Abstract
During purification of the selenium-dependent xanthine dehydrogenase (XDH) from Clostridium purinolyticum, another hydroxylase was uncovered that also contained selenium and exhibited similar spectral properties. This enzyme was purified to homogeneity. It uses purine, 2OH-purine, and hypoxanthine as substrates, and based on its substrate specificity, this selenoenzyme is termed purine hydroxylase (PH). The product of hydroxylation of purine by PH is xanthine. A concomitant release of selenium from the enzyme and loss of catalytic activity on treatment with cyanide indicates that selenium is essential for PH activity. Selenium-dependent XDH, also purified from C. purinolyticum, was found to be insensitive to oxygen during purification and to use both potassium ferricyanide and 2,6-dichloroindophenol as electron acceptors. Selenium is required for the xanthine-dependent reduction of 2,6-dichloroindophenol by XDH. Kinetic analyses of both enzymes revealed that xanthine is the preferred substrate for XDH and purine and hypoxanthine are preferred by PH. This characterization of these selenium-requiring hydroxylases involved in the interconversion of purines describes an extension of the pathway for purine fermentation in the purinolytic clostridia.
The fermentation of purines was first studied in depth nearly 60 years ago on two isolates, Clostridium acidi-urici and Clostridium cylindrosporum (1). These organisms were characterized with respect to their ability to decompose uric acid, xanthine, and guanine into the end products glycine, formate, carbon dioxide, and ammonia (1). The pathway for the fermentative degradation of purines to the above-mentioned end products was characterized initially by using radioisotope labels to determine end products and yields (2). One of the key enzymes in this pathway, xanthine dehydrogenase (XDH), was first purified by Bradshaw and Barker from C. cylindrosporum in 1960 (3). Since that time, much work has focused on the characterization of similar XDH enzymes from other bacterial sources (4–7) and characterization of these bacterial enzymes with respect to the well-studied xanthine oxidase (XO) from mammalian sources (8).
XDH is a member of a family of enzymes that has been termed the molybdenum hydroxylases. Some of the other members of this family include XO, nicotinic acid hydroxylase, aldehyde oxidase, and carbon monoxide dehydrogenase. In general, these enzymes contain FAD, molybdenum cofactor, and at least one Fe/S center (9). An additional sulfur atom, termed the cyanolyzable sulfur by Wahl and Rajagopalan (10), is required for activity of these enzymes. Treatment of the oxidized enzyme with cyanide inactivates XO activity with concomitant release of thiocyanate in a stoichiometric manner (11). Subsequent addition of sulfide at significantly high concentrations can restore enzyme activity (11). This mechanism of inactivation/reactivation was shown to be effective with similar molybdenum hydroxylase enzymes (12, 13). These studies demonstrated long ago that this family of enzymes contains a unique type of posttranslational modification at the active site. Biochemical evidence for the enzymatic addition of sulfur to the active site of enzymes such as XDH is lacking. However, genetic studies of the ma-l gene in Drosophila melanogaster (14) and the hxB genes in Aspergillus nidulans (15) implicate the products of these genes in this posttranslational modification, although these gene products have yet to be characterized biochemically.
More recent deduced structural data from aldehyde oxidase confirmed the presence of an additional sulfur atom, present in a modified cysteine residue closely associated with the molybdenum atom at the active site (16). Another structural study showed that an analogous selenium atom is present in a modified cysteine residue, termed selanylcysteine, in carbon monoxide dehydrogenase (17). Selenium present in a labile cofactor in nicotinic acid hydroxylase was shown to be coordinated with the molybdenum atom by using electron paramagnetic resonance studies (18). Activity of another molybdenum hydroxylase, clostridial XDH, was shown to be higher in cell extracts when cultures were supplemented with selenium (19), and characterization of a selenium-dependent XDH isolated from Eubacterium barkeri was reported recently (20). Activity of this XDH was found to be sensitive to treatment with cyanide, suggesting that the selenium atom is analogous to the long-studied cyanolyzable sulfur of XO from bovine milk (10). Clostridium purinolyticum, a purine-fermenting organism isolated by Andreesen and colleagues in 1981 (21), requires selenium for efficient fermentation of purines (19).
The present study describes the purification of XDH from C. purinolyticum and initial characterization of the enzyme. During purification of XDH, another selenium-containing hydroxylase was discovered, which can convert purine to xanthine by hydroxylation of either the 2- or the 6-position of the purine ring, and thus this enzyme is termed purine hydroxylase (PH). The purification and initial characterization of this selenium-containing enzyme are described.
Materials and Methods
Materials.
[75Se]Selenite was purchased from the University of Missouri Reactor Center, Columbus. Purine and xanthine derivatives were purchased from Sigma, ICN, or Fluka and were of the highest grade available. All other reagents were of the highest grade available.
Growth of C. purinolyticum.
