Abstract
The lipophilic yeast Malassezia globosa is one of the major constituents of the mycoflora of the skin of patients with atopic dermatitis (AD). We compared the genotypes of M. globosa colonizing the skin surface of 32 AD patients and 20 healthy individuals for polymorphism of the intergenic spacer (IGS) 1 region of the rRNA gene. Sequence analysis demonstrated that M. globosa was divided into four major groups, which corresponded to the sources of the samples, on the phylogenetic tree. Of the four groups, two were from AD patients and one was from healthy subjects. The remaining group included samples from both AD patients and healthy subjects. In addition, the IGS 1 region of M. globosa contained short sequence repeats: (CT)n, and (GT)n. The number of sequence repeats also differed between the IGS 1 of M. globosa from AD patients and that from healthy subjects. These findings suggest that a specific genotype of M. globosa may play a significant role in AD, although M. globosa commonly colonizes both AD patients and healthy subjects.
Malassezia species are lipophilic yeasts that are part of the normal human cutaneous commensal flora; they are isolated from sebaceous gland-rich areas of the skin, particularly on the chest, back, and head. They are also associated with several cutaneous diseases, including atopic dermatitis (AD), folliculitis, pityriasis versicolor, and seborrheic dermatitis (1, 7). In a taxonomic revision in 1996, the genus Malassezia was classified into seven different species: M. furfur, M. globosa, M. restricta, M. obtusa, M. pachydermatis, M. slooffiae, and M. sympodialis (9). Recently, we described an eighth species, M. dermatis, which was isolated from Japanese patients with AD (27). Since the taxonomic revision of the genus Malassezia, several studies have examined the distribution of the newly defined species of Malassezia on healthy human skin and lesions of skin diseases (2, 10, 21). However, culture media or sampling techniques often affect analyses of the Malassezia microflora. In a previous study, we used a nonculture method as an alternative to fungal culture to analyze the distribution of cutaneous Malassezia species (25). M. globosa and M. restricta were detected in approximately 90% of AD patients, and M. furfur and M. sympodialis were detected in approximately 40% of the subjects. In healthy subjects, M. globosa, M. restricta, and M. sympodialis were detected in approximately 40 to 60% of the subjects; M. furfur was found in only 4% of the subjects; and no other Malassezia species were detected. Therefore, these four species are common inhabitants of the skin of both AD patients and healthy individuals. In addition, while anti-Malassezia immunoglobulin E (IgE) antibody was detected in more than 90% of AD patients, no antibody was found in healthy subjects. Based on these results, M. globosa and M. restricta are thought to play a significant mycological role in AD. M. globosa is also part of the major microflora on the skin of healthy individuals. We used the intergenic spacer (IGS) region of the rRNA gene to investigate the genotypes of M. globosa colonizing the skin of AD patients and healthy subjects. The fungal rRNA gene consists of 5S, 5.8S, 18S (small subunit), and 26S (large subunit) subunits (Fig. 1). Two other regions are positioned between the subunits: the internal transcribed spacer (ITS) and the IGS. These two regions are further divided into two subregions. The 18S and 26S ribosomal DNAs (rDNAs) and ITS regions have been widely utilized in studies of molecular systematics and to identify microorganisms (17, 23). The IGS regions have higher rates of divergence than other subunits or regions. Some authors (5, 20, 24, 26) have demonstrated that the sequence of the IGS region shows remarkable intraspecies diversity.
FIG. 1.
Schematic representation of the rRNA gene in the type strain (CBS 7966) of M. globosa.
In this study, we compared the levels of DNA sequence divergence among the IGS regions of M. globosa, which is the key candidate allergen in AD, obtained from the skin of AD patients and from healthy subjects.
MATERIALS AND METHODS
Sequencing the IGS region of M. globosa stock strains.
