Abstract
The protease (PR) from human immunodeficiency virus (HIV) is essential for viral replication: this aspartyl protease, active only as a dimer, is responsible for cleavage of the viral polyprotein precursors (Gag and Gag-Pol), to release the functional mature proteins. In this work, we have studied the structure-function relationships of the HIV PR by combining a genetic test to detect proteolytic activity in Escherichia coli and a bacterial two-hybrid assay to analyze PR dimerization. We showed that a drug-resistant PR variant isolated from a patient receiving highly active antiretroviral therapy is impaired in its dimerization capability and, as a consequence, is proteolytically inactive. We further showed that the polypeptide regions adjacent to the PR coding sequence in the Gag-Pol polyprotein precursor, and in particular, the transframe polypeptide (TF), located at the N terminus of PR, can facilitate the dimerization of this variant PR and restore its enzymatic activity. We propose that the TF protein could help to compensate for folding and/or dimerization defects in PR arising from certain mutations within the PR coding sequence and might therefore function to buffer genetic variations in PR.
Human immunodeficiency virus (HIV), the causative agent of AIDS, is a complex retrovirus. HIV proteins are translated as large precursor polyproteins, Gag and Gag-Pol, which are then processed to generate mature viral proteins that assemble in infectious particles. This processing is performed by a virally encoded aspartic protease (PR), HIV PR, which is excised autocatalytically from the Gag-Pol precursor (10, 29). Because of its importance in the viral cycle, HIV PR has been chosen as a primary target for antiviral drug design (28, 30).
The 99-residue-long HIV PR is active as a homodimer: the active site of the enzyme is formed at the dimer interface, with each monomer contributing a catalytically essential aspartic acid (Asp 25) (9, 15, 31). The activation of HIV PR, that is, the dimerization and autocatalytic release from the Gag-Pol, is a critical step in the viral cycle (16). Yet, the precise molecular mechanisms leading to PR activation are still unclear (17, 18). Folding and dimerization of the PR domain are required to form an active site that will then cleave the peptide bond flanking the N terminus of the mature PR (20). This initial cleavage is accompanied by a large increase in proteolytic activity of PR that can then process the Gag and Gag-Pol polyproteins at specific sites to release the mature viral proteins (19, 33).
The PR domain within the Gag-Pol precursor is flanked by the transframe polypeptide (TF) (also known as p6*) at its N terminus and by the reverse transcriptase (RT) at its C terminus. Several studies have highlighted the critical role of the transframe protein in the regulation of PR activity. First, it was shown that deletion of TF sequence improves the rate of autoprocessing of a Gag-Pol precursor, suggesting that TF could play the role of a proregion similar to zymogen (23). Although TF does not exhibit a stable secondary and/or tertiary structure (3), it was shown to destabilize the folded dimeric structure of PR when expressed as a TF PR precursor (1, 24, 36). In addition, it was suggested that TF could be a competitive inhibitor of mature PR activity: the free carboxy-terminal end of TF might block the substrate binding cleft of PR, to prevent nonspecific degradation of viral proteins within the released particles (24).
In this study, we analyzed the structure-function relationships of PR by combining two bacterial genetic tools, an Escherichia coli genetic assay for proteolytic activity and a bacterial two-hybrid (BACTH) system, both based on Bordetella pertussis adenylate cyclase (AC). We showed that a drug-resistant PR variant isolated from a patient receiving highly active antiretroviral therapy (HAART) is impaired in its dimerization capability and, as a consequence, is proteolytically inactive. We further showed that the polypeptide regions adjacent to the PR coding sequence in the Gag-Pol polyprotein precursor, and, in particular, the TF polypeptide, located at the N terminus of PR, can facilitate the dimerization of this variant PR and restore its enzymatic activity.
We propose that in vivo, TF could play a dual role: on the one hand, it could negatively regulate the proteolytic activity of wild-type PR to prevent early Gag-Pol precursor processing, and on the other hand, it might facilitate the folding and/or dimerization of certain drug-resistant PR mutants that could be intrinsically less stable than the wild-type PR (34).
MATERIALS AND METHODS
Strain and growth media.
DHT1 (F− glnV44[AS] recA1 endA1 gyrA96 [Nalr] thi1 hsdR17 spoT1 rfbD1 cya-854 ilv 691::Tn10) is an AC-deficient (cya) strain constructed by cotransduction of cya-854 and ilv 691::Tn10 mutations into DH1 (7). Transformation of DHT1 was performed by standard techniques (27). Growth medium used was the rich Luria-Bertani (LB) medium. Antibiotic concentrations were as follows: ampicillin (100 μg/ml) and kanamycin (50 μg/ml). Screening for the ability to ferment sugar was performed on MacConkey agar plates containing 1% maltose (21). Saquinavir mesylate (invirase [Roche] dissolved in ethanol at a concentration of 10 mM) was directly diluted into bacterial growth media at the indicated concentrations.
Plasmid construction.
All in vitro DNA manipulations were performed according to standard protocols (27) using E. coli XL1-Blue strain (Stratagene) as recipient cells. The plasmids used in this study are described in Tables 1 and 2. All the plasmid constructs were verified by sequencing. Plasmid pST18C, used in the two-hybrid assay, was constructed by hybridizing two oligonucleotides, ST1 (CTGTTGACAATTAATCATCGGCTCGTATAATGTACGCAGTTCTTCACACAGGAAACAGCTATGACCATGATTACGCCA) and ST2 (AGCTTGGCGTAATCAGTGTCATAGCTGTTTCCTGTGTGAAGAACTGCGTACATTATACGAGCCGATGATAATTGTCAACAG), and ligating them into pUT18C (14) linearized by PvuII and HindIII. Plasmids pKACPRD25N and pKACB3D25N express fusion proteins AC-N22-PRD25N-C29 and AC-N22-B3D25N-C29, respectively. The D25N mutation, which abolishes the catalytic activity of PR (16), was inserted using the PCR overlap extension method (2) using pKACPR-L and pKACB3-L as templates and primers P3, P4, P17 (GAAGCTCTATTAaATACAGGAGCAGATGATACAGTATTAG), and P18 (CATCTGCTCCTGTATtTAATAGAGCTTCCTTTAGTTGC). The same method was used to introduce the D25N mutation in pKACTFPR and pKACTFB3, except that P19 (ggggcggctagcTTTTTTAGGGAAGATCTGGC) was used instead of P3. Plasmids pKACTFPR and pKACTFB3 were constructed by amplifying DNA sequences from pHNI and B3 viral clone (7), respectively, using P19 and P4 as primers. The PCR products were then digested by NheI and KpnI and inserted into the corresponding sites of pKACp5.