C. purinolyticum was cultured anaerobically in a 330-liter fermentor in a glycine-yeast extract-minimal salts medium as described (22) with minimal modifications. For 75Se-labeled 2-liter cultures sodium selenite at either 0.1 μM (PH) or 0.5 μM (XDH) was added and [75Se]selenite was added at 1.0 μCi/ml.
Enzyme Assay.
Enzyme assays were performed essentially as described for XDH from C. cylindrosporum (3). The enzymatic reduction of either potassium ferricyanide (FeCN) (2 mM) or 2,6-dichloroindophenol (DCIP) (0.2 mM) was followed by a decrease in absorbance at 420 nm or 600 nm, respectively, in a Milton Roy 3000 Diode array spectrophotometer. Standard assays were performed at 25°C in the presence of substrate at the indicated concentration for each experiment in 50 mM potassium phosphate, pH 7.5 in cuvettes open to air. The average rate of reduction of an electron acceptor was recorded for 2 min, and enzyme activities were calculated by using extinction coefficients of 1,050 M-1 for FeCN and 22,000 M-1 for DCIP. Activities are given as μmol·min-1⋅mg-1 protein.
Purification of XDH.
All steps of purification were performed at room temperature under air, because significant oxygen-insensitive xanthine-dependent reduction of FeCN was catalyzed by cell extracts. Approximately 100 g of frozen cells (stored −80°C) were thawed in 400 ml of buffer A (50 mM Tricine, pH 8.0/0.5 mM EDTA/0.1 mM PMSF). Cells were broken by passage through a French pressure cell operating at 6,000 psi, and the resulting extract was clarified by centrifugation at 20,000 × g for 30 min at 4°C. Protamine sulfate was added to give 0.125%, stirred slowly for 15 min, and centrifuged at 20,000 × g for 30 min at 4°C. This extract was mixed with a similarly prepared extract from a 2-liter culture labeled with [75Se]selenite harvested the same day. These mixed extracts were applied to a DEAE-Sepharose column (Amersham Pharmacia), which was preequilibrated with buffer A. XDH-containing fractions were eluted in a linear gradient from 0 to 0.5 M KCl, pooled, adjusted to 1 M (NH4)2SO4 by slow addition of the solid and loaded on a phenyl-Sepharose column (Amersham Pharmacia) equilibrated with 1 M (NH4)2SO4 in buffer A. A reverse gradient from 1.0 to 0.0 M (NH4)2SO4 was applied, and fractions containing XDH activity were pooled and concentrated by centrifugation using Centriprep 15 concentrators (Amicon). After overnight dialysis against buffer B (50 mM potassium phosphate, pH 7.0/0.1 mM PMSF/0.5 mM EDTA), the enzyme sample was passed over a SP-Sepharose column but under these conditions the enzyme failed to bind. Therefore, the sample was chromatographed on a DEAE-Sepharose column preequilibrated with buffer B and the proteins were eluted with a linear gradient of 50–500 mM potassium phosphate (monobasic), phosphate being an alternate anion from the initial anion exchange step. Active fractions were concentrated as before, and after overnight dialysis in buffer C (50 mM potassium phosphate, pH 7.5/0.1 mM PMSF/0.5 mM EDTA/0.15 M KCl) were chromatographed on Sepharose CL-6B (60 cm × 1.5 cm) in buffer C. Active fractions were pooled, dialyzed against buffer A, and then loaded on a preparative HPLC anion exchange column (DEAE). After an initial wash with 0.045 M sodium citrate (citrate as the anion for exchange), a linear gradient from 45 to 100 mM sodium citrate was used to elute XDH. Fractions were concentrated, dialyzed against buffer D (20 mM sodium phosphate, pH 6.5/0.1 mM PMSF/0.5 mM EDTA) and applied to a preparative CM-HPLC column (TosoHaas CM-5PW, Montgomeryville, PA) and eluted with a linear gradient of 0–0.5 M NaCl. Active XDH fractions, which were eluted, had an A280nm/A460nm ratio of 4.5:1, and analysis of these fractions by SDS/PAGE confirmed the homogeneity of the XDH sample. The pooled fractions were dialyzed against storage buffer (50 mM Tricine, pH 7.5/5% glycerol), and aliquots were quick-frozen and stored in liquid nitrogen.
Purification of PH.
Purification of PH on DEAE-Sepharose followed by chromatography on phenyl-Sepharose was similar to that described above for purification of XDH. In fact, the presence of a selenium-containing PH enzyme was detected only after several additional purification steps had been carried out.
