified samples revealed the presence of large complexes a few hundred nanometers across, mainly consisting of a mesh-like network of filaments 6 nm thick. Several lines of evidence indicated that such complexes were HIV-1 RTCs. They were not found in uninfected samples, they were specifically immunolabeled by anti-IN and anti-Vpr antibodies, and they contained nucleic acids, findings in agreement with the confocal microscopy data.
Most complexes observed by EM had a size larger than the one previously calculated for HIV-1 RTCs by gel filtration chromatography (56 nm in diameter) (31) but consistent with a partially condensed DNA molecule of ca. 1.5 μm (the size of the retroviral vector used) and with the high sedimentation rate that we have found. RTCs isolated 3, 4, and 16 h after acute infection appear to have a similar structure, although a detailed comparative analysis could not be performed due to the low number of complexes detected 16 h postinfection. Overall, our findings are in agreement with recent data obtained by scanning EM of HIV-1 RTCs in infected cells (29). Our results and those of McDonald et al. (29) suggest that major structural rearrangements are likely to occur in the early phases, shortly after virus entry into the cytoplasm, and support the view that reverse transcription is coupled with virus uncoating (19, 20). Consistently, HIV-1 replication can be reduced by intracellular expression of single-chain variable antibody fragments against RT and IN, indicating that these proteins are accessible in the context of an RTC (28, 40). Further, more-subtle rearrangements are likely to occur after reverse transcription is completed to form the intasome, a nucleoprotein structure at the viral DNA ends that contains IN as well as cellular proteins and presumably primes PICs for integration into cellular DNA (10, 11, 17, 46, 47).
EM observations indicated that proteins are associated with the nucleic acids in RTCs. Naked double-stranded DNA or RNA molecules are ca. 2 nm thick. Since the thickness of the RTC filaments is ca. 6 nm, proteins associated with the nucleic acids may not be larger than 2 to 3 nm or ca. 10 to 20 kDa. RTCs were not consistently immunolabeled with an MAb directed against the proximal zinc finger region of NC (14) and the presence of gold particles scattered throughout the grid suggests that NC proteins have dissociated from the complexes during sample preparation. It also suggests that other small proteins, in addition to NC, coat the viral nucleic acids within the intracellular RTC. Remarkably, incubation of the samples at pH 10 in high salt or in uranyl sulfate at pH 4.4 did not induce dissociation of all of the proteins from the nucleic acids, suggesting that at least some proteins may be tightly bound to them. We are currently trying to identify such protein(s). Nucleic acids were clearly revealed only by proteinase K digestion of the RTCs and by deproteination with guanidine thiocyanate. Micrococcal nuclease protection assays indicated that the viral genome was partially resistant to digestion compared to naked DNA and that the strong-stop DNA is more resistant than positive- and negative-strand DNAs. This is consistent with the presence of proteins bound throughout the viral genome and a forming intasome at the viral DNA ends (10, 46). It is also consistent with the ability of HIV-1 RTCs to survive for at least 24 to 48 h in infected cells, a relatively long time compared to most eukaryotic mRNAs (24, 36).
The detailed nature of the interactions between nucleic acids and proteins in the 6-nm filaments is not clear and deserves further investigation. Morphologically, the 6-nm filaments are similar to histone H1-depleted thin chromatin fibers (48). However, we tend to exclude that the 6-nm filaments are organized into nucleosomes like the thin chromatin fiber for several reasons. Chromatin fibers have an average thickness of 10 to 11 nm, which is about twice the size of the filaments observed in the RTCs. Unlike thin chromatin fibers, treatment of RTCs with DNase I never produced beads-on-a-string filaments (44, 48). Thus, our results suggest that proteins tightly coat the nucleic acid filament.
Our data suggest that reverse transcription is carried out in a relatively loose nucleoprotein complex. This would present two challenges for reverse transcription. Nucleic acids must be protected from cellular nucleases during DNA synthesis, and RT protein must have access and remain bound to the RNA or DNA templates during polymerization. Our EM observations indicate that nucleic acids in the RTCs are associated with small proteins, and nuclease protection assays indicated that the viral genome is partially resistant to micrococcal nuclease attack. Hence, it is likely that nucleic acids within the 6-nm filaments are sufficiently protected also from cellular nucleases. It is not clear how easily RT can access its template. Displacement of proteins from chromatin is not an absolute requirement for polymerase-mediated elongation. In prokaryotes, RNA polymerases can transcribe completely through one or more nucleosomes, although this induces pausing (33). Partial steric hindrance of the nucleic acids may explain the low polymerization rate of RT in vivo, which in turn may be the price to pay to avoid RTC degradation.
A consensus is emerging on the early events of retrovirus infection based on biochemical and confocal and EM studies (19, 20, 26, 29). The viral core would appear to remain intact for a short period of time after virus-cell fusion. Reverse transcription can start within the intracellular core, but its disassembly appears to be essential to allow reverse transcription to progress. After the disassembly of the core, reverse transcription would proceed to completion in a large nucleoprotein complex. In this respect, it is tempting to speculate that the recently described Lv-1 restriction phenotype, which inhibits reverse transcription and targets the p2 domain of p24CA (2, 12), acts by preventing the correct uncoating of the viral core. The method described here should improve our ability to investigate further these early events in the life cycle of HIV-1.
Acknowledgments
We thank Robin Weiss for helpful discussions and for critically reading the manuscript. We thank Yasuhiro Takeuchi and Massimo Pizzato for advice on the confocal microscopy technique, Bernard Roques for the anti-NC antibody, and Didier Trono and Adrian Thrasher for HIV-1 vector plasmids.