Two stock strains, CBS 7996 (type strain of M. globosa) and CBS 8745, were purchased from Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands). They were maintained on modified Leeming and Notman agar (LNA; 20 g of glucose, 50 g of malt extract, 1 g of polypeptone, 20 g of bile salts) (OXOID, Hampshire, United Kingdom), 1% Tween 40, 0.2% glycerol, and 50 μg of chloramphenicol per ml (Sankyo, Tokyo, Japan) at 32°C. Genomic DNA was extracted by the method of Makimura et al. (18). The IGS region containing 5S rDNA was amplified from each strain by using primers 26S-F and P1R, shown in Table 1. The reactions were performed in a final reaction mixture (50 μl) containing 10 pmol of each primer; 200 μM each dATP, dTTP, dGTP, and dCTP; 2.5 mM MgCl2; 0.5 U of Takara Ex Taq polymerase (Takara, Shiga, Japan); and 10× reaction buffer (Takara). Amplification reactions were performed in a GeneAmp PCR system 9700 (PE Applied Biosystems, Foster, Calif.) using the following cycling parameters: 94°C for 1 min; followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 2 min; followed by a final extension at 72°C for 10 min. The PCR product was sequenced with an ABI PRISM cycle sequencing kit (PE Applied Biosystems) using the primers shown in Table 1 in accordance with the manufacturer's instructions.
TABLE 1.
Primers used to amplify and sequence the IGS region
| Primer | Sequence (5′ to 3′) | Corresponding position in IGS sequence of strain CBS 7966 |
|---|---|---|
| Amplification | ||
| 26S-F | ATCCTTTGCAGACGACTTGA | 3′ end of 26S rDNA |
| P1R | ACTGGCAGGATCAACCAGAT | 5′ end of 18S rDNA |
| Sequencing | ||
| Forward | ||
| 26S-F | ATCCTTTGCAGACGACTTGA | 3′ end of 26S rDNA |
| gb-F2 | CCGATCTGCGAAGTTAAGCA | 483-502 |
| gb-F3 | GATCATAGCCTCATCATGTGCA | 997-1018 |
| gb-F4 | GAATACGTGACAATTTGTGTGG | 1380-1401 |
| gb-F5 | GTCGCACTGGAGAAAGATGT | 1765-1784 |
| Reverse | ||
| P1R | ACTGGCAGGATCAACCAGAT | 5′ end of 18S rDNA |
| gb-R2 | ACATCTTTCTCCAGTGCGAC | 1784-1765 |
| gb-R3 | CCACACAAATTGTCACGTATTC | 1401-1380 |
| gb-R4 | TGCACATGATGAGGCTATGATC | 1018-997 |
| gb-R5 | TGCTTAACTTCGCAGATCGG | 502-483 |
Subjects.
Thirty-six AD outpatients (24 males and 12 females; 20 to 64 years of age; mean age, 33.3 ± 10.5 years) at Tokyo Medical University Hospital and 30 healthy students (10 males and 20 females; 19 to 25 years of age; mean age, 20.9 ± 1.4 years) at Meiji Pharmaceutical University were involved in this study. AD was diagnosed according to the criteria of Hanifin and Rajka (11), and samples were collected from erythematous lesions on the face and neck. Routine skin care, including intermittent applications of mild steroid ointment or petrolatum, was administered before sampling. Written informed consent was obtained from each subject.
Sequencing the IGS 1 region from patient samples.
Malassezia samples were collected by applying a 3- by 3-cm transparent OpSite dressing (Smith and Nephew Medical Ltd., Hull, United Kingdom), and the fungal DNA was extracted from the OpSite dressing as described previously (25). Briefly, the collected dressing was placed in 1 ml of lysing solution (100 mM Tris-HCl [pH 8.0], 30 mM EDTA [pH 8.0], 0.5% sodium dodecyl sulfate) and incubated at 100°C for 15 min. After deproteinization, DNA was precipitated with 2-propanol and Ethatimate (Nippon Gen, Toyama, Japan). The DNA pellet was resuspended in 30 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). The DNA extracted (10 μl) from each sample was added to 40 μl of PCR master mixture, which consisted of 5 μl of 10× PCR buffer (Takara), 4 μl of 200 μM deoxynucleoside triphosphates, 10 pmol of each primer, and 0.5 U of Takara Ex Taq DNA polymerase (Takara). PCR was performed with an initial denaturation at 94°C for 1 min; followed by 30 cycles of 30 s at 94°C, 1 min at 54°C, and 30 s at 72°C; followed by a final extension at 72°C for 10 min with primers gb-F (GCTTTCGAGTGCATACCACAC) and gb-R (GGAAATAGGATGAGAGAAAC). The PCR products were cloned with a TA cloning kit (Invitrogen Corp., Carlsbad, Calif.), and three positive clones were sequenced with an ABI PRISM cycle sequencing kit (PE Applied Biosystems) and Sequence Rx Enhancer solution A (GIBCO BRL, Life Technologies, Rockville, Md.) in accordance with the manufacturers' instructions.