After elution from phenyl-Sepharose, a stepwise (NH4)2SO4 precipitation was performed by slow addition of (NH4)2SO4 to give the desired level of saturation, followed by centrifugation at 3,000 × g for 10 min to remove insoluble proteins. Three different precipitations were carried out, at 40%, 50%, and 70% of saturation. Only at 70% of saturation did XDH activity precipitate. This concentrated sample then was resuspended in 20 ml of buffer E (50 mM Tricine, pH 8.0/150 mM KCl/0.1 mM PMSF/0.5 mM EDTA). This sample was chromatographed on a CL6B-Sepharose column (Amersham Pharmacia) pre-equilibrated with buffer E. Active fractions (XDH) were concentrated by ultrafiltration (Amicon YM10) and loaded on a preparative HPLC anion exchange column (DEAE), which subsequently was washed with buffer A. Active XDH was eluted by using a sodium citrate gradient from 0 to 100 mM. At this step, it was determined that XDH separates from another enzyme with significant absorbance at 350 and 460 nm. After an initial check for substrate specificity, the other enzyme was termed purine hydroxylase (PH), because it reduced DCIP in the presence of purine. Therefore this PH fraction was further purified as follows.
Active PH samples were pooled and concentrated by ultrafiltration (Amicon centriprep 15) and chromatographed on a SW4000 HPLC gel filtration column equilibrated in buffer A with 250 mM KCl. After dialysis against buffer A, active fractions were chromatographed on an HPLC-DEAE column using a gradient of potassium phosphate (monobasic) from 0.2 to 0.3 M. Pooled fractions then were subjected to SW3000 HPLC gel filtration in buffer A with 0.2 M potassium phosphate (monobasic). Active fractions of PH from this step had an absorbance ratio at A280nm/A460nm of 4.0:1 and were judged to be homogeneous by SDS/PAGE. Purified PH subsequently was dialyzed overnight in storage buffer, quick-frozen in liquid nitrogen, and stored at −80°C.
Spectrophotometric Methods.
Spectra of purified XDH and PH were recorded by using a diode array spectrophotometer integrated into a Series 1050 Hewlett–Packard HPLC system.
Native Molecular Weight Determination.
Approximately 20 μg of either XDH or PH was applied to a TosoHaas G3000SW HPLC column (7.5 mm × 60 cm) and chromatographed at 0.5 ml/min in 50 mM potassium phosphate, pH 7.5, 250 mM NaCl. Native molecular weight standards (Bio-Rad) used to calibrate the column included bovine thyroglobulin (670,000), bovine gamma globulin (158,000), chicken ovalbumin (44,000), horse myoglobin (17,000), and vitamin B12 (1350).
N-Terminal Amino Acid Sequence Determination.
N-terminal amino acid sequence was determined as described (23), after separation on SDS/PAGE and passive elution from the gel.
HPLC Separation of Hydroxylated Purine Derivatives.
Separation of purine derivatives for analysis of PH products was carried out by reverse-phase HPLC chromatography on a C18 column (Jones chromatography). An isocratic elution of purine, 2OH-purine, 6OH-purine (hypoxanthine), and xanthine was optimized at the flow rate of 0.75 ml/min in an aqueous buffer containing 0.1% triflouroacetic acid, 0.5% methanol, and 1.0% acetonitrile. For PH-catalyzed reactions, 100-μl aliquots of 1-ml reaction mixtures (see enzyme assay) were added to 0.9 ml of aqueous 0.1% triflouroacetic acid, degassed under argon, and injected into the column. As a reference a standard aqueous solution containing purine, xanthine, hypoxanthine, and 2OH-purine (50 μM each) was injected. Each purine derivative, which eluted at a slightly different retention time depending on the composition of the mixture loaded, was confirmed by online diode-array spectrophotometer for its characteristic spectrum.
Cyanide Inactivation Experiments.
Native XDH or PH was incubated with KCN as described (13). The extent of KCN inactivation of XDH or PH, carried out either anaerobically under argon or aerobically, was the same. After KCN treatment, release of 75Se as a low-molecular weight molecule was determined either by ultrafiltration (Amicon Microcon YM10) or by chromatography on a G-25 size exclusion matrix (Amersham Pharmacia). The amount of 75Se remaining in the separated protein fraction and the protein content were measured to determine the extent of loss of selenium into the small molecule fraction during incubation with KCN.
Results and Discussion
Purification of XDH.
Although purine fermentation by C. purinolyticum is a strictly anaerobic process (21), significant oxygen-insensitive XDH activity was detected under aerobic conditions with FeCN as electron acceptor. Using this assay, 1.1 mg of purified XDH was obtained from 100 g of C. purinolyticum cells, which represented a 349-fold purification of XDH present in the initial extract with a yield of 3.2% (data not shown). Among alternate electron acceptors tested, DCIP was reduced by XDH in a xanthine-dependent manner and this activity was also sensitive to cyanide. For subsequent analysis of XDH activity DCIP was used as electron acceptor. Neither NAD nor NADP served as electron acceptor either in air or under anaerobic conditions. These characteristics are similar to those previously reported for XDH isolated from C. cylindrosporum (3).
