Abstract
Ovine PBMCs (peripheral blood mononuclear cells) express PrPC [cellular PrP (prion-related protein)] and have the potential to harbour and release disease-associated forms of PrP during scrapie in sheep. Cell-surface PrPC expression by PBMCs, together with plasma PrPC levels, may contribute to the regulatory mechanisms that determine susceptibility and resistance to natural scrapie in sheep. Here, we have correlated cell-surface PrPC expression on normal ovine PBMCs by FACS with the presence of PrPC in plasma measured by capture–detector immunoassay. FACS showed similar levels of cell-surface PrPC on homozygous ARR (Ala136-Arg154-Arg171), ARQ (Ala136-Arg154-Gln171) and VRQ (Val136-Arg154-Gln171) PBMCs. Cell-surface ovine PrPC showed modulation of N-terminal epitopes, which was more evident on homozygous ARR cells. Ovine plasma PrPC levels showed genotypic variation and the protein displayed C-terminal epitopes not available in cell-surface PrPC. Homozygous VRQ sheep showed the highest plasma PrPC level and homozygous ARR animals the lowest. For comparison, similar analyses were performed on normal bovine PBMCs and plasma. PrPC levels in bovine plasma were approx. 4-fold higher than ovine homozygous ARQ plasma despite similar levels of PBMC cell-surface PrPC expression. Immunoassays using C-terminal-specific anti-PrP monoclonal antibodies as capture and detector reagents revealed the highest level of PrPC in both ovine and bovine plasma, whilst lower levels were detected using N-terminal-specific monoclonal antibody FH11 as the capture reagent. This suggested that a proportion of plasma PrPC was N-terminally truncated. Our results indicate that the increased susceptibility to natural scrapie displayed by homozygous VRQ sheep correlates with a higher level of plasma PrPC.
Keywords: blood, epitope, immunoassay, polymorphism, cellular prion-related protein (PrPC), transmissible spongiform encephalopathy (TSE)
Abbreviations: ARQ, Ala136-Arg154-Gln171; ARR, Ala136-Arg154-Arg171; VRQ, Val136-Arg154-Gln171; BCA, bicinchoninic acid; BSE, bovine spongiform encephalopathy; CJD, Creutzfeldt–Jakob disease; vCJD, variant CJD; CNS, central nervous system; GPI, glycosylphosphatidylinositol; PBMC, peripheral blood mononuclear cell; PrP, prion-related protein; PrPC, cellular PrP; PrPSc, scrapie PrP; TSE, transmissible spongiform encephalopathy
INTRODUCTION
Prion diseases, such as scrapie in sheep, BSE (bovine spongiform encephalopathy) in cattle and CJD (Creutzfeldt–Jakob disease) in humans, are transmissible chronic neurodegenerative disorders. These diseases are characterized by the accumulation of PrPSc [scrapie PrP (prion-related protein)], an abnormal isomer of the host protein PrPC (cellular PrP). The two isomers of PrP are covalently identical but differ in secondary structure. PrPC is predominantly α-helical (42%) with little β-sheet (3%), whereas PrPSc has considerably more β-sheet content (43%) and a similar α-helical content (30%) [1–3]. These observations indicate that during conversion of PrPC into PrPSc, a major refolding event occurs that results in a more extensive β-sheet conformation. The protein-only hypothesis postulates that the transmissible prion agent consists solely of proteinaceous material [4]. Consequently, it is proposed that PrPSc forms part, or all, of the infectious prion agent and that this abnormal isomer is responsible for the modification of the normal cellular form, PrPC. Recombinant PrP refolded under oxidizing conditions yields predominantly α-helical protein, whereas refolding under reducing conditions generates a form with a higher β-sheet content [5,6]. The β-sheet form of recombinant PrP displays characteristics similar to PrPSc, which include partial resistance to proteolytic digestion and the propensity to form insoluble amorphous aggregates [7]. Recently, a β-rich form of mouse recombinant PrP (amino acid residues 89–230) has been shown to be infectious in mice that overexpress this protein [8,9].
The major polymorphisms in ovine PrP associated with differences in susceptibility to natural scrapie in sheep occur in the C-terminal portion of the molecule at amino acid residues 136, 171 and, to a lesser extent, 154. VRQ (Val136-Arg154-Gln171) or ARQ (Ala136-Arg154-Gln171) animals show susceptibility to scrapie, while those that express ARR (Ala136-Arg154-Arg171) show resistance [10,11]. All three polymorphic sites are located within, or close to, that region of PrP that undergoes the major conformational change associated with conversion of PrPC into PrPSc during prion disease [12]. Our computational modelling of ovine PrP shows that A136V results in an increase in the β-sheet content of PrP [13]. In addition, a hydrogen bond is seen between Gln171 and Arg167 that is not present in the ARR allele. The resultant loss of β-strand length and absence of a hydrogen bond between residues 171 and 167 collectively result in the loss of stability of the β-sheet region and probably lead to a loss in the potential for β-sheet formation in the Arg171 allele. This is also suggested by our recent observations, which show that after copper treatment of ovine PrP, the VRQ allelic form displays a greater increase in β-sheet content, while the ARR allelic form remains relatively structurally unchanged [14]. These results suggest that polymorphisms in ovine PrP not only affect the stability of the molecule but also its amyloidogenic potential [15,16].
The main site of PrPC protein expression occurs in the CNS (central nervous system) and to a lesser extent the peripheral lymphoid system. Prion infectivity and PrPSc may accumulate at both of these sites during the progression of prion disease. In natural scrapie of sheep the oral route is believed to be the main portal of entry of the infectious agent [17]. Following oral exposure to TSE (transmissible spongiform encephalopathy)-infected material, prion infectivity [18,19] and PrPSc [20,21] can be detected in gastrointestinal lymphoid tissue. However, the rapid accumulation of PrPSc in other gut and non-gut lymphoid tissues during scrapie disease has suggested lymphatic and haematogenous spread of prions [22]. As a consequence, peripheral blood is regarded as a possible reservoir of prion infectivity in scrapie-infected sheep and other TSE-affected individuals. Recent evidence has shown that prion disease can be transmitted through transfusion of whole blood, or buffy coat, from natural scrapie-infected, or BSE-experimentally infected sheep, into recipient sheep [23,24]. Similarly, prion infectivity has been detected in whole blood and in buffy coat from mice infected with a human-derived strain of vCJD (variant CJD) during both the preclinical and clinical phases of disease [25]. The presence of detectable infectivity in blood of experimentally infected animals, coupled with the observations that PrPSc can be detected in peripheral lymphoid tissue of human patients with vCJD, has reinforced concerns that human blood supplies may be contaminated with prion infectivity [26]. As a consequence, there is considerable interest in determining the biology of blood PrP.
PBMCs (peripheral blood mononuclear cells) of sheep [27,28] and other species [29] express PrPC on their cell surface, which has the potential for conversion into a disease-associated form of PrP. These cells therefore have the potential to carry, or harbour, and release disease-associated forms of PrP during prion disease, thereby contributing to the deposition of PrPSc on resident lymphoid tissue cells such as follicular dendritic cells. Cell-surface PrPC expression by PBMCs together with plasma PrPC levels may be regulatory determinants of susceptibility and resistance to natural scrapie in sheep. We have recently shown that PrPC expressed on the surface of ovine PBMCs displays conformational variation between scrapie-susceptible and -resistant genotypes, and between different susceptible allelic variants [30]. In our studies reported here, we have correlated the level of cell-surface PrPC on blood cells from scrapie-free homozygous ARR, ARQ and VRQ sheep with the level of PrPC found in plasma of these animals. Our results have shown for the first time that genotypic differences exist regarding levels of ovine plasma PrPC. Plasma from homozygous VRQ sheep was characterized by higher levels of PrPC than plasma from homozygous ARR and ARQ sheep. Since ovine VRQ PrP has a high potential to form β-sheet structure, we speculate that the elevated levels of PrPC seen in plasma from homozygous VRQ sheep contribute to the greater susceptibility to scrapie seen in this ovine genotype.