Molecular phylogenetic analysis.
The sequences of the IGS 1 region were aligned using Clustal W (28). For neighbor-joining analysis (22), the distances between sequences were calculated with Kimura's two-parameter model (13). A bootstrap analysis was conducted with 100 replicates (8).
Formation of chimeric molecules.
To confirm whether chimeric molecules formed under the PCR conditions used in this study, mixed genomic DNA from the eight known Malassezia species was used for PCR coamplification of the IGS region. Then, the IGS amplified from the mixed genomes was cloned, 30 clones were selected at random, and their sequences were determined.
Nucleotide sequence accession number.
The nucleotide sequences determined in this study have been deposited with the DNA Data Bank of Japan (DDBJ) under accession no. AB099877, AB099878 (CBS 7966), AB099879, and AB099880 (CBS 8745).
RESULTS
IGS 1 sequences of M. globosa. (i) Stock strains.
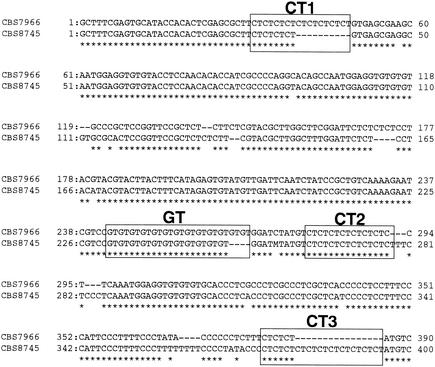
Complete sequences of the IGS region, including 5S rDNA, were determined for two CBS stock strains of M. globosa. Their sequences ranged from 2288 to 2300 bp long. The M. globosa IGS 1 regions were 444 to 454 bp long, while the IGS 2 regions were 1,716 to 1,738 bp long. The IGS 1 and 2 regions of this microorganism showed 12.6, and 6.1% dissimilarity, respectively. Therefore, IGS 1 is more suitable than IGS 2 for differentiating closely related strains. M. globosa IGS 1 had four short sequence repeats (SSRs) of (CT)n, (CT)n, (CT)n, and (GT)n at positions 29 to 49, 278 to 291, 380 to 485, and 242 to 267 in the IGS sequence of strain CBS 7996 (type strain of M. globosa), respectively. Alignments of IGS 1 of two strains of M. globosa are shown in Fig. 2. Because M. globosa had three (CT)ns in its IGS 1 region, these are referred to as (CT1)n, (CT2)n, and (CT3)n in this article.
FIG. 2.
Alignment of the DNA sequences of the IGS 1 region of M. globosa CBS 7966 and CBS 8745.
(ii) Samples from the subjects.
M. globosa DNA was found in 32 of 36 AD patients and 20 of 30 healthy subjects. Under the PCR conditions described above, 420- to 467-bp fragments were amplified and analyzed.
(iii) SSRs.
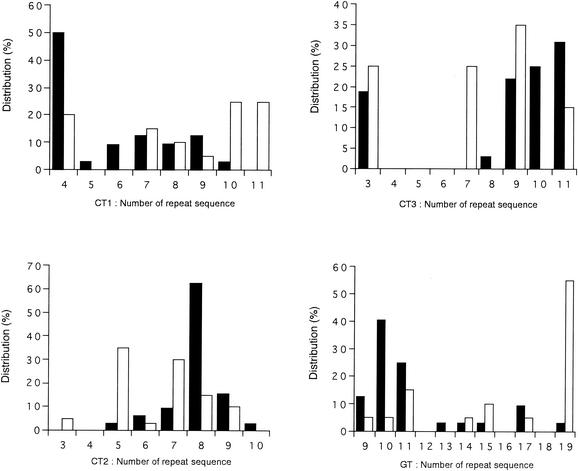
The number of sequence repeats of (GT)n and (CT)n in the IGS 1 region is shown in Fig. 3. The number of SSRs in the IGS 1 region of samples from healthy subjects was more variable than in samples from AD patients for (CT)n. The number of sequence repeats in the IGS 1 region ranged from 4 to 11 for (CT1)n, 3 to 10 for (CT2)n, and 3 to 11 for (CT3)n, and there were 4 in 50%, 8 in 60%, and 9 to 11 in 80% of the samples from AD patients. For (GT)n, the respective numbers of repeats in 70 to 80% of the SSRs in the IGS 1 region derived from AD patients and healthy subjects were 9 to 11 and 15 to 19, respectively.