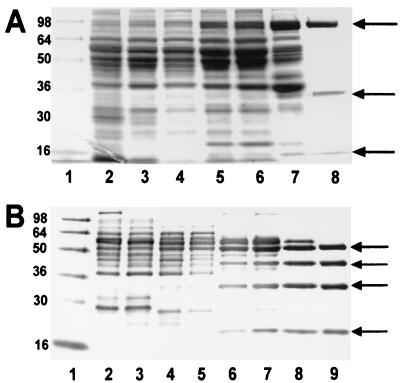
The purified XDH enzyme, as judged by SDS/PAGE (Fig. 1A), consists of three subunits of approximately 80 kDa, 35 kDa, and 16 kDa. Native XDH, estimated by HPLC on a SW3000 gel filtration column (TosoHaas), was calculated to be 529 kDa, based on a calibration of the column with molecular weight standards (Bio-Rad). Assuming an equimolar ratio of the three subunits from SDS/PAGE analysis, a α4β4γ4 configuration is suggested for the native protein. Related molybdenum hydroxylases also have been shown to contain subunits of similar sizes, including the selenium-dependent XDH from Eubacterium barkeri (20).
Figure 1.
(A) SDS/PAGE analysis of the purification of XDH. Lane 1, marker (Novex See Blue); lane 2, 10 μg C. purinolyticum extract; lane 3, 10 μg DEAE-1 pool; lane 4, 10 μg phenyl-Sepharose pool; lane 5, 10 μg DEAE-2 pool; lane 6, 10 μg CL6B pool; lane 7, 10 μg DEAE-3 pool; lane 8, 5 μg CM pool (purified XDH). Arrows indicate large, medium, and small subunits. (B) SDS/PAGE analysis of the purification of PH. Lane 1, marker (Novex See Blue); lane 2, 10 μg C. purinolyticum extract; lane 3, 10 μg DEAE-1 pool; lane 4, 10 μg phenyl-Sepharose NH4SO4 precipitation pool; lane 5, 10 μg CL6B pool; lane 6, 10 μg DEAE-2 pool; lane 7, 10 μg SW4000XL pool; lane 8, 10 μg DEAE-3 pool; lane 9, 5 μg SW3000XL pool (purified PH). Arrows indicate four subunits. Numbers to the left in both A and B represent molecular mass in kDa of proteins present in the standard.
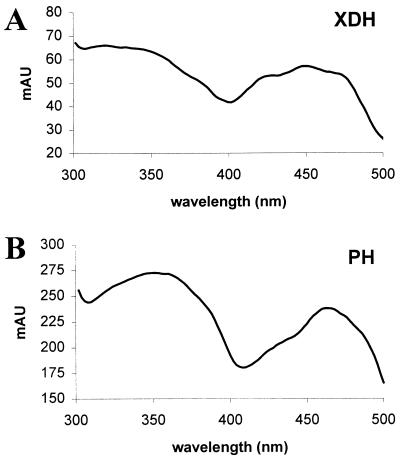
Although many enzymes isolated from anaerobes are subject to oxygen inactivation, XDH from C. purinolyticum was stable both during purification and as the isolated enzyme in the presence of air. Enzyme stored at −70°C was stable for at least 6 months. The A280nm/A460nm ratio of purified enzyme was determined to be 4.5:1 (Fig. 2A). This ratio is regarded as indicative of the purity of XDH and related molybdenum hydroxylases. The electronic absorption spectrum shown in Fig. 2A is typical of the oxidized form of XDH enzymes and other molybdenum hydroxylases (24, 25).
Figure 2.
Absorption spectra of purified XDH (A) and purified PH (B).
Purification of PH.
During one of the later XDH purification procedures, a contaminating protein that exhibited absorbance at 350 nm and 460 nm was detected in active XDH fractions (see Materials and Methods). Because xanthine was not a substrate for this enzyme, other purine substrates and alternate electron acceptors were tested. The unknown enzyme was found to reduce DCIP in the presence of purine, 2OH-purine, and hypoxanthine as substrates. This enzyme, PH, was purified to apparent homogeneity and analyzed in parallel with XDH with regard to its general properties. SDS/PAGE analysis, after the final gel filtration chromatographic step, revealed PH consists of four subunits, approximately 54 kDa, 44 kDa, 32 kDa, and 20 kDa (Fig. 1B). The native enzyme, calculated by gel filtration analysis to be approximately 590 kDa, appears to consist of a α4β4γ4δ4 structure.
The calculated yield of PH was only 0.6% based on specific enzyme activity measurements (data not shown). However, after the final dialysis against storage buffer the selenium content per 150 kDa was 0.3:1 whereas after the gel filtration step but before final dialysis the selenium content was 1:1. This apparent loss of 70% of the labile selenium during dialysis correlates directly with a concomitant loss of catalytic activity from 3.1 units to 1.0 unit of activity. Thus a calculated actual yield of 6 mg of Se-containing plus inactive forms corresponds to about 3% of the total PH activity present in the extract (data not shown). The amount of PH in the cell thus appears to be about 6-fold higher than the levels of XDH. The loss of selenium during dialysis was attributed to the lowering of the ionic strength of the buffer and represents a significant difference in the stability of the labile selenium in XDH as compared with PH.