MATERIALS AND METHODS
Generation of ovine and bovine recombinant PrP
Recombinant PrP was purified from BL21(DE3)pLysS bacteria transformed with the prokaryotic expression vector pET-23b (Novagen) that contained the open reading frame-coding sequence of full-length ovine ARR, ARQ or VRQ PrP (amino acid residues 25–232), or bovine PrP (amino acid residues 25–240) in a method adapted from Hornemann et al. [31]. Briefly, transformed bacteria were grown at 37 °C in Luria–Bertani medium supplemented with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol, and induced overnight with 1 mM isopropyl β-D-thiogalactoside. Bacteria were harvested by centrifugation at 4000 g for 15 min at 4 °C, resuspended in 20 mM Tris/HCl, 50 mM NaCl, 1 mM EDTA, 0.1 mM PMSF and 200 μg/ml lysozyme and incubated at 37 °C for 1 h. DNase was added to 10 μg/ml and incubated for a further 1 h, deoxycholic acid was then added to 1 mg/ml and incubation continued for a final 1 h. Samples were centrifuged at 13000 g for 20 min and the inclusion body pellet was resuspended in a buffer consisting of 8 M urea and 20 mM Tris/HCl (pH 8.0) supplemented with 2-mercaptoethanol at 14.3 mM. The soluble fraction collected after centrifugation at 13000 g for 30 min was applied to a nickel-ion-charged Sepharose column (Chelating Sepharose Fast Flow; Amersham Biosciences). The column was washed with 1 column volume of 20 mM Tris/HCl (pH 8.0), 8 M urea, 200 mM NaCl, 10 mM imidazole and then an excess of 20 mM Tris/HCl (pH 8.0) and 8 M urea. PrP protein was eluted with 20 mM Tris/HCl and 8 M urea (pH 4.5) and reduced with 10 mM dithiothreitol. PrP was further purified by application to a cation-exchange column [SP-Sepharose (sulfopropyl-Sephadex) Fast Flow; Amersham Biosciences] and eluted with 20 mM Tris/HCl and 9 M urea containing 200 mM NaCl. Eluted PrP was oxidized using copper sulfate (five times molar concentration of PrP) and refolded by dialysis into 50 mM sodium acetate buffer (pH 5.5) containing 10 mM EDTA, followed by extensive dialysis into the same buffer without EDTA. Recombinant PrP proteins were verified by MS to confirm the correct protein sequence and the presence of a disulfide bond. Oxidized and refolded recombinant PrP was stored at −80 °C.
Anti-PrP monoclonal antibodies
The N-terminal-specific monoclonal antibody FH11 [32] was purchased from the TSE Resource Centre (Institute for Animal Health, Compton, Berks., U.K.). The N-terminal-specific monoclonal antibody SAF32 [33] was purchased from SPI-Bio. Monoclonal antibody 6H4 [34] was a gift from Prionics. The N-terminal-specific anti-PrP monoclonal antibody T325 was generated from Prnp−/− mice immunized with mouse brain homogenate [35]. The C-terminal-specific anti-PrP monoclonal antibodies A516, V24 and V26 were generated from Prnp−/− mice immunized with ovine recombinant PrP [35]. Monoclonal antibody 245 was generated from Prnp−/− mice immunized with copper-refolded full-length murine recombinant PrP (amino acid residues 23–231) [36]. Biotinylated monoclonal antibodies were prepared for use as detector antibodies as follows. One milligram of each purified monoclonal antibody was washed four times with 50 mM sodium bicarbonate buffer (pH 9.3) using YM-30 concentrators (Fisher UK, catalogue no. FDR-563-020L). The non-diffusible material was then labelled overnight at 4 °C using N,N-dimethylformamide (Sigma, catalogue no. D-4551) and biotinamidocaproate N-hydroxysuccinimide ester (Sigma, catalogue no. B-2643). Biotinylated antibodies were purified by adsorption on to the membranes of YM-30 concentrators and isolated using elution buffer (50 mM Tris/HCl, pH 7.8, 0.9% sodium chloride and 0.1% sodium azide, pH 7.8). The immunoreactivity of biotinylated monoclonal antibodies was tested by direct ELISA using ovine recombinant PrP as antigen, prior to storage at 4 °C.
Isolation of ovine PBMCs and platelet-free plasma
Peripheral blood from 6-month-old lambs and 3.5–4-year-old female Cheviot sheep (New Zealand scrapie-free flock; ADAS, Mepal, Cambs., U.K.) or from 8-year-old normal cattle (ADAS) was collected into EDTA tubes by venepuncture from live animals, transported on ice and routinely stored at 4 °C overnight. A buffy coat was prepared by centrifugation at 1400 g for 20 min at 4 °C and the harvested cells were layered on to NycoPrep™ Animal (density 1.077 g/ml; osmolarity 265 mOsmol), and centrifuged at 600 g for 15 min at 20 °C. Mononuclear cells were recovered from the density medium interface and washed three times in FACS buffer (PBS containing 1% heat-inactivated foetal calf serum plus 0.1% sodium azide) prior to immunofluorescence staining. Plasma samples were centrifuged at 1400 g for 15 min at 20 °C to remove all cellular debris, aliquoted and stored at −80 °C before use.
Immunofluorescence staining of PBMCs
Cell-surface phenotype was assessed using aliquots of 1×106 cells incubated with anti-PrP monoclonal antibody culture supernatant, purified anti-PrP monoclonal antibody used at 5 μg/ml, or normal mouse serum at 1:1000 as control, for 20 min at 4 °C followed by three washes in FACS buffer and incubation with goat anti-mouse IgG–biotin (Sigma, catalogue no. B-7264) at 1:1000 or goat anti-mouse IgG1–biotin (Caltag, catalogue no. M32115), at 1:500, for 20 min at 4 °C. Cells were washed three times with FACS buffer and subsequently incubated with 0.25 μg of streptavidin–phycoerythrin (Pharmingen, catalogue no. 554061) for 20 min at 4 °C. Cells were finally washed three times with FACS buffer and analysed for cell-surface fluorescence using a FACSCalibur® (Becton Dickinson, Mountain View, CA, U.S.A.). Ten thousand cells were analysed per sample with dead cells excluded on the basis of forward and side light scatter.
Western blot analysis of ovine PBMCs and brain material
Cell lysates were prepared by rapid thawing of scrapie-free PBMC pellets followed by resuspension in either ice-cold PBS or lysis buffer (10 mM Tris/HCl, pH 8.0, 10 mM EDTA, 100 mM NaCl, 0.5% Nonidet P40 and 0.5% sodium deoxycholate) containing 4 units/ml benzonase (Sigma, catalogue no. E-1014). For Western blot analysis of ovine PBMCs, cell lysates were subjected to SDS/PAGE run under reducing conditions (1×106 cells/track) and subsequently transferred on to nitrocellulose membranes by semi-dry blotting. For Western blot analysis of scrapie-free ovine brain material, cerebellum tissue was homogenized in ice-cold PBS (pH 7.4). Then, 20 μg of total protein was resolved by SDS/PAGE run under reducing conditions and subsequently transferred to nitrocellulose membranes by semi-dry blotting. Membranes were blocked overnight at 4 °C with TBS-T (10 mM Tris/HCl, pH 7.8, 100 mM NaCl and 0.05% Tween 20) containing 5% (w/v) non-fat milk, and subsequently washed three times with TBS-T. Membranes were incubated with purified SAF32 (250 ng/ml) for 2 h at 20 °C and then washed five times with TBS-T. This was followed by incubation with goat anti-mouse IgG–horseradish peroxidase (Sigma, catalogue no. A-3673) at 1:2000 for 2 h at 20 °C and five washes with TBS-T. All the antibody dilutions were prepared in 1% non-fat milk in TBS-T. PrP bands were detected by enhanced chemiluminescence (ECL®; Amersham Biosciences).