TABLE 1.
Plasmids used in the E. coli assay for proteolytic activities
| Plasmid | Expressed protein | Description of plasmid constructiona
|
Reference | ||
|---|---|---|---|---|---|
| Template | Primer
|
||||
| Name | Sequence (5′→3′) | ||||
| pUCPR-Sb | N7-PR-C7 | pNHI | P1 | gcggtcgactcatatgGGAACTGTATCCTTTAAC | 7 |
| pUCB3-Sb | N7-B3-C7 | B3 viral DNA | P2 | cgcggatccAGTTTCAATAGGAC | 7 |
| pUCPR-Lb | N22-PR-C29 | pKACPR-L | P5 | ggggaagcttggctagcGGTAGAGACAACAAC | This work |
| pUCB3-Lb | N22-B3-C29 | pKACB3-L | P6 | gcgcccggatccccttacccTTCTTCTGTCAATGG | This work |
| pUCPR-L/Sb | N22-PR-C7 | pKACPR-L | P5 | ggggaagcttggctagcGGTAGAGACAACAAC | This work |
| pUCB3-L/Sb | N22-B3-C7 | pKACB3-L | P2 | cgcggatccAGTTTCAATAGGAC | This work |
| pUCPR-S/Lb | N7-PR-C29 | pKACPR-L | P1 | gcggtcgactcatatgGGAACTGTATCCTTTAAC | This work |
| pUCB3-S/Lb | N7-B3-C29 | pKACB3-L | P6 | gcgcccggatccccttacccTTCTTCTGTCAATGG | This work |
| pUC-TFPRb | TF-PR-C7 | pNHI | P16 | gcgcgcgcggtcgacgTTTTTTAGGGAAGATCTGGC | This work |
| pUC-TFB3b | TF-B3-C7 | B3 viral DNA | P2 | cgcggatccAGTTTCAATAGGAC | This work |
| pKACPR-Lc | AC-N22-PR-C29 | pNHI | P3 | ggggctagcGGTAGAGACAACAACTCC | 7 |
| pKACB3-Lc | AC-N22-B3-C29 | B3 viral DNA | P4 | cccggtaccTTCTTCTGTCAATGGCC | 7 |
| pKACPR-Sc | AC-N7-PR-C7 | pKACPR-L | P7 | ggggctagcGGAACTGTATCCTTT | This work |
| pKACB3-Sc | AC-N7-B3-C7 | pKACB3-L | P8 | cccggtaccAGTTTCAATAGGAC | This work |
| pKACPR-L/Sc | AC-N22-PR-C7 | pKACPR-L | P3 | ggggctagcGGTAGAGACAACAACTCC | This work |
| pKACB3-L/Sc | AC-N22-B3-C7 | pKACB3-L | P8 | cccggtaccAGTTTCAATAGGAC | This work |
| pKACPR-S/Lc | AC-N7-PR-C29 | pKACPR-L | P7 | ggggctagcGGAACTGTATCCTTT | This work |
| pKACB3-S/Lc | AC-N7-B3-C29 | pKACB3-L | P4 | cccggtaccTTCTTCTGTCAATGGCC | This work |
The plasmids were constructed by PCR amplification of DNA sequences using the indicated templates with corresponding primers (capital letters correspond to HIV Gag-Pol sequence, and underlined sequences correspond to the restriction sites used for subcloning of PCR products). The PCR products were then digested and inserted between the corresponding sites of pUC19 (26) or pKACp5 (7). The plasmids pUCPR-S, pUCB3-S, pKACPR-L, and pKACB3-L were previously named pUCHIV, pUCB3, pKACPR, and pKACB3, respectively, in the work of Dautin et al. (7).
Product inserted between corresponding sites of pUC19.
Product inserted between corresponding sites of pKACp5.
TABLE 2.
Plasmids used in two-hybrid assays
| Plasmid | Expressed protein | Description of plasmid constructiona
|
||
|---|---|---|---|---|
| Template | Primer
|
|||
| Name | Sequence (5′→3′) | |||
| pKT25PR-Sb | T25-N7-PRD25N-C7 | pKACPRD25N | P9 | gcgggatcctcatatgGGAACTGTATCCTTTAAC |
| pKT25B3-Sb | T25-N7-B3D25N-C7 | pKACB3D25N | P10 | cgcggtaccAGTTTCAATAGGAC |
| pKT25PR-Lb | T25-N22-PRD25N-C29 | pKACPRD25N | P11 | gcgggatcctcatatgGGTAGAGACAACAA |
| pKT25B3-Lb | T25-N22-B3D25N-C29 | pKACB3D25N | P12 | cgcggtaccTTCTTCTGTCAATGGCCATTG |
| pKT25PR-L/Sb | T25-N22-PRD25N-C7 | pKACPRD25N | P11 | gcgggtacctcatatgGGTAGAGACAACAA |
| pKT25B3-L/Sb | T25-N22-B3D25N-C7 | pKACB3D25N | P10 | cgcggtaccAGTTTCAATAGGAC |
| pKT25TFPRb | T25-TF-PRD25N-C7 | pKACTFPRD25N | P15 | ggggatccgTTTTTTAGGGAAGATC |
| pKT25TFB3b | T25-TF-B3D25N-C7 | pKACTFB3D25N | P10 | cgcggtaccAGTTTCAATAGGAC |
| pST18CPR-Sc | T18-N7-PRD25N-C7 | pKACPRD25N | P1 | gcggtcgactcatatgGGAACTGTATCCTTTAAC |
| pST18CB3-Sc | T18-N7-B3D25N-C7 | pKACB3D25N | P2 | cgcggatccAGTTTCAATAGGAC |
| pST18CPR-Lc | T18-N22-PRD25N-C29 | pKACPRD25N | P13 | gcggtcgactcatatgGGTAGAGACAACAAC |
| pST18CB3-Lc | T18-N22-B3D25N-C29 | pKACB3D25N | P6 | gcgcccggatccccttacccTTCTTCTGTCAATGG |
| pST18CPR-L/Sc | T18-N22-PRD25N-C7 | pKACPRD25N | P13 | gcggtcgactcatatgGGTAGAGACAACAAC |
| pST18CB3-L/Sc | T18-N22-B3D25N-C7 | pKACB3D25N | P2 | cgcggatccAGTTTCAATAGGAC |
| pST18CTFPRc | T18-TF-PRD25N-C7 | pKACTFPRD25N | P16 | gggtcgacgTTTTTTAGGGAAGAT |
| pST18CTFB3c | T18-TF-B3D25N-C7 | pKACTFB3D25N | P2 | cgcggatccAGTTTCAATAGGAC |
The plasmids were constructed by PCR amplification of DNA sequences using the indicated templates with corresponding primers (capital letters correspond to HIV Gag-Pol sequence, and underlined sequences correspond to the restriction sites used for subcloning of PCR products). The PCR products were then digested and inserted between the corresponding sites of pKT25 (7) or pST18C. Plasmid pST18C expresses the T18 fragment of AC (aa 225 to 400) under the control of a constitutive promoter (see Materials and Methods).