The electronic absorption spectrum of purified PH, shown in Fig. 2B, is similar to that of XDH. An A280nm/A460nm ratio for PH is 4.0:1. This ratio is lower than the usual value of 4.5:1 found for other purified enzymes within this family and may represent an altered tertiary structure with regard to the active site and flavin cofactor of PH.
N-Terminal Amino Acid Sequence for XDH and PH.
Table 1 summarizes the data from the N-terminal sequence analysis of each of the subunits of XDH and PH. Limited database searches did reveal some similarity of amino acid sequences of the XDH subunits with other members of the molybdenum hydroxylase family (data not shown), but no significant similarity was found to the amino acid sequences of PH subunits.
Table 1.
Amino-terminal amino acid sequence for XDH and PH
| Subunit | Molecular weight, approx. | Amino-terminal sequence |
|---|---|---|
| XDH | ||
| Large | 80,000 | MKIVNKPYXKXDXM |
| Medium | 35,000 | MITIKEYIVPKXIEE |
| Small | 16,000 | MIINVKINGLKKTL |
| PH | ||
| Large | 54,000 | XFDVIGKSILRVDGL |
| Medium 1 | 42,000 | MKKRGKGVAXTFYGT |
| Medium 2 | 34,000 | MLLKXVFXAEXLEEA |
| Small | 20,000 | MKRINLKVNYKDVXV |
In the sequences X represents ambiguous sample for which no amino acid was determined.
Substrate Specificity and Kinetic Analysis of XDH.
Additional good substrates for XDH were 1-methylxanthine, hypoxanthine, allopurinol, and 2OH-purine (Table 2). A similar substrate specificity has been reported for other clostridial XDH enzymes (3, 20). The lack of reactivity with 8-methylxanthine, adenine, and guanine is not surprising. Based on analysis of similar XDH enzymes (3), hydroxylation occurs primarily at the 8-position regardless of the substrate.
Table 2.
Substrate specificity of XDH
| Substrate | Substrate concentration, mM | Specific activity, μmol· min−1⋅mg−1 |
|---|---|---|
| Xanthine (control) | 0.5 | 2.6 |
| Hypoxanthine | 1.0 | 1.2 |
| 2OH-purine | 2.0 | 2.3 |
| Allopurinol | 2.0 | 1.5 |
| 1-Methylxanthine | 2.0 | 2.6 |
| 7-Methylxanthine | 2.0 | ND |
| 8-Methylxanthine | 2.0 | ND |
| Purine | 2.0 | 0.7 |
| Adenine | 2.0 | ND |
| Guanine | 2.0 | ND |
| Nicotinic acid | 2.0 | ND |
ND, activity not detectable.
The substrate specificity of XDH, determined by kinetic analyses at varying substrate concentrations, is shown in Table 3. As expected, the approximate Km value calculated for xanthine was low (4.5 μM), and XDH activity with xanthine yielded the highest Vmax value. Hypoxanthine had a reasonably high Vmax of 1.2 μmol·min-1⋅mg-1, but also a much higher Km value of 0.5 mM (Table 3). By comparison of the relative Vmax/Km ratios for the three substrates shown in Table 3, it is apparent that XDH prefers xanthine as a substrate and may act primarily on xanthine in vivo.
Table 3.
Kinetic parameters of XDH
| Substrate | Km, μM | Vmax, μmol· min−1⋅mg−1 | Vmax/Km |
|---|---|---|---|
| Xanthine | 4.5 | 2.7 | 0.6 |
| Hypoxanthine | 500 | 1.2 | 0.0024 |
| Allopurinol | 1000 | 2.0 | 0.0020 |
The approximate Km value was estimated by using a Michaelis–Menten plot and confirmed experimentally after this determination. The Vmax was estimated from a Michaelis–Menten plot.
Substrate Specificity and Kinetic Analysis of PH.
Purine, hypoxanthine, 2OH-purine, and allopurinol all served as good substrates for PH (Table 4). As observed with XDH, NAD and NADP were ineffective electron acceptors for PH. Neither xanthine nor 2,6-diaminopurine was used as substrate by PH, suggesting the site of hydroxylation is either at the 2- or 6-position in the purine ring. Benzaldehyde was used as a substrate at relatively high concentrations (with respect to purine or purine derivatives). To assess which of these substrates are most likely to be acted on by PH in vivo, values for Km and Vmax were determined.
Table 4.
Substrate specificity of PH
| Substrate | Substrate concentration, mM | Specific activity, μmol· min−1⋅mg−1 |
|---|---|---|
| Purine | 0.1 | 1.0 |
| Hypoxanthine | 0.1 | 1.6 |
| 2OH-purine | 0.1 | 1.2 |
| Allopurinol | 0.5 | 1.3 |
| Benzaldehyde | 1.0 | 0.7 |
| Adenine | 1.0 | ND |
| Nicotinic acid | 2.0 | ND |
| Xanthine | 2.0 | ND |
| 2,6-Diaminopurine | 2.0 | ND |
ND, activity not detectable.