Direct ELISA
Recombinant PrP protein was coated on to 96-well flat-bottomed plates for 16 h at 4 °C. Excess protein was removed and wells were blocked with PBS containing 5% non-fat milk for 90 min at 20 °C. Plates were washed three times with PBS-T (PBS containing 0.1% Tween 80). Purified anti-PrP monoclonal antibodies (FH11, SAF32, A516 or V24) at 1.0 μg/ml (diluted in PBS) were added to the plates and incubated for 1 h at 20 °C followed by three washes with PBS-T. Goat anti-mouse IgG–biotin (Sigma, catalogue no. B-7264) at 1:3000 was added for 1 h at 20 °C followed by three washes with PBS-T. Avidin–alkaline phosphatase (Sigma, catalogue no. A-7294) at 1:3000 dilution was added for 1 h at 20 °C. Plates were washed three times in PBS-T and once with ELISA buffer (0.05 M glycine, 0.03 M NaOH and 0.25 mM each of ZnCl2 and MgCl2) before addition of the substrate p-nitrophenyl phosphate (Sigma, catalogue no. N-2765) at 0.5 mg/ml in ELISA buffer for up to 1 h at 20 °C. ELISA plates were read at 415 nm on a Bio-Rad 680 microplate reader.
Capture–detector immunoassay
Capture antibody (FH11, A516 or 6H4) was coated routinely at 1.0 μg/well in 96-well flat-bottomed plates for 16 h at 4 °C. Excess antibody was removed and wells were blocked with PBS containing 5% non-fat milk for 2 h at 20 °C. Plates were washed three times with PBS-T and appropriate dilutions of either ovine or bovine plasma, or ovine or bovine recombinant PrP diluted in a buffer (10 mM Tris/HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, 0.5% Nonidet P40 and 0.5% sodium deoxycholate) were captured for 1 h at 20 °C. Plates were washed three times with PBS-T and captured PrP was detected by biotinylated monoclonal antibody (245, V24 or V26) at 50 ng/well for 1 h at 20 °C. Avidin–alkaline phosphatase (Sigma, catalogue no. A-7294) at 1:3000 dilution was added for 1 h at 20 °C. Plates were washed three times in PBS-T and once with ELISA buffer (0.05 M glycine, 0.03 M NaOH and 0.25 mM each of ZnCl2 and MgCl2) before addition of the substrate p-nitrophenyl phosphate (Sigma, catalogue no. N-2765) at 0.5 mg/ml in ELISA buffer for up to 90 min at 20 °C. ELISA plates were read at 415 nm on a Bio-Rad 680 microplate reader.
Protein quantification
Total protein concentration in plasma was measured using the BCA (bicinchoninic acid) assay (Pierce). Samples were initially diluted in PBS (pH 7.4), at 1:50, 1:100 and 1:200 dilutions plus a diluent-only blank. A 2-fold dilution series of BSA was prepared ranging from 800 μg/ml down to 0 μg/ml to act as a standard. Each dilution was dispensed into triplicate wells in a 96-well flat-bottomed plate (5 μl/well). An aliquot of 95 μl of BCA working reagent (prepared by mixing 1 part solution A and 49 parts solution B) was added to each well and the plates were incubated for 90 min at 20 °C. Absorbance was read at 570 nm using a Bio-Rad 680 microplate reader. Protein concentration was estimated by calculation from the standard curve using Microplate Manager software (Bio-Rad).
Statistical analysis
Statistical analysis of the data was performed by one-way ANOVA together with Tukey HSD (honestly significant difference) for post hoc analysis. Analyses were conducted using the SPSS v.13 package.
Nomenclature
Amino acid residue numbers refer to the ovine PrP sequence unless otherwise stated.
RESULTS
Reactivity of anti-PrP monoclonal antibodies with ovine and bovine PrP
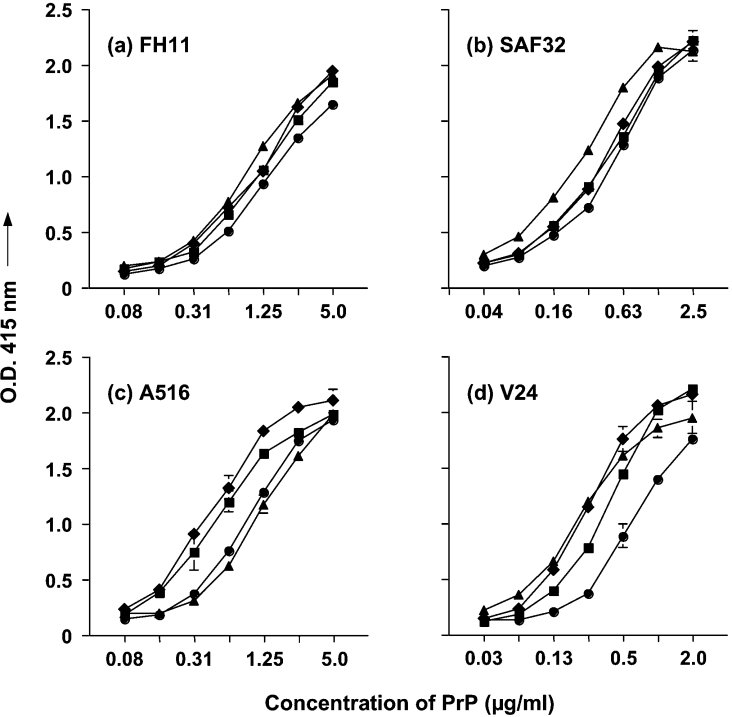
We have utilized a panel of N-terminal- and C-terminal-specific anti-PrP monoclonal antibodies in order to investigate the expression of PrPC on ovine and bovine blood cells, and the level of this protein in plasma. ELISA using ovine and bovine recombinant PrP attached directly to plastic plates was carried out in order to show the reactivity of the anti-PrP monoclonal antibodies with the two species forms of prion protein. Figure 1 shows that the N-terminal-specific monoclonal antibodies FH11 and SAF32, together with C-terminal-specific monoclonal antibodies A516 and V24, all reacted efficiently with ovine recombinant ARR, ARQ and VRQ, and also bovine recombinant PrP. Ovine recombinant VRQ protein was least well recognized by the C-terminal-specific monoclonal antibodies and this may reflect the compact nature of the C-terminal domain of this protein and a relative inaccessibility of epitopes within this region of the molecule [13]. Bovine recombinant PrP was recognized less efficiently than ovine PrP by the C-terminal-specific monoclonal antibody A516, which may reflect a conformational or genotypic difference between the two ruminant forms of prion protein in the binding region of this antibody [35].
Figure 1. Immunological detection of recombinant PrP by direct ELISA.
Full-length ovine recombinant ARR (■), ARQ (◆), VRQ (●) or bovine (▲) recombinant PrP was coated directly on to ELISA plates as described in the Materials and methods section. Recombinant PrP was detected by 1.0 μg/ml purified monoclonal antibody (a) FH11, (b) SAF32, (c) A516 and (d) V24. Results shown are means±S.D. for triplicate wells.