Product inserted between corresponding sites of pKT25.
Product inserted between corresponding sites of pST18C.
Analytical methods.
β-Galactosidase assays were performed on toluene-treated bacterial suspensions, as described by Pardee et al. (22). One unit of activity corresponds to 1 nmol of o-nitrophenyl β-d-galactoside hydrolyzed per min at 28°C. Cyclic AMP (cAMP) measurements were done by an enzyme-linked immunosorbent assay, as described previously (13). The data presented here represent the average values obtained for at least three independent cultures.
Western blot analysis.
Western blot analysis was performed on whole-cell bacteria from overnight cultures. The cell suspensions were precipitated by trichloroacetic acid (10% final concentration, kept for 10 min at 4°C). After centrifugation (10 min, 11,200 × g), pellets were resuspended in 8 M urea-20 mM HEPES (pH 7.5), neutralized with 1 M Tris-base (pH 8.8), dissolved in sodium dodecyl sulfate-gel loading buffer and heated for 5 min at 100°C. Proteins were then separated by migration on a sodium dodecyl sulfate-15% polyacrylamide gel, and electrotransferred on nitrocellulose membrane (27). After saturation of the membrane with milk, AC polypeptides were detected with the mouse monoclonal anti-AC antibody 5G12 (kindly provided by Mohammed El-Azami El-Idrissi, Institut Pasteur) according to standard protocols (27).
RESULTS
Assay of HIV PR activity in E. coli.
We have previously designed a genetic test that permits an easy in vivo assay of the proteolytic activity of HIV PR in E. coli (7). It is based on the specific proteolysis of a signaling enzyme, the B. pertussis AC, modified by an in-frame insertion of a PR processing site, p5 (TVSFNFPQITLW). The resulting ACp5 protein retains enzymatic activity and confers a Cya+ phenotype to an E. coli cya strain (i.e., lacking endogenous AC). When the PR was coexpressed in the same cells, it cleaved and inactivated ACp5, and the host cells became Cya−. Cya− and Cya+ cells can be easily distinguished on appropriate indicator media, like MacConkey or LB-X-Gal. Addition of a specific HIV PR inhibitor (saquinavir) blocked the PR-mediated inactivation of ACp5 and thus restored a Cya+ phenotype of the host cells. With this test (the trans assay), we were able to reveal the proteolytic activity of different drug-resistant variants of PR isolated from patients receiving HAART, with the exception of a an indinavir-resistant variant, called B3 (with mutations: M46I, V77I, V82T), found to be inactive, i.e., unable to cleave the ACp5 molecule.
Yet, the B3 variant was proteolytically active when tested in a different setup of the AC-based genetic assay (the cis assay), in which the full PR protein surrounded by its adjacent processing sites was inserted in frame into the AC polypeptide. The resulting AC-PR fusion protein, when expressed in E. coli cya, underwent an autoproteolytic cleavage, thus inactivating the AC enzyme (host cells are Cya−). Addition of HIV PR-specific inhibitor (saquinavir) prevented autoproteolysis and, therefore, inactivation of AC: host cells exhibited a Cya+ phenotype.
Initially, we hypothesized that, in the case of B3 variant, the discrepancy between the results obtained with the two different set ups of the system, might reflect an intrinsic property of this drug-resistant variant to perform differently cis and trans proteolytic cleavages. Actually, as shown in Fig. 1, the HIV PRs (wild type or B3 variant) used in cis and trans proteolysis were not identical at their flanking sequences. The PR precursor expressed in the trans system contained short flanking sequences of 7 residues at both N and C termini (N7 and C7), surrounding the mature PR in the Gag-Pol precursor. In contrast, the PR precursor inserted within AC in the cis design contained extended flanking sequences of 22 and 29 residues at the N and C termini, respectively (N22 and C29). We have therefore examined whether such variations in flanking sequences could explain the differences in the proteolytic activity of the B3 variant observed between the cis and the trans genetic assays.
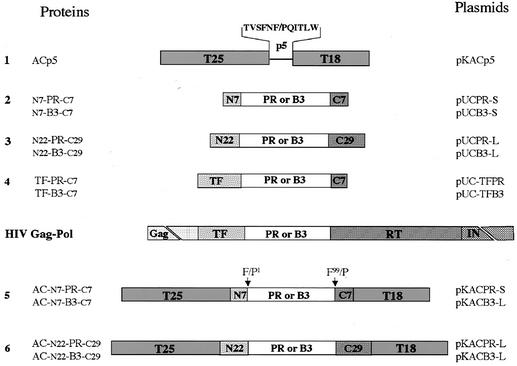
FIG. 1.
Schematic representation of AC proteins and HIV PRs used in this work. (Panel 1) In ACp5 protein, a specific proteolytic site of HIV PR, named p5 (amino acid sequence in one-letter code), was inserted in frame between the two complementary fragments of the catalytic domain of B. pertussis AC, T25 (residues 1 to 224) and T18 (residues 225 to 400), represented as grey rectangles. (Panels 2 to 4) Wild-type HIV PR and/or B3 variant (mature polypeptide of 99) are represented as white rectangles. The dotted rectangles (N7, N22, and TF) represent the various flanking segments deriving from the TF protein coding regions of 7, 22, and 68 residues (full TF protein), respectively. The hatched rectangles represent the RT-derived polypeptides (C7 or C29). HIV Gag-Pol represents the schematic organization of the HIV polyprotein precursor Gag-Pol. (Panels 5 and 6) Wild type or B3 variant of HIV PR, flanked by short (N7/C7) or long (N22/C29) flanking sequences, were inserted in frame between T25 and T18 fragments of AC.
PR flanking sequences are essential for the proteolytic activity of the B3 variant.
To test this hypothesis, we constructed a vector, pUCB3-L (Fig. 1), that expressed B3 PR flanked by the extended sequences N22 and C29 (N22-B3-C29). In fact, the original patient's blood sample (B3) was found to contain a mixture of two PR variants, a triple mutant (M46I/V77I/V82T), and a double mutant V77I/V82T. These two variants exhibited the same phenotype in our bacterial assays (i.e., inactive in trans and active in cis, data not shown). Thus, to simplify B3 structure-function analysis, all experiments were performed using the B3 double mutant rather than with the original triple mutant.