Both purine and hypoxanthine had low Km values, and rates of reduction of DCIP were significant (Table 5). 2OH-purine and allopurinol also were substrates for PH, yielding Vmax/Km ratios of 0.026 and 0.033, respectively. Nonetheless, the high Vmax/Km ratio for either purine or hypoxanthine suggests that one of these compounds is the primary substrate for PH in vivo. Benzaldehyde, a good substrate for aldehyde oxidase, was a much poorer substrate for PH based on a relatively high determined km (200 μM) value, as well as a low Vmax/Km ratio of 0.004. This relatively poor activity with benzaldehyde distinguishes PH from aldehyde oxidase enzymes previously studied, one of which has been reported to use purine and its derivatives as substrates (26). Because PH used purine derivatives hydroxylated at either the 2- or 6-position, further investigation targeted determination of the specific site of hydroxylation in the end products of reaction with purine and its derivatives.
Table 5.
Kinetic parameters of PH
| Substrate | Km, μM | Vmax, μmol· min−1⋅mg−1 | Vmax/Km |
|---|---|---|---|
| Purine | 4.0 | 1.0 | 0.25 |
| Hypoxanthine | 5.0 | 2.0 | 0.40 |
| 2OH-purine | 50 | 1.3 | 0.026 |
| Allopurinol | 40 | 1.3 | 0.033 |
| Benzaldehyde | 200 | 0.7 | 0.004 |
The approximate Km value was estimated by using a Michaelis–Menten plot and confirmed experimentally after this determination. The Vmax was estimated from a Michaelis–Menten plot.
HPLC Analysis of the Products of Hydroxylation by PH.
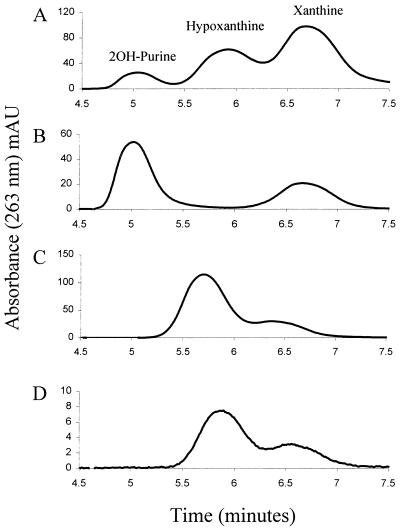
An isocratic elution of purine and its hydroxylated derivatives was developed by using a reverse-phase HPLC chromatography under acidic conditions. Analysis of a standard solution of purine, 2OH-purine, hypoxanthine, and xanthine (25 μM of each) using this technique is shown in Fig. 3A. Elution of each purine derivative was followed by relative absorbance at 263 nm. Because each compound has a different absorption maximum and molar extinction coefficient, the information given in Fig. 3 is qualitative. The individual spectrum of each peak was recorded to confirm that it was identical to one of the four standard compounds injected into the column (data not shown). Purine eluted as a broad peak beginning at about 9 min and thus is not included in Fig. 3.
Figure 3.
HPLC analysis of purine and hydroxylated derivatives. Retention time is plotted versus absorbance at 263 nm, at which all of the tested purine derivatives have significant absorbance. Peaks that represent each compound from the standard are marked in A. (A) Standard mixture of purine, 2OH-purine, hypoxanthine, and xanthine. (B) PH incubated with 2OH-purine. (C) PH incubated with hypoxanthine. (D) PH incubated with purine. See Materials and Methods for further details.
Addition of either hypoxanthine or 2OH-purine as a substrate for PH at a final concentration of 100 μM resulted in the formation of xanthine (Fig. 3 B and C). Addition of 100 μM purine to PH resulted in significant production of both hypoxanthine and xanthine (Fig. 3D). These results demonstrate that PH can hydroxylate both the 2- and 6-positions of purine but the lack of significant formation of 2OH-purine from purine (Fig. 3D) suggests there is some bias with regard to which site is preferentially hydroxylated. Apparently, the 6-position is acted on first (giving hypoxanthine as a product), and only after hydroxylation of the 6-position is the 2-position then acted on as a substrate, yielding xanthine as the final product. Indeed hypoxanthine was found to be a better substrate for PH when compared with 2OH-purine in kinetic analysis (Table 5), indicating the 2-position is more readily hydroxylated when the 6-position already has been substituted.
Inactivation of XDH and PH by Cyanide.
A so-called cyanolyzable sulfur (or analogous selenium) exists in the active site of many molybdenum hydroxylases (11, 12, 20) and the nature of this cyanolyzable sulfur has been determined in structural studies for two enzymes (16, 17). In carbon monoxide dehydrogenase, a selenium atom was modeled as part of a modified cysteine and reported to be a selanylcysteine residue (17). Andreesen and coworkers (20) could inactivate XDH purified from E. barkeri by incubation with cyanide, and the enzyme could be reactivated by addition of high concentrations of selenide. Because XDH and PH from C. purinolyticum are labeled with 75Se in vivo, it is possible that the selenium is labile and comparable to the cyanolyzable sulfur of XO. To test the possibility that the 75Se was present as a labile selenium cofactor, both [75Se]XDH and [75Se]PH were treated with cyanide.