The predominant form of ovine blood cell PrPC is di-glycosylated
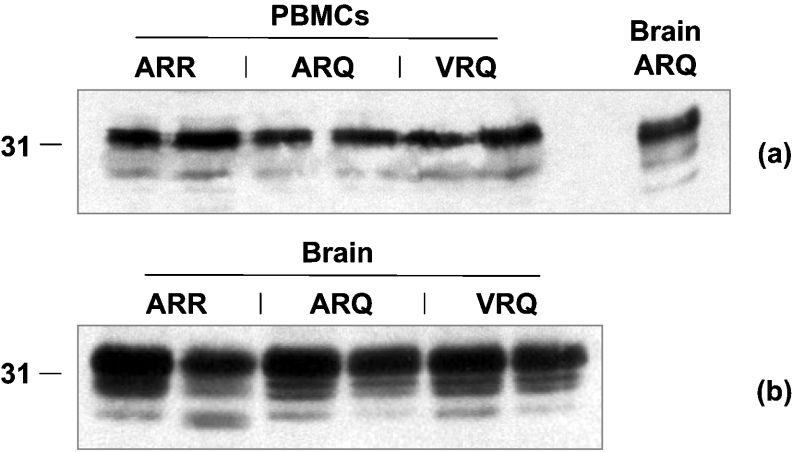
In order to investigate the level of PrPC associated with ovine blood cells we first performed a Western blot analysis of PBMCs from homozygous ARR, ARQ and VRQ sheep using the N-terminal-specific anti-PrP monoclonal antibody SAF32. Figure 2(a) shows that blood cells from all three PrP genotypes of sheep expressed similar levels of total PrPC. In addition, in all cases the predominant form of the protein was the di-glycosylated moiety. Normal brain homogenate from an ARQ homozygous sheep was used to compare the relative levels of each of the glycoform types of PrPC. Comparison of the PrPC glycoform band patterns showed that the un-glycosylated form of PrPC was less evident in blood cells than in CNS material. However, these results show that blood cells from sheep do express significant levels of PrPC protein, which is a potential source of plasma PrPC. In addition, there does not appear to be any significant genotypic difference in the level of total PrPC protein expression by ovine blood cells. Figure 2(b) shows that there does not appear to be any significant genotypic difference in the level of total PrPC protein expression by ovine brain tissue.
Figure 2. Western blot detection of ovine PrPC.
(a) PBMCs (including homozygous ARQ brain material as a positive control) or (b) brain tissue from homozygous ARR, ARQ or VRQ sheep were prepared as described in the Materials and methods section and analysed by SDS/PAGE and Western blotting using monoclonal antibody SAF32. Molecular-mass marker (kDa) is shown on the left-hand side.
Reactivity of N-terminal anti-PrP monoclonal antibodies with cell-surface ovine PrPC
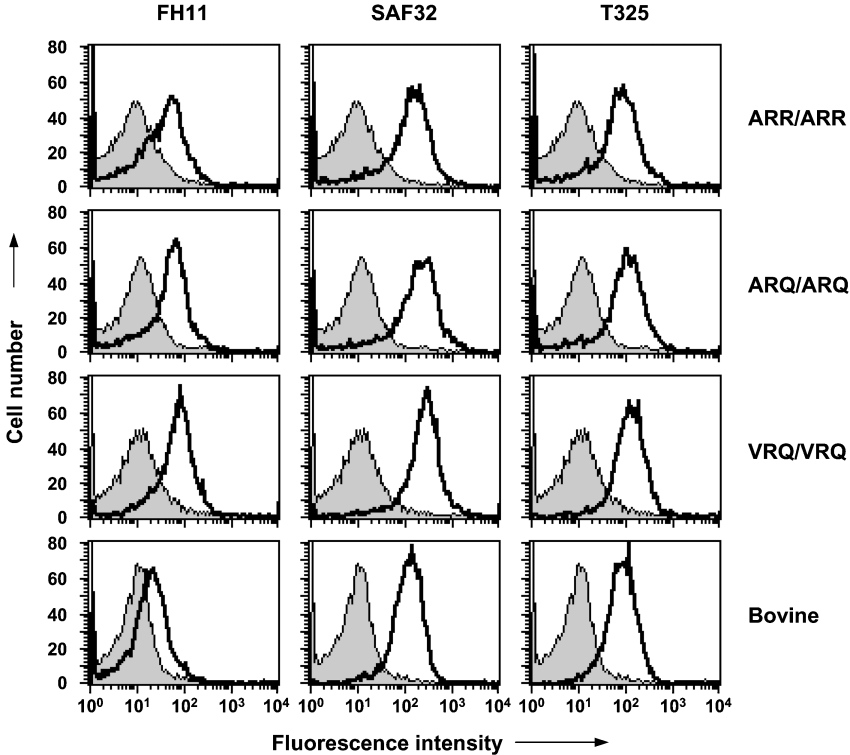
In order to correlate prion protein expression on ovine blood cells with that present in plasma we first investigated the expression of PrPC on PBMCs from scrapie-free sheep by FACS. We investigated PrPC expression with N-terminal-specific anti-PrP monoclonal antibodies since there is genotypic heterogeneity in the C-terminal region of cell-surface ovine PrPC. Figure 3 shows representative FACS profiles for PBMCs from homozygous ARR, ARQ or VRQ sheep reacted with the N-terminal-specific monoclonal antibodies FH11, SAF32 and T325. Cells from all three genotypes of sheep tested typically showed a monophasic FACS profile, although occasionally a biphasic profile was seen with some of the N-terminal-specific monoclonal antibodies. Figure 3 also shows that bovine PBMCs displayed monophasic FACS profiles with monoclonal antibodies FH11, SAF32 and T325. The most noticeable difference between the ruminant PBMCs was the low reactivity of bovine PBMCs with monoclonal antibody FH11 compared with that seen by ovine cells.
Figure 3. Reactivity of N-terminal-specific anti-PrP monoclonal antibodies with ovine and bovine PBMCs.
Cell-surface PrPC expression was analysed by FACS with N-terminal-specific anti-PrP monoclonal antibodies as described in the Materials and methods section. Profiles shown are representative of five out of five sheep. Shaded peak represents control fluorescence; black line represents FH11, SAF32 or T325 fluorescence.
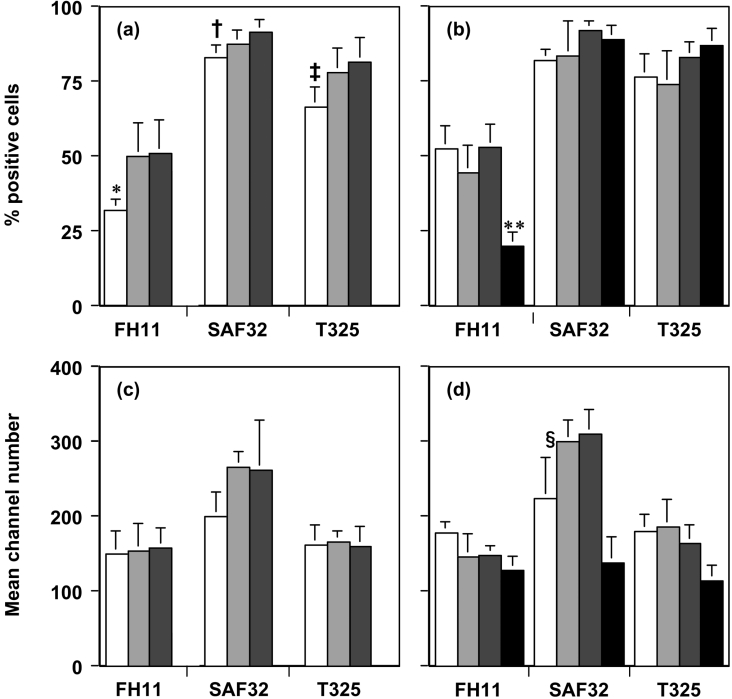
Figure 4(a) shows the percentages of lamb PBMCs that were detected by the N-terminal-specific monoclonal antibodies FH11, SAF32 and T325. All three N-terminal antibodies tested were able to detect some differences between the three allelic variants of cell-surface ovine PrPC. Monoclonal antibody FH11 showed the least reactivity with PBMCs from young lambs from all three ovine PrP genotypes, while SAF32 and T325 showed similar percentages (Figure 4a). FH11 did demonstrate a significant difference between the ARR samples in comparison with the ARQ and VRQ samples (P<0.05) and there were significant differences between the ARR and the VRQ data (P<0.05) using both SAF32 and T325 when PBMCs were tested from young lambs. Figure 4(b) shows that the N-terminal-specific monoclonal antibodies reacted with similar percentages of cells from all the genotypes of ovine PBMCs from adult sheep. The level of cellular reactivity on bovine PBMCs using FH11 was significantly less (P<0.001) than the reactivity shown with SAF32 and T325 (Figure 4b). The epitope recognized by monoclonal antibody FH11 is sited around residues 54–58 [37], while SAF32 binds the sequence QPHGGW located between residues 59 and 89 [33] and monoclonal antibody T325 binds around residues 47–90 [30]. One reason for the decreased reactivity of monoclonal antibody FH11 with both ovine and bovine PBMCs is that truncation of cell-surface PrP may occur with the subsequent loss of the epitope for this particular antibody. Collectively, these results show that scrapie-susceptible and -resistant genotypes of ovine PBMCs and bovine PBMCs express cell-surface PrPC with freely accessible epitopes within the flexible N-terminal region of the protein. There was no significant difference in fluorescence intensity between the three ovine allelic variants using monoclonal antibodies FH11, SAF32 and T325 when tested on PBMCs from lambs (Figure 4c) or adult sheep and bovine PBMCs (Figure 4d). However, adult ARR sheep PBMCs gave significantly less fluorescence (P<0.05) in comparison with ARQ and VRQ when tested with SAF32 (Figure 4d).