This work was funded by a Wellcome Trust Career Development Research Fellowship (to A.F.) and by the MRC.
REFERENCES
- 1.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J.-L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besnier, C., Y. Takeuchi, and G. Towers. 1925. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediated the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 5.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinskaya, A., B. Brichacek, A. Mann, and M. Stevenson. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 188:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charenau, P., M. Alizon, and F. Clavel. 1992. Second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M.-Y., C. F. Garon, and T. S. Papas. 1980. Native ribonucleoprotein is an efficient transcriptional complex of avian myeloblastosis virus. Proc. Natl. Acad. Sci. USA 77:1269-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H., and A. Engelman. 1998. The barrier to autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H., S.-Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J. Biol. Chem. 274:17346-17358. [DOI] [PubMed] [Google Scholar]
- 12.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 1919. 2002. Cellular inhibitors with Fv-1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demiason, C., K. Parsley, G. Brouns, M. Scherr, K. Battmer, C. Kinnon, M. Grez, and A. J. Thrasher. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a HIV-1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13:803-813. [DOI] [PubMed] [Google Scholar]
- 14.De Rocquigny, H., A. Caneparo, C.-Z. Dong, T. Delaunay, and B. P. Roques. 2000. Generation of monoclonal antibodies specifically directed against the proximal zinc finger of HIV type 1 NCp7. AIDS Res. Hum. Retrovir. 16:1259-1267. [DOI] [PubMed] [Google Scholar]
- 15.Eichinger, D. J., and J. D. Boeke. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54:955-966. [DOI] [PubMed] [Google Scholar]
- 16.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 18.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency type-1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forshley, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garfinkel, D. J., J. D. Boeke, and G. R. Fink. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:507-517. [DOI] [PubMed] [Google Scholar]
- 22.Goff, S. P. 2001. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3:517-528. [DOI] [PubMed] [Google Scholar]
- 23.Heinzinger, N. K., M. I. Bukrinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in non-dividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrick, D., R. Parker, and A. Jacobson. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karageorgos, L., P. Li, and C. Burrell. 1993. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res. Hum. Retrovir. 9:817-823. [DOI] [PubMed] [Google Scholar]
- 27.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 28.Levy-Mintz, P., L. Duan, H. Zhang, B. Hu, G. Dornadula, M. Zhu, J. Kulkosky, D. Bizub-Bender, A. M. Skalka, and R. J. Pomerantz. 1996. Intracellular expression of single-chain variable fragments to inhibit early stages of the viral life cycle by targeting human immunodeficiency virus type 1 integrase. J. Virol. 70:8821-8832. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 29.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McEwen, C. R. 1967. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal. Biochem. 20:114-149. [DOI] [PubMed] [Google Scholar]
- 31.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type-1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 33.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471-475. [DOI] [PubMed] [Google Scholar]
- 34.Peliska, J. A., S. Balasubramanian, D. P. Giedroc, and S. J. Benkovic. 1994. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalysed DNA strand transfer reactions and modulates RNase H activity. Biochemistry 33:13817-13823. [DOI] [PubMed] [Google Scholar]
- 35.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizzato, M., S. A. Marlow, E. D. Blair, and Y. Takeuchi. 1999. Initial binding of murine leukemia virus particles to cells does not require specific env-receptor interaction. J. Virol. 33:8599-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reicin, A., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in HIV-1 virion morphogenesis and early steps in the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartzberg, P., J. Colicelli, M. L. Gordon, and S. P. Goff. 1984. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virus and reverse transcriptase. J. Virol. 49:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaheen, F., L. Duan, M. Zhu, O. Bagasra, and R. J. Pomerantz. 1996. Targeting human immunodeficiency virus type 1 reverse transcriptase by intracellular expression of single-chain variable fragments to inhibit early stages of the viral life cycle. J. Virol. 70:3392-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, D. E., T. T. Perkins, and S. Chu. 1996. Dynamic scaling of DNA diffusion coefficients. Macromolecules 29:1372-1373. [Google Scholar]
- 42.Strambio de Castilla, C., and E. Hunter. 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 66:7021-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 44.Thoma, F., and T. Koller. 1977. Influence of histone H1 on chromatin structure. Cell 12:101-107. [DOI] [PubMed] [Google Scholar]
- 45.Vanderplasschen, A., and G. L. Smith. 1997. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J. Virol. 71:4032-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei, S.-Q., K. Mizuuchi, and R. Craigie. 1997. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 16:7511-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, S.-Q., K. Mizuuchi, and R. Craigie. 1998. Footprints on the viral DNA ends in Moloney murine leukemia virus pre-integration complexes reflect a specific association with integrase. Proc. Natl. Acad. Sci. USA 95:10535-10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worcel, A., and C. Benyajati. 1977. Higher order coiling of DNA in chromatin. Cell 12:83-100. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, H., G. Dornadula, and R. J. Pomerantz. 1996. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J. Virol. 70:2809-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, H., G. Dornadula, and R. J. Pomerantz. 1998. Natural endogenous reverse transcription of HIV-1. J. Reprod. Immunol. 41:255-260. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, H., G. Dornadula, J. Orenstein, and R. J. Pomerantz. 2000. Morphologic changes in human immunodeficiency virus type 1 virions secondary to intravirion reverse transcription. J. Hum. Virol. 3:165-172. [PubMed] [Google Scholar]
- 52.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]