XDH catalytic activity dropped to about 25% of the initial value over a period of 4 h when incubated with 15 mM KCN under conditions similar to those reported for removal of cyanolyzable sulfur (or analogous selenium) from similar enzymes (13, 20). More than 50% of the 75Se was separated from the inactivated XDH by ultrafiltration (cutoff approximately 10 kDa) and appeared in the filtrate.
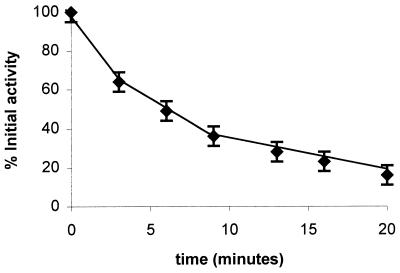
PH was more sensitive than XDH to KCN, with significant loss of catalytic activity occurring within 5 min of incubation with 1 mM KCN (Fig. 4). A sample of inactivated [75Se]PH, after chromatography 2 × on a Sephadex G-25 spin column (Amersham Pharmacia), contained less that 10% of the initial selenium present in the native enzyme. From these results it is clear that the selenium present in PH is also of the labile type and is easily released by treatment with cyanide.
Figure 4.
Inactivation of PH by KCN. Specific activity of PH was determined by using hypoxanthine as the substrate at a concentration of 100 μM and DCIP as electron acceptor at 50 μM as described in Materials and Methods. KCN was added to give 1 mM final concentration to PH in storage buffer and samples were taken at the indicated times. The value plotted represents the fraction of the initial activity. Three independent experiments were carried out and the error bars represent one SD from the average value.
In contrast to XDH, exposure of PH to low ionic strength led to a loss in selenium and a corresponding loss in enzyme activity. To test whether the rate of inactivation by KCN was influenced by ionic strength, partially purified PH was incubated with KCN in 0.25 M sodium citrate, 50 mM Tricine, instead of 50 mM Tricine, pH 7.5, 5% glycerol used previously. A higher concentration of KCN (20 mM) was required to inactivate the enzyme to about 25% of its initial activity over a time period of 4 h under these conditions. The selenium that was released from the inactivated enzyme appeared in the small molecule fraction as found before (data not shown). Based on these observations, XDH and PH contain selenium in the form of a labile cofactor that is analogous to the cyanolyzable sulfur characterized in related molybdenum hydroxylase enzymes.
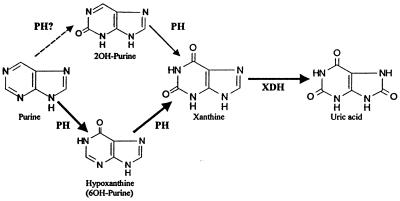
PH Adds an Enzyme to the Pathway for Purine Fermentation.
Previously, elucidation of the pathway for the fermentation of purines has centered mainly on the study of a key enzyme, XDH. Analysis of the products of hydroxylation of purines by a purified clostridial XDH revealed that XDH preferentially hydroxylates the 8-position of the purine ring (3). This purified XDH from C. cylindrosporum also clearly showed a specificity for xanthine as a substrate when careful examination of the kinetic constants are taken into account (3).
The present work describes an enzyme, PH, which can hydroxylate either the 2- or the 6-position of the purine ring. Kinetic analyses of both XDH and PH reveal that the best substrate for PH, based on the Vmax/Km ratio (Table 5), is either purine or hypoxanthine. By similar analysis the best substrate for XDH is xanthine (Table 3). A pathway that shows schematically the reactions that appear to be catalyzed preferentially by these two enzymes is shown in Fig. 5. Because analysis of the reaction products of PH yielded only hypoxanthine and xanthine (no 2OH-purine, Fig. 3), the enzyme may actively differentiate between an initial hydroxylation at the 2- or the 6-position, and only hydroxylate the 2-position after hydroxylation of the 6-position. This scheme is corroborated by the lower apparent affinity for 2OH-purine as a substrate (Table 5).
Figure 5.
Pathway for hydroxylation of purine to uric acid in C. purinolyticum. Each compound is labeled and the arrows are labeled with respect to which enzyme (XDH or PH) is responsible for each reaction.
Future work should focus on analysis of the cofactor compositions of XDH and PH, as well as the exact nature, location, and mode of incorporation of the labile selenium cofactor. Elucidation of the reaction mechanism for PH may lead to an understanding of how this enzyme can efficiently hydroxylate either the 2- or 6-position of the purine ring to yield xanthine as its final product.