Figure 4. Quantification of N-terminal PrPC FACS analysis.
(a, b) Percentage of positive cells (means±S.D.) and (c, d) mean channel number (means±S.D.) are shown for PBMCs from homozygous ARR (white bar) (n=5); ARQ (light grey bar) (n=5) and VRQ (dark grey bar) (n=5) lambs (a, c) or PBMCs from homozygous ARR (white bar) (n=3); ARQ (light grey bar) (n=5) and VRQ (dark grey bar) (n=4) adult sheep (b, d) or bovine PBMCs (black bar) (n=8) (b, d) stained with N-terminal-specific anti-PrP monoclonal antibodies FH11, SAF32 or T325. *P<0.05 (comparison with ARQ and VRQ); †P<0.05 (comparison with VRQ); ‡P<0.05 (comparison with VRQ); **P<0.001 (comparison with SAF32 and T325); §P<0.05 (comparison with ARQ and VRQ).
Buried epitopes in the C-terminal region of ovine and bovine cell-surface PrPC
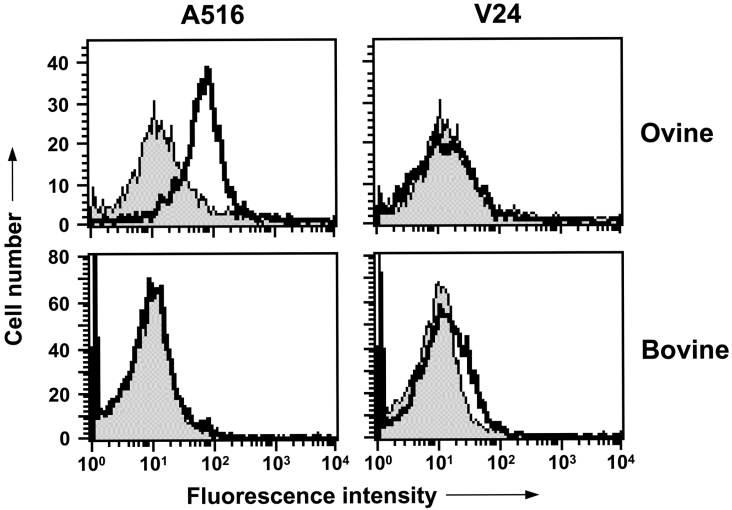
The structure of the C-terminal globular domain of ovine and bovine PrP is predicted to comprise three α-helices, inter-dispersed by a short antiparallel β-sheet region similar to that described for other species forms of PrP [13,16,38–40]. There is a close association between helix-1, the C-terminal region of helix-2 and the N-terminal region of helix-3, and this central core is bound by an intra-molecular disulfide bond. The ordered structure of the C-terminal region of PrPC may lead to epitopes being buried or obscured in the native cell-surface form of the protein. This was investigated by FACS analysis with the C-terminal-specific monoclonal antibodies A516, V24 and V26.
The data in Figure 5 show that monoclonal antibody A516, which binds in the region of helix-1 [35], reacted efficiently with ovine cell-surface PrPC. However, despite the fact that ovine PBMCs expressed significant levels of cell-surface PrPC, monoclonal antibody V24 failed to react with cells from homozygous ARR, ARQ or VRQ sheep. Monoclonal antibody V26 also failed to react with ovine PBMCs (results not shown). Monoclonal antibodies A516 and V24 both failed to bind to bovine cell-surface PrPC (Figure 5) as did monoclonal antibody V26 (results not shown). The failure of monoclonal antibodies V24 and V26 to bind to cell-surface PrPC supports the view that specific epitopes in the C-terminal region of cell-surface PrPC are buried or obscured, which may be a consequence of homodimeric or heterodimeric structures of PrP with other molecules [41]. The epitope for monoclonal antibody V26 is located in the C-terminal region of ovine PrP around residues 217–232 [30]. The failure of this antibody to react with cell-surface PrPC may be due to steric hindrance by the GPI (glycosylphosphatidylinositol) anchor.
Figure 5. Reactivity of C-terminal-specific anti-PrP monoclonal antibodies with ovine and bovine PBMCs.
Cell-surface PrPC expression was analysed by FACS with C-terminal-specific anti-PrP monoclonal antibodies as described in the Materials and methods section. Profiles shown are representative of five out of five homozygous VRQ sheep and eight out of eight cattle. Shaded peak represents control fluorescence; black line represents A516 or V24 fluorescence.
Quantification of ruminant plasma PrPC levels
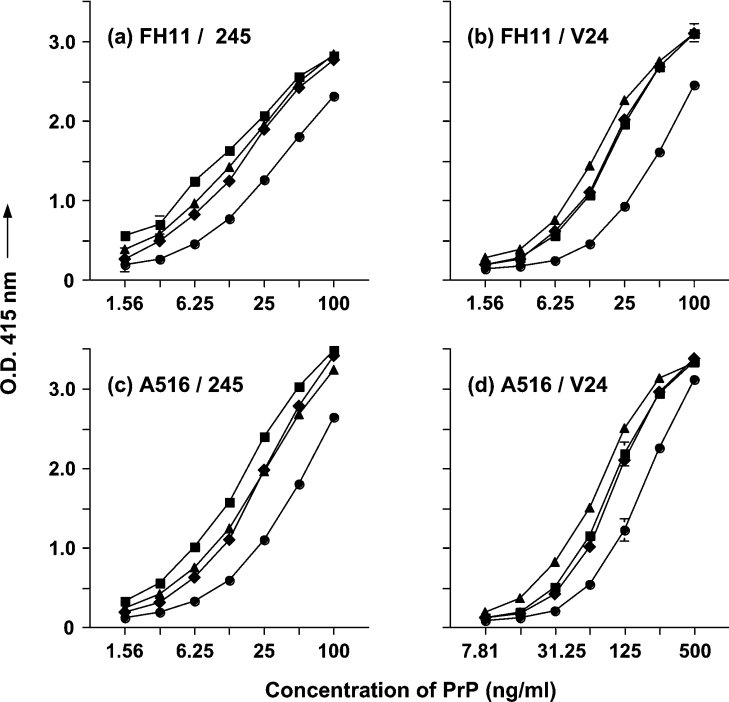
In order to quantify the level of PrPC present in the plasma of sheep and cattle we developed a sensitive capture–detector immunoassay that utilized different pairs of anti-PrP monoclonal antibodies. The monoclonal antibodies were reactive with either the N-terminal or C-terminal regions of the ovine or bovine prion protein, which would allow detection of N-terminal intact and truncated PrPC. Figure 6 shows that the combination of the N-terminal-specific monoclonal antibody FH11 and the C-terminal-specific antibody 245 (Figure 6a) or V24 (Figure 6b) efficiently recognized ovine recombinant ARR, ARQ and VRQ PrP, and bovine recombinant PrP. With respect to ovine PrP, recombinant ARR and ARQ were recognized more efficiently than VRQ recombinant protein. This may reflect the more compact nature of the ovine PrP VRQ compared with ARQ and ARR protein. Similar data were obtained when the C-terminal-specific monoclonal antibody A516 was used as the capture antibody with either antibody 245 (Figure 6c) or V24 (Figure 6d) as detector. Furthermore, monoclonal antibody V26 was effective as the detector antibody with either FH11 or A516 as the capture antibody (results not shown). The sensitivity of the capture–detector immunoassay was in the order of approx. 1 nM of recombinant PrP.