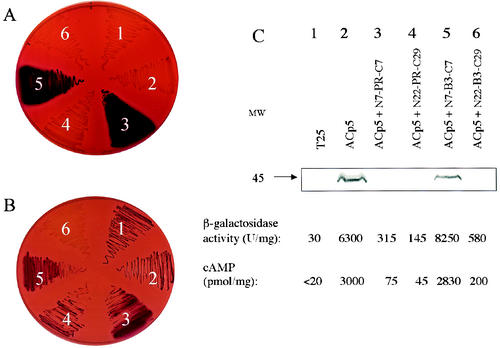
When the pUCB3-L vector was cotransformed with plasmid pKACp5, which expresses ACp5, in the E. coli cya strain DHT1, the resulting transformants displayed a Cya− phenotype (Fig. 2A, panel 4). Low levels of β-galactosidase activity and cAMP were measured in these cells (Fig. 2C, lane 6), compared to cells cotransformed with pKACp5 and pUCB3-S (i.e., coexpression of ACp5 and N7-B3-C7) (Fig. 2C, lane 5), indicating that B3, when fused to extended sequences N22 and C29, could cleave the ACp5 molecule. In the presence of the HIV PR inhibitor saquinavir (100 μM), ACp5 cleavage was prevented and cells were Cya+ (Fig. 2B, panel 4). Western blot analysis confirmed that ACp5 was not cleaved by N7-B3-C7 but was fully degraded by N22-B3-C29 (Fig. 2C, lanes 5 and 6). Conversely, when B3 surrounded by short flanking sequences (N7 and C7) was inserted in frame into AC, the resulting fusion protein was unable to autoproteolyze. DHT1 cells expressing the corresponding AC-N7-B3-C7 hybrid protein (Fig. 1), exhibited a Cya+ phenotype (red colonies on MacConkey-maltose, Fig. 3A, panel 4), expressed β-galactosidase and produced cAMP (Fig. 3C, lane 9). Western blot analysis indicated that this AC-N7-B3-C7 fusion protein was not autoproteolyzed, in contrast to AC-N22-B3-C29 hybrid which contained B3 surrounded by long flanking sequences (Fig. 3C, lanes 7 and 9). Altogether, these results show that the extended N22 and C29 sequences are necessary for the proteolytic activity of B3 in both cis and trans assays.
FIG. 2.
Assays of HIV PR and B3 proteolytic activities in E. coli. (A and B) DHT1 cells were cotransformed with plasmids expressing ACp5 and N7-PR-C7 (section 1), ACp5 and N22-PR-C29 (section 2), ACp5 and N7-B3-C7 (section 3), or ACp5 and N22-B3-C29 (section 4) or with empty vector (pUC19) and plasmids expressing ACp5 (section 5) or T25 (section 6). Cotransformants were plated on MacConkey-maltose plus kanamycin and ampicillin (A) or on the same medium supplemented with 100 μM saquinavir (B) and grown for 48 h at 30°C. (C) Recombinant ACp5 (apparent molecular mass of 45 kDa), in DHT1 cells expressing the indicated proteins, was detected by Western blot analysis as described in Materials and Methods. β-Galactosidase activities and cAMP levels in the corresponding samples were measured as described in Materials and Methods.
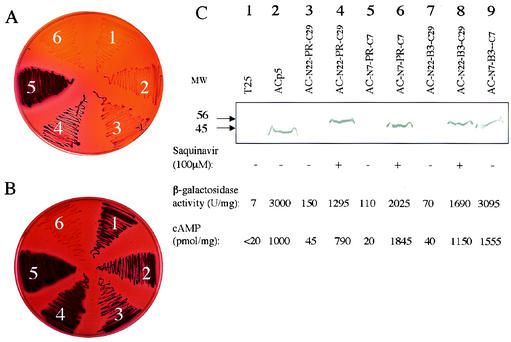
FIG. 3.
Assay of HIV PR and B3 variant autoproteolysis in E. coli. (A and B) DHT1 cells were transformed with plasmids expressing AC-N22-PR-C29 (section 1), AC-N7-PR-C7 (section 2), AC-N22-B3-C29 (section 3), AC-N7-B3-C7 (section 4), ACp5 (section 5), or T25 (section 6) and plated on MacConkey-maltose plus kanamycin (A) or on the same medium supplemented with 100 μM saquinavir (B) and grown for 48 h at 30°C. (C) Recombinant ACs (ACp5, apparent molecular mass of 45 kDa; ACPR or ACB3, apparent molecular mass of 56 kDa), in DHT1 cells expressing the indicated proteins, were detected by Western blot analysis as described in Materials and Methods. β-Galactosidase activities and cAMP levels in the corresponding samples were measured as described in Materials and Methods.
In contrast, both long (N22-PR-C29) and short (N7-PR-C7) forms of the wild-type HIV PR were proteolytically active (Fig. 2 and 3): DHT1 cells coexpressing ACp5 and N7-PR-C7 or ACp5 and N22-PR-C29 exhibited a Cya− phenotype (Fig. 2A, panels 1 and 2), indicating that ACp5 was cleaved and inactivated by both forms of HIV PR. A similar Cya− phenotype was observed when DHT1 cells expressed AC hybrid proteins, AC-N22-PR-C29 or AC-N7-PR-C7, indicating that both AC-PR fusion proteins were autoproteolyzed and inactivated (Fig. 3A, panels 1 and 2). Western blot analysis confirmed that in all cases recombinant ACs were inactivated by wild-type PR (Fig. 2C and 3C).
To further determine whether both flanking sequences N22 and C29 were equally essential for the B3 PR activity, we tested in E. coli hybrid PRs with only one of these two polypeptide extensions. As shown in Fig. 4 (lane 2), N22-B3-C7 was proteolytically active against ACp5, whereas N7-B3-C29 exhibited a lower proteolytic activity (Fig. 4, lane 3), suggesting that the N-terminal flanking sequence was important for B3 activity. As expected, both N22-PR-C7 and N7-PR-C29 were able to inactivate ACp5, in agreement with the above observations (Fig. 4, lanes 5 and 6). Finally, both wild-type and B3 PRs with either N or C-terminal extensions were also tested for autoproteolytic activity, i.e., after insertion into AC. As shown in Fig. 4, lanes 8 and 9, when expressed in DHT1, both AC-N22-B3-C7 and AC-N7-B3-C29, were autoproteolytically cleaved, as revealed by the Cya− phenotype of transformed cells. This suggests that, in the cis configuration, the C-terminal flanking sequence (C29) might also enhance the autoproteolytic activity of the B3 variant. Similar constructs with wild-type HIV PR, AC-N22-PR-C7 and AC-N7-PR-C29, were both able to autoproteolyze (Fig. 4, lanes 11 and 12).