Acknowledgments
We thank Dr. Rodney Levine (Laboratory of Biochemistry, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD) for his work in the determination of the N-terminal amino acid sequence of the subunits for both XDH and PH.
Abbreviations
- XDH
xanthine dehydrogenase
- PH
purine hydroxylase
- FeCN
potassium ferricyanide
- DCIP
2,6-dichloroindophenol
- XO
xanthine oxidase
References
- 1.Barker H A, Beck J V. J Biol Chem. 1941;141:3–27. [Google Scholar]
- 2.Rabinowitz J C, Barker H A. J Biol Chem. 1955;218:147–160. [PubMed] [Google Scholar]
- 3.Bradshaw W H, Barker H A. J Biol Chem. 1960;235:3620–3629. [Google Scholar]
- 4.Perez-Vicente R, Alamillo J M, Cardenas J, Pineda M. Biochim Biophys Acta. 1992;1117:159–166. doi: 10.1016/0304-4165(92)90074-5. [DOI] [PubMed] [Google Scholar]
- 5.Scazzocchio C, Sealy-Lewis H M. Eur J Biochem. 1978;91:99–109. doi: 10.1111/j.1432-1033.1978.tb20942.x. [DOI] [PubMed] [Google Scholar]
- 6.Schräder T, Rienhöfer A, Andreesen J R. Eur J Biochem. 1999;264:862–871. doi: 10.1046/j.1432-1327.1999.00678.x. [DOI] [PubMed] [Google Scholar]
- 7.Wagner R, Andreesen J R. Arch Microbiol. 1979;121:255–260. doi: 10.1007/BF00425064. [DOI] [PubMed] [Google Scholar]
- 8.Hunt J, Massey V. J Biol Chem. 1994;269:18904–18914. [PubMed] [Google Scholar]
- 9.Hille R, Rétey J, Bartlewski-Hof U, Reichenbecher W, Schink B. FEMS Microbiol Rev. 1998;22:489–501. doi: 10.1111/j.1574-6976.1998.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 10.Wahl R C, Rajagopalan K V. J Biol Chem. 1982;257:1354–1359. [PubMed] [Google Scholar]
- 11.Massey V, Edmondson D. J Biol Chem. 1970;245:6595–6598. [PubMed] [Google Scholar]
- 12.Wahl R C, Hageman R V, Rajagopalan K V. Arch Biochem Biophys. 1984;230:264–273. doi: 10.1016/0003-9861(84)90107-3. [DOI] [PubMed] [Google Scholar]
- 13.Coughlan M P, Johnson J L, Rajagopalan K V. J Biol Chem. 1980;255:2694–2699. [PubMed] [Google Scholar]
- 14.Wahl R C, Warner C K, Finnerty V, Rajagopalan K V. J Biol Chem. 1982;257:3958–3962. [PubMed] [Google Scholar]
- 15.Amrani L, Cecchetto G, Scazzocchio C, Glatigny A. Mol Microbiol. 1999;31:1065–1073. doi: 10.1046/j.1365-2958.1999.01242.x. [DOI] [PubMed] [Google Scholar]
- 16.Romao M J, Huber R. Biochem Soc Trans. 1997;25:755–757. doi: 10.1042/bst0250755. [DOI] [PubMed] [Google Scholar]
- 17.Dobbek H, Gremer L, Meyer O, Huber R. Proc Natl Acad Sci USA. 1999;96:8884–8889. doi: 10.1073/pnas.96.16.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gladyshev V N, Khangulov S V, Stadtman T C. Proc Natl Acad Sci USA. 1994;91:232–236. doi: 10.1073/pnas.91.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dürre P, Andreesen J R. J Bacteriol. 1983;154:192–199. doi: 10.1128/jb.154.1.192-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schräder T, Rienhöfer A, Andreesen J R. Eur J Biochem. 1999;264:862–871. doi: 10.1046/j.1432-1327.1999.00678.x. [DOI] [PubMed] [Google Scholar]
- 21.Dürre P, Andersch W, Andreesen J R. Int J Syst Bacteriol. 1981;31:184–194. [Google Scholar]
- 22.Garcia G E, Stadtman T C. J Bacteriol. 1991;173:2093–2098. doi: 10.1128/jb.173.6.2093-2098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladyshev V N, Khangulov S V, Stadtman T C. Biochemistry. 1996;35:212–223. doi: 10.1021/bi951793i. [DOI] [PubMed] [Google Scholar]
- 24.Kim K S, Ro Y T, Kim Y M. J Bacteriol. 1989;171:958–964. doi: 10.1128/jb.171.2.958-964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmann M, Tshisuaka B, Fetzner S, Roger P, Lingens F. J Biol Chem. 1994;269:11254–11260. [PubMed] [Google Scholar]
- 26.Hall W W, Krenitsky T A. Arch Biochem Biophys. 1986;251:36–46. doi: 10.1016/0003-9861(86)90048-2. [DOI] [PubMed] [Google Scholar]