Figure 6. Capture–detector immunoassay reactivity of monoclonal antibodies with recombinant PrP.
ELISA plates were coated with capture antibody FH11 (a, b) or A516 (c, d) as described in the Materials and methods section. Full-length ovine recombinant ARR (■), ARQ (◆), VRQ (●) or bovine (▲) recombinant PrP were detected by biotinylated monoclonal antibody 245 (a, c) or V24 (b, d). Results shown are means±S.D. for triplicate wells.
In order to determine the level of PrPC in ovine or bovine plasma, EDTA-treated blood was collected from New Zealand-derived scrapie-free sheep or normal cattle. Platelet-free plasma was prepared by centrifugation and PrPC was measured by capture–detector immunoassay using the different combinations of anti-PrP monoclonal antibodies described above. The quantity of PrPC in plasma was calculated from the absorbance readings obtained from the immunoassay by reference to the genotype-specific standard curves as shown in Figure 6.
Tables 1a and 1b show the levels of PrPC detected in the plasma from adult and young scrapie-free homozygous ARR, ARQ and VRQ sheep. Homozygous VRQ sheep showed the highest level of plasma PrPC, which was 61.2 ng/ml for adult sheep (Table 1a) and 63.0 ng/ml for young lambs (Table 1b), when assessed using the C-terminal-specific monoclonal antibody A516 for capture and monoclonal antibody V24 as the detector reagent. Lower levels of plasma PrPC were seen in homozygous ARR and ARQ sheep, which were less than 50% of that seen in both homozygous VRQ adult sheep and young lambs. Similar trends were seen when monoclonal antibody A516 was used for capture with antibody V26 as the detector reagent (results not shown). The level of plasma PrPC in homozygous ARR sheep was significantly less than that detected in homozygous VRQ sheep when assessed by monoclonal antibody A516 for capture in combination with monoclonal antibody V24 (Table 1a) or V26 (results not shown) as detector (P<0.001 and P=0.006, respectively). These results show that the level of ovine plasma prion protein shows genotypic variation.
Table 1. Quantification of PrPC in plasma from sheep and lambs.
Quantification of ovine plasma PrPC levels by capture–detector ELISA. Plasma PrPC levels in homozygous ARR, ARQ or VRQ adult sheep (a) or young lambs (b) were determined as described in the Materials and methods section. Capture monoclonal antibodies were FH11 or A516 and detector monoclonal antibodies were biotinylated V24 or 245. The results shown are means±S.D. for five animals analysed for each genotype and age group.
| (a) | |||
|---|---|---|---|
| Plasma PrPC (ng/ml) | |||
| Antibody pair | ARR/ARR | ARQ/ARQ | VRQ/VRQ |
| FH11/V24 | 3.0±4.0 | 6.6±3.4 | 31.8±6.7* |
| FH11/245 | 3.6±0.8 | 6.4±2.2 | 5.8±3.0† |
| A516/V24 | 13.8±2.6 | 19.8±13.1 | 61.2±11.6‡ |
| A516/245 | 5.4±0.6 | 16.8±10.0 | 29.6±11.6†§ |
| (b) | |||
| Plasma PrPC (ng/ml) | |||
| Antibody pair | ARR/ARR | ARQ/ARQ | VRQ/VRQ |
| FH11/V24 | 17.2±3.0 | 11.4±2.3 | 39.4±1.4* |
| FH11/245 | 0.1±0.2† | 0.2±0.4† | 5.4±1.2† |
| A516/V24 | 38.4±10.7 | 22.2±10.1 | 63.0±31.4∥ |
| A516/245 | <0.1† | 0.8±1.3† | 9.8±1.3† |
*P≤0.003 (comparison with ARR and ARQ).
†P≤0.008 (comparison with monoclonal antibody V24 as detector).
‡P<0.001 (comparison with ARR and ARQ).
§P=0.005 (comparison with ARR).
∥P=0.019 (comparison with ARQ).
When the level of ovine plasma PrPC was measured using the N-terminal-specific antibody FH11 for capture, significantly less PrPC protein was detected compared with that seen when the C-terminal-specific monoclonal antibody A516 was used. When monoclonal antibody FH11 was used for capture and monoclonal antibody V24 as the detector reagent, the level of VRQ plasma PrPC was found to be 31.8 ng/ml (Table 1a) and 39.4 ng/ml (Table 1b) in adult and young sheep respectively. This was approx. 50% less than that seen when monoclonal antibody A516 was used for capture. Furthermore, PrPC levels were similarly reduced in plasma from homozygous ARR and ARQ sheep when monoclonal antibody FH11 was used for capture. These observations suggest that a substantial portion of plasma PrPC is truncated in the region of the octapeptide repeat domain in plasma from all three PrP genotypes of sheep tested. The level of plasma PrPC in homozygous ARR sheep was significantly less than that detected in homozygous VRQ sheep when assessed by monoclonal antibody FH11 for capture in combination with monoclonal antibody V24 (P=0.003) (Table 1a). Similar trends were seen when monoclonal antibody FH11 was used for capture with monoclonal antibody V26 as the detector reagent (results not shown).
When monoclonal antibody FH11 or A516 was used for capture with monoclonal antibody 245 as the detector, generally lower levels of PrPC protein were detected compared with that seen when monoclonal antibody V24 was used as detector reagent. Monoclonal antibody 245 recognizes un-glycosylated and mono-glycosylated PrPC but does not react with di-glycosylated protein unlike monoclonal antibodies V24 and V26, which react with all three glycoforms of PrPC ([36]; and A.M. Thackray, L. Hopkins and R. Bujdoso, unpublished work). These results are therefore consistent with the Western blot shown in Figure 2 that showed that the predominant form of PrPC in the PBMCs of sheep is di-glycosylated protein and that un-glycosylated and mono-glycosylated bands are relatively minor molecular species.
Similar levels of total protein were present in plasma from homozygous ARR, ARQ and VRQ sheep (96.5±12.1, 88.1±12.6 and 96.2±13.7 ng/ml respectively; n=5; values are means±S.D.). This supports the view that the difference in plasma PrPC levels between these three ovine genotypes was a specific feature of prion protein metabolism and not a consequence of genotypic variation in plasma total protein level.
Bovine plasma PrPC levels were assessed using monoclonal antibody FH11 or 6H4 for capture and monoclonal antibody V24 or V26 as detector, and compared with that seen in plasma from homozygous ARQ sheep. The data in Table 2 show that the level of bovine plasma PrPC was approx. 4-fold greater than that seen in ovine ARQ plasma (P<0.001). Lower levels of bovine and ovine plasma PrPC were detected when monoclonal antibody FH11 was used as capture reagent instead of monoclonal antibody 6H4. This indicated that in both ovine and bovine plasma, a significant proportion of PrPC is N-terminally truncated. These results also show that the C-terminal epitopes recognized by monoclonal antibodies V24 and V26, which are buried or obscured in both ovine and bovine cell-surface PrPC, are accessible within PrPC in plasma.