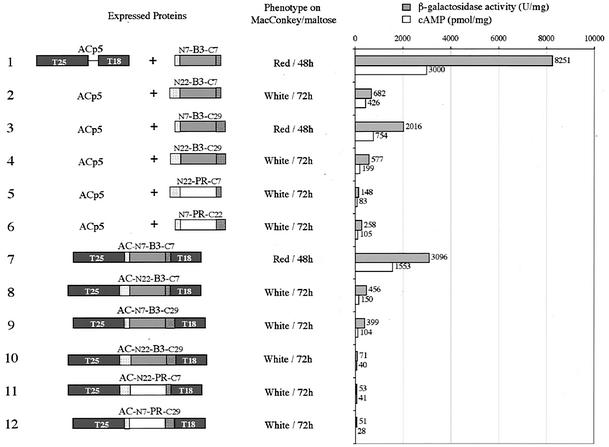
FIG. 4.
Role of flanking regions on HIV PR and B3 proteolytic activity in E. coli. The AC molecules and HIV (or B3) PRs are schematized as in Fig. 1. The phenotype of DHT1 cells expressing the indicated proteins was scored on MacConkey-maltose plates plus kanamycin and ampicillin (lanes 1 to 6) or plus kanamycin (lanes 7 to 12). β-Galactosidase activities and cAMP levels were measured on liquid cultures grown overnight at 30°C in LB supplemented with appropriate antibiotics, as described in Materials and Methods. The plasmids expressing the indicated hybrid proteins were as follows: lane 1, pKACp5 + pUCB3-S; lane 2, pKACp5 + pUCB3-L/S; lane 3, pKACp5 + pUCB3-S/L; lane 4, pKACp5 + pUCB3-L; lane 5, pKACp5 + pUCHIV-L/S; lane 6, pKACp5 + pUCHIV-S/L; lane 7, pKACB3-S; lane 8, pKACB3-L/S; lane 9, pKACB3-S/L; lane 10, pKACB3-L; lane 11, pKACPR-L/S; 12, pKACPR-S/L.
We conclude that the sequence of 22 residues upstream from the mature PR, N22, which encompasses the C-terminal part of the TF, is essential to confer proteolytic activity to B3 PR variant. Besides, the C-terminal extension (C29) might also contribute to some extent to B3 activity.
HIV PR B3 variant with short flanking sequences is unable to dimerize in E. coli.
To further define the role of the flanking sequences on B3 activity, we examined if these segments might modulate B3 dimerization.
Indeed, it has been shown previously that some drug-resistant mutations can drastically affect the dimerization ability of the HIV PR (34). In particular, Xie et al. (34) showed that HIV PR mutants V82F, V82F/I84V, V82T/I84V and L90M, exhibited reduced dimer stability compared to the wild-type enzyme. As the B3 variant carries, among other mutations, a V82T change, we hypothesized that this variant could have a lower dimer stability and thus a decreased proteolytic activity.
To check this hypothesis, we used a BACTH system that permits the detection of protein-protein interactions in E. coli. This system, which has been used successfully to analyze interactions between a large variety of polypeptides (8), is based on the functional complementation between two complementary fragments of B. pertussis AC (designated T25 and T18, encompassing residues 1 to 224 and 225 to 400 of AC, respectively) (13). When the fragments are coexpressed in E. coli cya cells as separated polypeptides, they are not able to associate and cannot reconstitute a functional AC enzyme. When these fragments are fused to two interacting proteins, their association results in the reconstitution of AC enzyme that can complement the cya defect of the E. coli host cells (cells become Cya+). Interaction between hybrid proteins can be monitored easily on indicator plates and quantified by measuring cAMP levels and/or β-galactosidase activity (13, 14).
To test whether the BACTH system could reveal HIV PR homodimerization in E. coli, the PR coding region with its short flanking segments (i.e., N7-PR-C7) was fused in frame to the C termini of T25 and T18 fragments (expressed by two compatible plasmids, pKT25 and pST18C). To avoid the autoproteolytic cleavage of the hybrid proteins, the essential Asp residue at position 25 of the HIV PR was converted to an Asn residue in all two-hybrid constructions.
When the fused proteins, T25-N7-PRD25N-C7 and T18-N7-PRD25N-C7, were expressed separately in DHT1 cells (data not shown) or when T25-N7-PRD25N-C7 was coexpressed with T18 (Fig. 5, lane 2), transformants exhibited a Cya− phenotype (i.e., white colonies on MacConkey-maltose medium and background levels of cAMP and β-galactosidase activities). When T25-N7-PRD25N-C7 and T18-N7-PRD25N-C7 were coexpressed in DHT1, transformants exhibited a Cya+ phenotype (i.e., red colonies on MacConkey-maltose plates), indicating that the two hybrid proteins interacted. Measurements of cAMP levels and β-galactosidase activities in liquid cultures confirmed that functional complementation occurred between these two hybrid proteins (Fig. 5, lane 4). These data indicate that the BACTH system is suitable to reveal the HIV PR dimerization in E. coli. Next, the dimerization of the B3 variant was tested. As shown in Fig. 5 lane 8, DHT1 cells coexpressing the T25-N7-B3D25N-C7 and T18-N7-B3D25N-C7 hybrid proteins exhibited a Cya− phenotype and expressed only background levels of β-galactosidase or cAMP. This shows that the B3 PR with its short flanking sequences cannot efficiently dimerize in E. coli.
FIG. 5.