FIG. 3.
Distribution of SSRs in the IGS 1 region of the M. globosa rRNA gene. Solid bars, M. globosa obtained from AD patients; open bars, M. globosa obtained from healthy subjects.
(iv) Phylogenetic analysis.
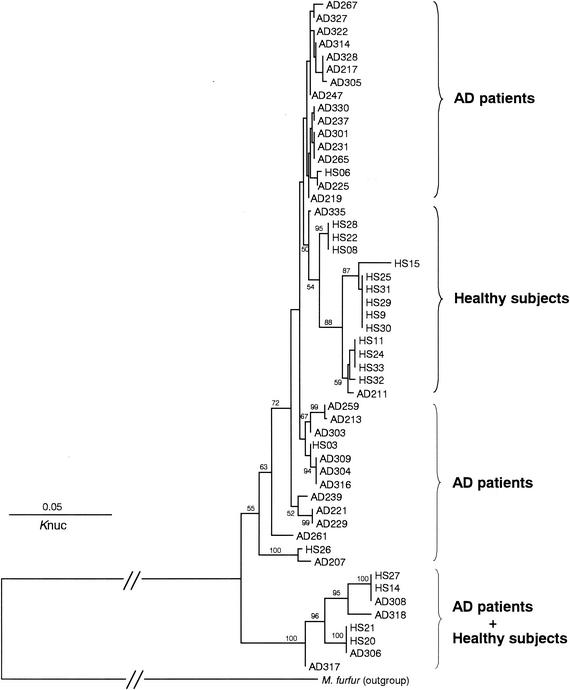
In each patient or healthy subject, three clone sequences were determined, and they were identical without exception. A phylogenetic tree constructed from 52 IGS 1 sequences is shown in Fig. 4. The tree consists of four major groups, which correspond to the sources of the samples (AD patients or healthy subjects). Two groups were from AD patients, and one was from healthy subjects. The remaining group included samples from both AD patients and healthy subjects. The IGS 1 sequences derived from the healthy subjects were more diverse than those from AD patients. The levels of IGS 1 sequence similarity within samples collected from AD patients and from healthy subjects were 94.5% ± 3.5% and 89.9% ± 3.5%, respectively.
FIG. 4.
Phylogenetic tree of M. globosa colonizing the skin surface of AD patients and healthy subjects based on DNA sequences of the IGS 1 region. AD, patient with atopic dermatitis; HS, healthy subject. The numbers show the confidence level from 100 replicate bootstrap samplings (frequencies of <50% are not shown). Knuc, Kimura's parameter (10).
Formation of chimeric molecules.
Thirty clones were chosen at random and sequenced. No clone was identified as a chimeric molecule.
DISCUSSION
One member of the genus Malassezia, M. globosa, commonly colonizes the skin of both AD patients and healthy subjects. The major antigen for IgE antibodies in AD patients is a glycoprotein (Malg46b) from M. globosa (14, 15). This paper describes differences between the IGS 1 genotypes of M. globosa colonizing the skin surface of both populations. The rRNA gene is a marker that reflects the phylogenetic evolution of microorganisms and has been widely used for taxonomy and identification (17, 23). While the taxonomic significance of the 18S and 26S rDNA and ITS regions is known, that of the IGS region is unclear. Previously, we demonstrated that the DNA sequence of the IGS region showed remarkable intraspecies diversity in the pathogenic yeasts Cryptococcus neoformans and Trichosporon asahii (24, 26). While analyzing the IGS sequences of several yeasts from humans, we found SSRs in the M. globosa IGS sequence. Due to their high variability, SSRs are widely used to study the molecular epidemiology of pathogenic microorganisms (3, 4, 19, 29). As far as we know, M. globosa is the only yeast from humans that has these SSRs. The IGS 1 sequences of M. globosa isolates obtained from AD patients and healthy individuals were almost identical in the two groups, with the exception of the four SSRs, which could be used to distinguish between microorganisms from AD patients and those from healthy individuals. When a phylogenetic tree was constructed from IGS sequences excluding the SSRs, the M. globosa sequences obtained from AD patients and healthy subjects intermingled.