Table 2. Quantification of PrPC in plasma from ovine and bovine blood.
Quantification of bovine plasma PrPC levels by capture–detector immunoassay. Bovine and homozygous ovine ARQ plasma PrPC levels were determined as described in the Materials and methods section. Capture monoclonal antibodies were 6H4 or FH11 and detector monoclonal antibodies were biotinylated V24 or V26. The results are means±S.D. (n≥5). *P≤0.001 (comparison with ovine homozygous ARQ).
| Plasma PrPC (ng/ml) | ||
|---|---|---|
| Antibody pair | ARQ/ARQ | Bovine |
| FH11/V24 | 8.2±7.8 | 24.3±3.5* |
| 6H4/V24 | 49.2±25.1 | 198.0±10.3* |
| 6H4/V26 | 46.2±21.2 | 270.6±13.6* |
DISCUSSION
In the present study, we have investigated the level of PrPC expression by blood cells from sheep of different PrP genotypes and have correlated this with the level of PrPC in plasma in order to further our understanding on the biology of ruminant blood PrP. Similar percentages of PBMCs from scrapie-susceptible and -resistant sheep expressed cell-surface PrPC and did so with similar fluorescence intensity when assessed with N-terminal-specific anti-PrP monoclonal antibodies. Less than 2-fold variations in cell-surface fluorescence intensity were seen between PBMCs from homozygous ARR, ARQ and VRQ sheep using a panel of N-terminal-specific anti-PrP monoclonal antibodies. Some genotypic variation in young animals was seen when monoclonal antibody FH11 was used to analyse cell-surface PrPC expression. In addition, PBMCs from homozygous ARR, ARQ and VRQ sheep all showed less reactivity with monoclonal antibody FH11 compared with that seen with monoclonal antibodies SAF32 and T325. This phenomenon is similar to that reported by Halliday et al. [42]. The epitope for monoclonal antibody FH11 is located closer to the N-terminus of the PrP molecule compared with the epitopes recognized by monoclonal antibodies SAF32 and T325, which are situated within the octapeptide repeat region of the molecule. The failure of monoclonal antibody FH11 to react with similar percentages of cells compared with monoclonal antibodies SAF32 and T325 may be a consequence of truncation of cell-surface PrPC. Truncated PrP molecules have been described on the surface of neuroblastoma cells [43]. Alternatively, the intact N-terminal region of cell-surface PrPC may show genotypic conformational variation as suggested previously [35]. PBMCs from young homozygous ARR lambs showed significantly less reactivity with monoclonal antibody FH11 compared with cells from homozygous VRQ lambs. This could imply that amino acid residue 136 of ovine PrPC may influence the folding of the N-terminal region. For example, on occasions PBMCs from Ala136 genotypes showed a biphasic FACS profile when reacted with N-terminal-specific anti-PrP monoclonal antibodies, whereas cells from the Val136 genotype showed a single uniform profile on each occasion.
Epitopes within the C-terminal domain of cell-surface ovine and bovine PrPC were also inaccessible to anti-PrP monoclonal antibody binding. While monoclonal antibody A516, which binds in the C-terminal region of helix-1, reacted with ovine PBMCs, monoclonal antibody V24 failed to react with either ovine or bovine PBMCs. Monoclonal antibody V24 appears to bind in the C-terminal region of helix-2, although peptide mapping has so far failed to identify its specific epitope (A. M. Thackray and R. Bujdoso, unpublished work). In addition, monoclonal antibody V26, which binds to the C-terminal region of ovine PrP (amino acid residues 217–232) [30] failed to bind to PrPC expressed on ovine or bovine PBMCs. PrPC is linked to the plasma membrane via a GPI anchor attached to the C-terminal end of the molecule. This may prevent binding of monoclonal antibody V26 to its epitope in native cell-surface PrPC as a consequence of steric hindrance. We have previously shown that the region between β-strand-2 and residue 171 was buried or obscured in cell-surface PrPC on PBMCs from scrapie-susceptible and -resistant sheep [35]. Interestingly, the region between β-strand-2 and residue 171, and the proposed epitope for monoclonal antibody V26 contain tyrosine dimers, in the form of YYR (amino acid residues 165–167) and YYQ (amino acid residues 228–230). These motifs are reportedly buried in native PrPC and more solvent-exposed in PrPSc [44]. The reactivity of monoclonal antibody A516 with ovine but not bovine cell-surface PrPC may reflect a distinct conformation of native bovine PrPC or a genotypic difference between the two ruminant forms of prion protein. Monoclonal antibody A516 appears to bind to an epitope located wholly, or in part, within amino acid residues 150–164 of ovine PrP, which incorporates helix-1. The only difference between the amino acid sequence of ovine and bovine PrPC in this particular region of the PrP protein is a tyrosine→histidine substitution at ovine PrP residue 148. Since monoclonal antibody A516 does bind to bovine recombinant PrP, this substitution would appear to have more influence on conformation in the native form of the molecule.
Although PBMCs from homozygous ARR, ARQ and VRQ sheep showed similar levels of total and cell-surface PrPC, geno-typic differences were seen in the level of PrPC present in plasma. The highest levels of ovine plasma PrPC were seen in homozyg-ous VRQ sheep and the lowest level was seen in plasma from homozygous ARR animals. PrPC levels in bovine plasma were approx. 4-fold higher than ovine ARQ/ARQ plasma despite similar levels of PBMC cell-surface PrPC expression. The highest level of plasma PrPC was detected in all three ovine PrP genotypes and in bovine plasma by capture–detector immunoassay using C-termi-nal-specific anti-PrP monoclonal antibodies as both capture and detector reagent. Interestingly, monoclonal antibodies V24 and V26, which both failed to react with cell-surface PrPC, were able to react with PrPC in both ovine and bovine plasma. This suggests that there are significant differences in the epitope repertoire between cell-surface and soluble PrPC, at least in the C-terminal region of the molecule. Lower levels of plasma PrPC were detected using monoclonal antibody FH11 as the capture reagent, which correlated with the reduced reactivity of this monoclonal antibody with cell-surface PrPC. At least two possibilities exist for the increased level of plasma PrPC in homozygous VRQ sheep compared with the Ala136 genotypes, in particular homozygous ARR animals. Firstly, there may be increased expression of PrPC protein at one or more tissue sites in homozygous VRQ animals. This was not found to be the case in ovine brain tissue, which is a major site of PrPC expression. Secondly, VRQ PrPC could be more resistant to metabolic breakdown than the ARQ and ARR forms of protein. Our own data and results obtained by others have shown that VRQ PrP is more compact than ARQ and ARR protein [13,15,16]. This suggests that scrapie-susceptible PrPC may have a longer cellular half-life than disease-resistant PrPC. The fact that the VRQ allelic variant is predisposed to more β-sheet formation compared with ARR [14] renders the protein more susceptible to accumulate in the cell in a disease-associated form. This would support the view that differences in the metabolism of allelic variants of ovine PrP contribute to the mechanism(s) that determine susceptibility and resistance of sheep to natural scrapie [15,16]. The finding that plasma PrPC levels in homozygous VRQ and ARQ sheep were of the order VRQ>ARQ, and that levels in both of these genotypes were elevated above that in homozygous ARR sheep, supports this hypothesis. In such a scheme, the thermodynamically less stable ARR allelic variant would be efficiently metabolized before it was able to accumulate in any significant amount in a disease-associated form. The level of bovine plasma PrPC was significantly greater than that of ovine PrPC. We have not been able to assess tissue expression levels of bovine compared with ovine PrPC and cannot therefore determine whether differences in prion protein synthesis or metabolism account for the different levels of plasma PrPC seen between the two species.