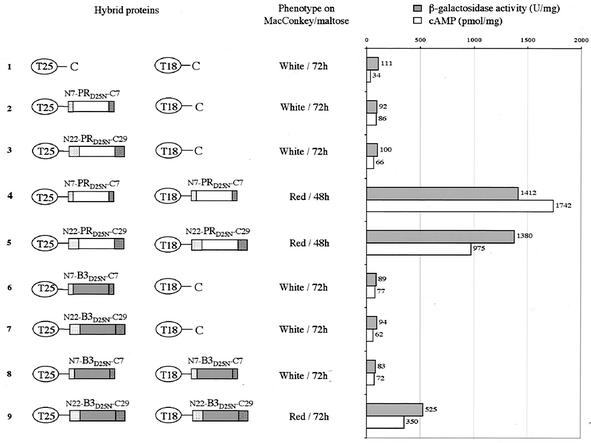
BACTH assay of HIV PR dimerization. The two AC fragments T25 and T18 are schematized by ovals, and the PR and B3 polypeptides are represented by white and grey rectangles, respectively. The N terminus HIV PR flanking sequences (TF derivatives: N7 and N22) are represented as dotted rectangles, whereas C terminus flanking sequences (RT derivatives: C7 and C29) are represented as hatched rectangles. The phenotype of DHT1 cells expressing the indicated pairs of proteins was scored on MacConkey-maltose plates plus kanamycin and ampicillin and supplemented with 0.5 mM isopropyl β-d-thiogalactoside (IPTG). β-Galactosidase activities and cAMP levels were measured on liquid cultures grown overnight at 30°C in LB supplemented with appropriate antibiotics and 0.5 mM IPTG. The plasmids expressing the indicated hybrid proteins were as follows: lane 1, pKT25 + pST18C; lane 2, pKT25HIV-S + pST18C; lane 3, pKT25PR-L + pST18C; lane 4, pKT25PR-S + pST18CPR-S; lane 5, pKT25PR-L + pST18CPR-L; lane 6, pKT25B3-S + pST18C; lane 7, pKT25B3-L + pST18C; lane 8, pKT25B3-S + pST18CB3-S; lane 9, pKT25B3-L + pST18CB3-L.
Extended flanking sequences facilitate B3 variant dimerization in E. coli.
The inability of N7-B3-C7 to dimerize in vivo might, therefore, explain its lack of proteolytic activity and suggests that the extended sequences N22 and C29 could restore B3 activity by promoting its dimerization in E. coli. To test this hypothesis, N22-B3-C29 was fused to the C termini of T25 and T18 and tested in two-hybrid assays. As shown in Fig. 5, lane 9, DHT1 cells coexpressing T25-N22-B3D25N-C29 and T18-N22-B3D25N-C29 synthesized cAMP, expressed β-galactosidase, and exhibited a red phenotype on MacConkey-maltose. This indicates that these two hybrid proteins are able to interact. Hence, N22-B3-C29 is able to dimerize in vivo, albeit less efficiently than wild-type PR. We can thus conclude that the extended sequences N22 and C29 can facilitate the dimerization of B3 PR and therefore are essential for B3 proteolytic activity in E. coli. These extended sequences have only a marginal effect on the dimerization of the wild-type HIV PR (compare Fig. 5, lanes 4 and 5), in good agreement with the results showing that they have no influence on its proteolytic activity.
To determine the relative contribution of the N22 and C29 flanking sequences on B3 PR dimerization, hybrid proteins with either one of these two polypeptide extensions were tested in the BACTH system. As shown in Fig. 6, lane 4, the N22-B3-C7 protein was able to dimerize efficiently whereas the dimerization of the N7-B3-C29 hybrid was weaker (Fig. 6, lane 5). The lesser effect of C29 extension on B3 dimerization could explain the reduced activity of N7-B3-C29 in the trans proteolytic assays (Fig. 4). Assays performed with the wild-type PR showed that both N22-PR-C7 and N7-PR-C29 were able to dimerize, as expected (Fig. 6, lanes 8 and 9).
FIG. 6.
Assay of HIV PR and B3 variant dimerization in E. coli. The T25 and T18 fragments and the HIV PRs are represented as in Fig. 4. The phenotype of DHT1 cells expressing indicated pairs of proteins was scored as described in Fig. 4 and β-galactosidase activities were measured as indicated in Fig. 4. The plasmids expressing the indicated hybrid proteins were as follows: lane 1, pKT25 + pST18C; lane 2, pKT25B3-L/S + pST18C; lane 3, pKT25B3-S/L + pST18C; lane 4, pKT25B3-L/S + pST18CB3-L/S; lane 5, pKT25B3-S/L + pST18CB3-S/L; lane 6, pKT25PR-L/S + pST18C; lane 7, pKT25PR-S/L + pST18C; lane 8, pKT25PR-L/S + pST18CPR-L/S; and lane 9, pKT25PR-S/L + pST18CPR-S/L.
Taken together, results from the two-hybrid assays indicate that the polypeptide sequence flanking the N terminus of mature PR is critical for the dimerization and subsequent proteolytic activity of the HIV PR B3 variant.
The full TF protein can restore the proteolytic activity to the drug-resistant B3 variant.
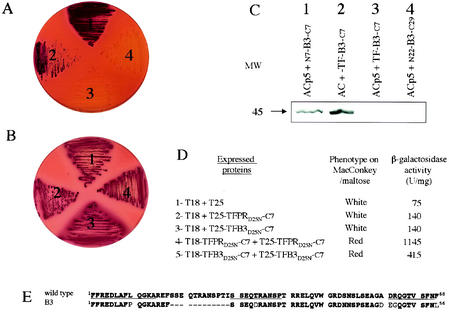
We examined whether the full TF could also favor the dimerization of B3 PR and therefore stimulate its enzymatic activity. As shown in Fig. 7A, panel 3, when a TF-B3-C7 fusion protein was coexpressed with ACp5 in DHT1, cells exhibited a Cya− phenotype, indicating that TF-B3-C7 was proteolytically active in vivo and able to cleave and inactivate the ACp5 target enzyme. Western blot analysis of cell extracts confirmed that ACp5 was specifically cleaved at the inserted p5 sequence by TF-B3-C7 (Fig. 7C, lane 3). Indeed, the wild-type AC, i.e., lacking the p5 site, was not cleaved by TF-B3-C7 (Fig. 7C, lane 2): DHT1 cells coexpressing AC and TF-B3-C7 were Cya+ (Fig. 7A, panel 2). The B3 PR with short flanking sequences (N7-B3-C7) was unable to degrade ACp5 (Fig. 7A, panel 1 and Fig. 7C, lane 1) as shown previously (Fig. 2A).
FIG. 7.
The full TF protein restores proteolytic activity of B3 variant. (A and B) DHT1 cells were cotransformed with plasmids expressing ACp5 and N7-B3-C7 (section 1), AC and TF-B3-C7 (section 2), ACp5 and TF-B3-C7 (section 3), and ACp5 and N22-B3-C29 (section 4); plated on MacConkey-maltose plus kanamycin and ampicillin, without (A) or with (B) 100 μM saquinavir; and grown 48 h at 30°C. (C) Recombinant ACp5 (apparent molecular mass of 45 kDa), in DHT1 cells coexpressing the indicated proteins, were detected by Western blot analysis as described in Materials and Methods. (D) The phenotype and β-galactosidase activities of DHT1 cells expressing the indicated pairs of proteins were determined as indicated in Fig. 4. The plasmids expressing the indicated hybrid proteins were as follows: lane 1, pKT25 + pST18C; lane 2, pST18C + pKT25-TFPR; lane 3, pST18C + pKT25-TFB3; lane 4, pKT25-TFPR + pST18C-TFPR; lane 5, pKT25-TFB3 + pST18C-TFB3. (E) Amino acid sequences of TF protein isolated from B3 variant and wild-type clones. The sequences underlined correspond to the consensus sequences defined by Beissinger et al. (3).