We used a PCR-based nonculture method to analyze the genotypes of M. globosa colonizing the skin surface of patients with AD, since M. globosa is difficult to isolate by culture methods. When using PCR-based approaches, the generation of chimeric sequences must also be considered, because pseudosequences may generate nonexistent genotypes of this microorganism. Wang and Wang (30) found that chimeric sequences occurred at a rate of 32% after 30 cycles of PCR amplification targeting the consensus sequence of the bacterial 16S rRNA gene by using mixed genomic DNA from eight bacterial species. We used M. globosa species-specific oligonucleotide primers targeting the IGS sequence, which is the most variable region in the rRNA gene. The primers used in this study did not amplify the DNA of other Malassezia species (data not shown). Although chimera molecules should not be generated theoretically, we also confirmed that no chimera molecules formed under our experimental conditions with genomic DNA from the eight known Malassezia species.
The M. globosa organisms originating from AD patients were phylogenetically different from the M. globosa organisms obtained from healthy subjects with respect to their IGS sequences, although M. globosa colonized both AD patients and healthy individuals at high frequency. Why do the genotypes in each population differ? The reason is unclear, but the genotypes might correspond to the physiological characteristics of this microorganism. First, we considered the possible influence of skin surface lipids, a mixture of secretions from the sebaceous glands and epidermal cells, consisting mainly of triglycerides, squalene, wax esters, cholesterol, ceramides, and free fatty acids (6). Although the lipid composition in AD patients is generally no different from that of healthy subjects, a significant decrease in ceramide 1 and differences in the concentrations of the related molecules linoleate and oleate have been reported (12, 31). Such differences in composition may affect the colonization of strains with different lipid requirements. Moreover, therapeutic agents used to treat AD may affect the selective colonization of the microorganism. The base ingredients in these ointments affect the growth of Malassezia species (16). While M. furfur can utilize white petrolatum, hydrophilic ointment, and heparinoid in hydrophilic ointment, M. globosa cannot utilize these ingredients. Therefore, active ingredients such as steroids and tacrolimus might affect the selective colonization of M. globosa. The antifungal drug susceptibility of this microorganism should also be considered. Since no patient in this study received antifungal therapy, this possibility can be excluded. Since the analysis of M. globosa genotypes in this study is based on a nonculture method, the significance of genotype differences will be elucidated by investigating their phenotypic and physiological characteristics with viable cells.
In conclusion, our IGS sequence analysis revealed differences in the genotypes of M. globosa colonizing the skin surface of AD patients and healthy subjects, suggesting that genotype should be taken into consideration when studying the relationship between M. globosa and AD.
Acknowledgments
This study was supported in part by a Grant for the Promotion of the Advancement of Education and Research in Graduate Schools from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and in part by IATRON Laboratories, Inc. (A.N.).
REFERENCES
- 1.Ashbee, H. R., and E. G. V. Evans. 2002. Immunology of diseases associated with Malassezia species. Clin. Microbiol. Rev. 15:21-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspiroz, C., L. A. Moreno, A. Rezusta, and C. Rubio. 1999. Differentiation of three biotypes of Malassezia species on human normal skin. Correspondence with M. globosa, M. sympodialis and M. restricta. Mycopathologia 145:69-74. [DOI] [PubMed] [Google Scholar]
- 3.Bart-Delabesse, E., J.-F. Humbert, E. Delabesse, and S. Bretagne. 1998. Microsatellite markers for typing Aspergillus fumigatus isolates. J. Clin. Microbiol. 36:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botterel, F., C. Desterke, C. Costa, and S. Bretagne. 2001. Analysis of microsatellite markers of Candida albicans used for rapid typing. J. Clin. Microbiol. 39:4076-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz, M. R., T. Boekhout, B. Theelen, and J. W. Fell. 2000. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast, Cryptococcus neoformans. Syst. Appl. Microbiol. 23:535-545. [DOI] [PubMed] [Google Scholar]
- 6.Downing, D. T., M. E. Stewart, and J. S. Strauss. 1999. Lipids of the epidermis and the sebaceous glands, p. 144-155. In I. M. Freedberg, A. Z. Eisen, K. Wolff, K. F. Austen, L. A. Goldsmith, S. I. Katz, and T. B. Fitzpatrick (ed.), Fitzpatrick's dermatology in general medicine, 5th ed. McGraw-Hill, New York, N.Y.