In addition, when TF-B3D25N-C7 was fused to T25 and T18 and tested in the BACTH system, complementation occurred: cells were red on MacConkey-maltose and expressed β-galactosidase (Fig. 7D, lane 5). These results demonstrate that the TF, when fused to B3, restores its dimerization and its proteolytic activity.
When wild-type PR was fused to TF, the obtained fusion protein (TF-PR-C7) was able to dimerize efficiently in E. coli (Fig. 7D, lane 4) and was proteolytically active against ACp5 (data not shown), indicating that, despite the presence of TF sequence, the wild-type PR was able to mature to an active form.
Interestingly, we noted that the TF protein isolated from the B3 HIV clone contained amino acid substitutions and a deletion of 11 residues (from 17 to 28) compared to the wild-type TF sequence (Fig. 7E). The high variability of TF sequence has already been reported (3, 4), although the influence of these variations on HIV PR activity has not been examined.
DISCUSSION
Formation of the PR dimer is a prerequisite for PR activity and represents a key step in HIV polyprotein maturation. Although numerous studies on wild-type PR have revealed a low dimer dissociation constant, with equilibrium constant (Keq) values below nanomolar levels (5, 6, 11, 12, 35), Xie and collaborators reported recently a weaker PR dimer stability at neutral pH, with Keq values in the micromolar range (34). Moreover, they showed that several drug-resistant PR mutants exhibited a reduced dimer stability compared to wild-type enzyme. In particular, PR variants with mutations at position 82 and/or 84 had equilibrium dissociation constants up to 20-fold higher than those for the wild type. Our present results are in good agreement with Xie et al.'s in vitro data. In the BACTH system, the wild-type PR was shown to dimerize efficiently in the cytosol of E. coli, whereas an indinavir-resistant PR variant, B3, which harbors a double mutation, V77I/V82T, was unable to form dimers. Assays of PR proteolytic activity in an E. coli genetic test based on the site specific cleavage and inactivation of ACp5 (an AC modified by insertion of a PR processing site; p5), confirmed that the dimerization defective B3 variant was inactive. The defect in dimerization and proteolytic activity of B3 variant seems to be mainly due to the V82T change, as another PR variant with a V77I single mutation was shown to be proteolytically active (variant B1 in reference 7).
More interestingly, we showed that the polypeptide regions adjacent to the PR coding sequence in the Gag-Pol polyprotein precursor, and in particular, the TF, located at the N terminus of PR, can facilitate the dimerization of this mutant PR and restore its proteolytic activity. When B3 was expressed as a fusion with the last 22 amino acids of TF, it was able to dimerize as revealed by the BACTH assay and was proteolytically active toward the ACp5 target in E. coli. Similarly B3, when expressed as a fusion with the entire TF, was able to dimerize and was active in the bacterial PR assay. Yet, the TF protein had little effect on the dimerization and proteolytic activity of wild-type PR.
Recently, Pettit and coauthors reported that PR variants carrying mutations within the dimer interface could be compensated to some extent when expressed as part of the Gag-Pol precursor (25). The authors suggested that extra-PR sequences could facilitate the dimerization of the defective PR variants. Although, the precise location of these regions was not reported, our present findings on the effect of the PR flanking regions are in good line with this hypothesis. Beside, various proteins encoded by Gag-Pol were shown to multimerize and might contribute also to the overall dimerization of the PR polypeptide.
Our present findings on the effect of the PR flanking regions are in good support of this hypothesis.
How could the flanking sequences of PR assist the dimerization of PR variants? One hypothesis is that these amino acid sequences can establish transient contacts with the PR polypeptide that could stabilize the dimer during the autoproteolytic reaction. A key structural element that maintains the dimeric state of PR consists of a four-stranded β-sheet made of the intertwined N- and C-terminal extremities of the mature PR polypeptide (9). During the autoproteolytic cleavage of PR, the N-terminal β-strand must transiently unfold from the β-sheet structure to reach the catalytic site where it will be processed. This intermediate, with only three β-strands, should be intrinsically less stable than the mature PR dimer. In the case of PR variants that have destabilizing amino acid substitutions, the maintenance of such an active dimeric intermediate during the autoproteolytic cleavage might be compromised. Additional contacts established between the TF sequence and PR protein might help to stabilize this transient intermediate. However, although the autoproteolytic release of PR from TF is assumed to be a triggering event in HIV polyprotein processing, the molecular mechanisms leading to PR activation, including the contribution of the TF protein, remain to be precisely determined.
Earlier studies have suggested that the TF may function as a negative regulator for PR folding and dimerization (19, 20, 23, 32). Louis et al. (19) proposed that TF might function as a proregion of PR that could destabilize the folded dimeric structure in the polyprotein by interacting with the nascent dimer interface and would thus prevent activation of the proteolytic activity until assembly of viral particle is completed. Our present results expand the potential role of TF in the regulation of PR activity. They suggest that TF could also help to compensate for defects in PR folding and dimerization arising from amino acid substitutions in the enzyme. In other words, the TF might function to “buffer” genetic variations in PR in a manner similar to the heat shock protein Hsp90, which chaperones the maturation of many regulatory proteins and was shown to buffer genetic variation in morphogenetic pathways (26). By contributing to the tolerance of PR to certain amino acid substitutions, the TF could allow the expansion of the genetic variability of this key HIV enzyme and, in particular, could permit the emergence of certain drug-resistant mutations.
Acknowledgments
We are grateful to Mohammed El-Azami El-Idrissi and Claude Leclerc for providing monoclonal antibody AC 5G12 and to Agnes Ullmann for critical reading of the manuscript.
Financial support came from the Institut Pasteur and the Centre National de la Recherche Scientifique (CNRS, URA 2185, Biologie Structurale et Agents Infectieux). N.D. was supported by fellowships from the Ministère de l'Education Nationale, de la Recherche et de la Technologie, and the Agence Nationale de Recherche sur le Sida (ANRS).