- 7.Faergemann, J. 2002. Atopic dermatitis and fungi. Clin. Microbiol. Rev. 15:545-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Guého, E., G. Midgley, and J. Guillot. 1996. The genus Malassezia with description of four new species. Antonie Leeuwenhoek 69:337-355. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, A. K., Y. Kohli, R. C. Summerbell, and J. Faergemann. 2001. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med. Mycol. 39:243-251. [DOI] [PubMed] [Google Scholar]
- 11.Hanifin, J. M., and G. Rajka. 1980. Diagnostic features of atopic dermatitis. Acta Dermato-Venereol. 92:4-47. [Google Scholar]
- 12.Hara, J., K. Higuchi, R. Okamoto, M. Kawashima, and G. Imokawa. 2000. High-expression of sphingomyelin deacylase is an important determinant of ceramide deficiency leading to barrier disruption in atopic dermatitis. J. Investig. Dermatol. 115:406-413. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, M. 1980. A simple method for estimation of evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 14.Koyama, T., T. Kanbe, A. Ishiguro, A. Kikuchi, and Y. Tomita. 2001. Antigenic components of Malassezia species for immunoglobulin E antibodies in sera of patients with atopic dermatitis. J. Dermatol. Sci. 26:201-208. [DOI] [PubMed] [Google Scholar]
- 15.Koyama, T., T. Kanbe, A. Ishiguro, A. Kikuchi, and Y. Tomita. 2000. Isolation and characterization of a major antigenic component of Malassezia globosa to IgE antibodies in sera of patients with atopic dermatitis. Microbiol. Immunol. 44:373-379. [DOI] [PubMed] [Google Scholar]
- 16.Koyama, T., T. Kanbe, A. Kikuchi, and Y. Tomita. 2002. Effects of topical vehicles on growth of the lipophilic Malassezia species. J. Dermatol. Sci. 29:166-170. [DOI] [PubMed] [Google Scholar]
- 17.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makimura, K., Y. S. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungal species by polymerase chain reaction (PCR). J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 19.Metzgar, D., D. Field, R. Haubrich, and C. Wills. 1998. Sequence analysis of a compound coding-region microsatellite in Candida albicans resolves homoplasies and provides a high-resolution tool for genotyping. FEMS Immunol. Med. Microbiol. 20:103-109. [DOI] [PubMed] [Google Scholar]
- 20.Mochizuki, T., M. Kawasaki, H. Ishizaki, R. Kano, A. Hasegawa, H. Tosaki, and M. Fujihiro. 2001. Molecular epidemiology of Arthroderma benhamiae, an emerging pathogen of dermatophytoses in Japan, by polymorphisms of the non-transcribed spacer region of the ribosomal DNA. J. Dermatol. Sci. 27:14-20. [DOI] [PubMed] [Google Scholar]
- 21.Nakabayashi, A., Y. Sei, and J. Guillot. 2000. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med. Mycol. 38:337-341. [DOI] [PubMed] [Google Scholar]
- 22.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 23.Sugita, T., A. Nishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugita, T., R. Ikeda, and T. Shinoda. 2001. Diversity among strains of Cryptococcus neoformans var. gattii as revealed by a sequence analysis of multiple genes and a chemotype analysis of capsular polysaccharide. Microbiol. Immunol. 45:757-768. [DOI] [PubMed] [Google Scholar]
- 25.Sugita, T., H. Suto, T. Unno, R. Tsuboi, H. Ogawa, T. Shinoda, and A. Nishikawa. 2001. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J. Clin. Microbiol. 39:3486-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugita, T., M. Nakajima, R. Ikeda, T. Matsushima, and T. Shinoda. 2002. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. J. Clin. Microbiol. 40:1826-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugita, T., M. Takashima, T. Shinoda, H. Suto, T. Unno, R. Tsuboi, H. Ogawa, and A. Nishikawa. 2002. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J. Clin. Microbiol. 40:1363-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Belkum, A. 1999. Short sequence repeats in microbial pathogenesis and evolution. Cell Mol. Life Sci. 56:729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, G. C.-Y., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto, A., S. Serizawa, M. Ito, and Y. Sato. 1991. Stratum corneum lipid abnormalities in atopic dermatitis. Arch. Dermatol. Res. 283:219-223. [DOI] [PubMed] [Google Scholar]