REFERENCES
- 1.Almog, N., R. Roller, G. Arad, L. Passi-Even, M. A. Wainberg, and M. Kotler. 1996. A p6POL-protease fusion is present in mature particles of human immunodeficiency virus type 1. J. Virol. 70:7228-7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaldi, M., M. Lepelletier, and V. Mejean. 1996. Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal. Biochem. 234:110-111. [DOI] [PubMed] [Google Scholar]
- 3.Beissinger, M., C. Paulus, P. Bayer, H. Wolf, P. Rösch, and R. Wagner. 1996. Sequence-specific resonance assignments of the 1H-NMR spectra and structurale characterization in solution of the HIV-1 transframe protein p6*. Eur. J. Biochem. 237:383-392. [DOI] [PubMed] [Google Scholar]
- 4.Candotti, D., C. Chappey, M. Rosenheim, P. M'Pele, J. M. Huraux, and H. Agut. 1994. High variability of the gag/pol transframe region among HIV-1 isolates. C. R. Acad. Sci. Ser. III 317:183-189. [PubMed] [Google Scholar]
- 5.Cheng, Y. S. E., F. H. Yin, S. Foundling, D. Blomstrom, and C. A. Kettner. 1990. Stability and activity of human immunodeficiency virus protease: Comparison of the natural dimer with a homologous, single-chain tethered dimer. Proc. Natl. Acad. Sci. USA 87:9660-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darke, P. L., S. P. Jordan, D. L. Hall, J. A. Zugay, J. A. Shafer, and L. C. Kuo. 1994. Dissociation and association of the HIV-1 protease dimer subunits: equilibria and rates. Biochemistry 33:98-105. [DOI] [PubMed] [Google Scholar]
- 7.Dautin, N., G. Karimova, A. Ullmann, and D. Ladant. 2000. A sensitive genetic screen for protease activity based on a cyclic AMP signaling cascade in Escherichia coli. J. Bacteriol. 182:7060-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dautin, N., G. Karimova, and D. Ladant. 2002. Bordetella pertussis adenylate cyclase toxin: a versatile screening tool. Toxicon 40:1383-1387. [DOI] [PubMed] [Google Scholar]
- 9.Erickson, J., D. J. Neidhart, J. VanDrie, D. J. Kempf, X. C. Wang, D. W. Norbeck, J. J. Plattner, J. W. Rittenhouse, et al. 1990. Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease. Science 249:527-533. [DOI] [PubMed] [Google Scholar]
- 10.Frankel, A. D., and J. A. Yong. 1998. HIV-1: fifteen proteins and a RNA. Annu. Rev. Biochem. 67:1-25. [DOI] [PubMed] [Google Scholar]
- 11.Grant, S. K., I. C. Deckman, J. S. Culp, M. D. Minnich, I. S. Brooks, P. Hensley, C. Debouck, and T. D. Meek. 1992. Use of protein unfolding studies to determine the conformational and dimeric stabilities of HIV-1 and SIV proteases. Biochemistry 31:9491-9501. [DOI] [PubMed] [Google Scholar]
- 12.Jordan, S. P., J. Zugay, P. L. Darke, and L. C. Kuo. 1992. Activity of human immunodeficiency virus protease as a function of solvent composition and enzyme concentration. J. Biol. Chem. 267:20028-20032. [PubMed] [Google Scholar]
- 13.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3:73-82. [PubMed] [Google Scholar]
- 15.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 16.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotler, M., G. Arad, and S. Hughes. 1992. Human immunodeficiency virus type 1 Gag-protease fusion proteins are enzymatically active. J. Virol. 66:6781-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis, J. M., N. T. Nashed, K. D. Parris, A. R. Kimmel, and D. M. Jerina. 1994. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the Gag-Pol polyprotein. Proc. Natl. Acad. Sci. USA 91:7970-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis, J. M., C. M. Clore, and A. M. Gronenborn. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 6:868-875. [DOI] [PubMed] [Google Scholar]
- 20.Louis, J. M., E. M. Wondrak, A. R. Kimmel, P. T. Wingfield, and N. T. Nashed. 1999. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 274:23437-23442. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic expression of inducibility in the synthesis of β-galactosidase of Escherichia coli. J. Mol. Biol. 1:165-168. [Google Scholar]
- 23.Partin, K., G. Zybarth, L. Ehrlich, M. Decrombrugghe, E. Wimmer, and C. Carter. 1991. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 88:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulus, C., S. Hellebrand, U. Tessmer, H. Wolf, H. G. Kräuslich, and R. Wagner. 1999. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J. Biol. Chem. 274:21539-21543. [DOI] [PubMed] [Google Scholar]
- 25.Pettit, S. C., S. Gulnik, L. Everitt, and A. H. Kaplan. 2003. The dimer interfaces of protease domains influence the activation of protease and the specificity of Gag-Pol cleavage. J. Virol. 77:366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford, S. L., and S. Lindquist. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396:336-342. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sommadossi, J. P. 1999. HIV protease inhibitors: pharmacologic and metabolic distinctions. AIDS 13:529-540. [PubMed] [Google Scholar]
- 29.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 30.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:543-585. [DOI] [PubMed] [Google Scholar]
- 31.Wlodawer, A., M. Miller, M. Jaskolski, B. K. Sathyanarayana, E. Baldwin, I. T. Weber, L. M., Selk, L. Clawson, J. Schneider, and S. B. Kent. 1989. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science 245:616-621. [DOI] [PubMed] [Google Scholar]
- 32.Wondrak, E. M., and J. M. Louis. 1996. Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease. Biochemistry 35:12957-12962. [DOI] [PubMed] [Google Scholar]
- 33.Wondrak, E. M., N. T. Nashed, M. T. Haber, D. M. Jerina, and J. M. Louis. 1996. A transient precursor of the HIV-1 protease. Isolation, characterization, and kinetics of maturation. J. Biol. Chem. 271:4477-4481. [DOI] [PubMed] [Google Scholar]
- 34.Xie, D., S. Gulnik, E. Gustchina, B. Yu, W. Shao, W. Qoronfleh, A. Nathan, and J. W. Erickson. 1999. Drug resistance mutations can affect dimer stability of HIV-1 protease at neutral pH. Protein Sci. 8:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Z. Y., R. A. Poorman, L. L. Maggiora, R. L. Heinrikson, and F. J. Kezdy. 1991. Dissociative inhibition of dimeric enzymes. Kinetic characterization of the inhibition of HIV-1 protease by its COOH-terminal tetrapeptide. J. Biol. Chem. 266:15591-15594. [PubMed] [Google Scholar]
- 36.Zybarth, G., and C. Carter. 1995. Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J. Virol. 69:3878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]