PrPC is attached to the extracellular surface of the plasma membrane by a GPI anchor, clustered within cholesterol- and sphingolipid-rich lipid rafts [45,46]. It still remains to be established how PrPC is released from the cell membrane. It has been proposed that soluble PrPC is produced by cleavage of its GPI anchor by an endogenous mammalian GPI-specific phospholipase [47,48] or by proteolytic cleavage mediated by zinc metalloproteases known as secretases or sheddases [49,50]. In addition, PrPC and PrPSc have been reported to be released from cells in association with exosomes, vesicles of endosomal origin released from cells after the fusion of multivesicular bodies with the cell surface [51]. Many cell types including lymphocytes, macrophages and dendritic cells generate exosomes. Transfer of cell membranes between cells is mediated by exosomes and they are thought to be responsible for the intercellular transfer of PrPC [52]. In our studies reported here, plasma PrPC could be detected by monoclonal antibody V26, which does not react with cell-surface PrPC expressed by scrapie-free PBMCs. This suggests that a significant proportion, if not all, of the ovine plasma PrPC is soluble with a freely accessible C-terminal region. Our results presented here show that there is allelic variation in the amount of ovine plasma PrPC, despite the fact that ovine blood cells appear to show similar levels of total and cell-surface PrPC. In addition, we have shown that a significant level of ovine and bovine plasma PrPC is refractory to detection with N-terminal-specific anti-PrP monoclonal antibodies. Whether this is a feature of disease-associated PrP in plasma from prion-infected individuals remains to be established.
Acknowledgments
This work was supported by grants from the BBSRC (Biotechnology and Biological Sciences Research Council) and from Defra (Department for Environment, Food and Rural Affairs, U.K.). L.H. is in receipt of a Defra PhD studentship. We thank Dr C.R. Birkett and the TSE Resource Centre for monoclonal antibody FH11.
References
- 1.Caughey B. W., Dong A., Bhat K. S., Ernst D., Hayes S. F., Caughey W. S. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 2.Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E., et al. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pergami P., Bramanti E., Ascoli G. A. Structural dependence of the cellular isoform of prion protein on solvent: spectroscopic characterization of an intermediate conformation. Biochem. Biophys. Res. Commun. 1999;264:972–978. doi: 10.1006/bbrc.1999.1430. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 5.Lu B. Y., Chang J. Y. Isolation of isoforms of mouse prion protein with PrPSC-like structural properties. Biochemistry. 2001;40:13390–13396. doi: 10.1021/bi011111t. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H., Stockel J., Mehlhorn I., Groth D., Baldwin M. A., Prusiner S. B., James T. L., Cohen F. E. Physical studies of conformational plasticity in a recombinant prion protein. Biochemistry. 1997;36:3543–3553. doi: 10.1021/bi961965r. [DOI] [PubMed] [Google Scholar]
- 7.Baskakov I. V., Legname G., Baldwin M. A., Prusiner S. B., Cohen F. E. Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 2002;277:21140–21148. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 8.Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 9.Legname G., Nguyen H. O., Baskakov I. V., Cohen F. E., Dearmond S. J., Prusiner S. B. Strain-specified characteristics of mouse synthetic prions. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clouscard C., Beaudry P., Elsen J. M., Milan D., Dussaucy M., Bounneau C., Schelcher F., Chatelain J., Launay J. M., Laplanche J. L. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. J. Gen. Virol. 1995;76:2097–2101. doi: 10.1099/0022-1317-76-8-2097. [DOI] [PubMed] [Google Scholar]
- 11.Goldmann W., Hunter N., Smith G., Foster J., Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 12.Bossers A., Belt P., Raymond G. J., Caughey B., de Vries R., Smits M. A. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujdoso R., Burke D. F., Thackray A. M. Structural differences between allelic variants of the ovine prion protein revealed by molecular dynamics simulations. Proteins. 2005;61:840–849. doi: 10.1002/prot.20755. [DOI] [PubMed] [Google Scholar]
- 14.Wong E., Thackray A. M., Bujdoso R. Copper induces increased beta-sheet content in the scrapie-susceptible ovine prion protein PrPVRQ compared with the resistant allelic variant PrPARR. Biochem. J. 2004;380:273–282. doi: 10.1042/BJ20031767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rezaei H., Choiset Y., Eghiaian F., Treguer E., Mentre P., Debey P., Grosclaude J., Haertle T. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J. Mol. Biol. 2002;322:799–814. doi: 10.1016/s0022-2836(02)00856-2. [DOI] [PubMed] [Google Scholar]
- 16.Eghiaian F., Grosclaude J., Lesceu S., Debey P., Doublet B., Treguer E., Rezaei H., Knossow M. Insight into the PrPC→PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10254–10259. doi: 10.1073/pnas.0400014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadlow W. J., Kennedy R. C., Race R. E. Natural infection of Suffolk sheep with scrapie virus. J. Infect. Dis. 1982;146:657–664. doi: 10.1093/infdis/146.5.657. [DOI] [PubMed] [Google Scholar]
- 18.Kimberlin R. H., Hall S. M., Walker C. A. Pathogenesis of mouse scrapie. Evidence for direct neural spread of infection to the CNS after injection of sciatic nerve. J. Neurol. Sci. 1983;61:315–325. doi: 10.1016/0022-510x(83)90165-x. [DOI] [PubMed] [Google Scholar]
- 19.Kimberlin R. H., Walker C. A. Pathogenesis of scrapie in mice after intragastric infection. Virus Res. 1989;12:213–220. doi: 10.1016/0168-1702(89)90040-3. [DOI] [PubMed] [Google Scholar]
- 20.Beekes M., McBride P. A. Early accumulation of pathological PrP in the enteric nervous system and gut-associated lymphoid tissue of hamsters orally infected with scrapie. Neurosci. Lett. 2000;278:181–184. doi: 10.1016/s0304-3940(99)00934-9. [DOI] [PubMed] [Google Scholar]
- 21.Maignien T., Lasmezas C. I., Beringue V., Dormont D., Deslys J. P. Pathogenesis of the oral route of infection of mice with scrapie and bovine spongiform encephalopathy agents. J. Gen. Virol. 1999;80:3035–3042. doi: 10.1099/0022-1317-80-11-3035. [DOI] [PubMed] [Google Scholar]
- 22.Heggebo R., Press C. M., Gunnes G., Ulvund M. J., Tranulis M. A., Lsverk T. Detection of PrPSc in lymphoid tissues of lambs experimentally exposed to the scrapie agent. J. Comp. Pathol. 2003;128:172–181. doi: 10.1053/jcpa.2002.0625. [DOI] [PubMed] [Google Scholar]
- 23.Houston F., Foster J. D., Chong A., Hunter N., Bostock C. J. Transmission of BSE by blood transfusion in sheep. Lancet. 2000;356:999–1000. doi: 10.1016/s0140-6736(00)02719-7. [DOI] [PubMed] [Google Scholar]
- 24.Hunter N., Foster J., Chong A., McCutcheon S., Parnham D., Eaton S., MacKenzie C., Houston F. Transmission of prion diseases by blood transfusion. J. Gen. Virol. 2002;83:2897–2905. doi: 10.1099/0022-1317-83-11-2897. [DOI] [PubMed] [Google Scholar]
- 25.Cervenakova L., Yakovleva O., McKenzie C., Kolchinsky S., McShane L., Drohan W. N., Brown P. Similar levels of infectivity in the blood of mice infected with human-derived vCJD and GSS strains of transmissible spongiform encephalopathy. Transfusion. 2003;43:1687–1694. doi: 10.1046/j.0041-1132.2003.00586.x. [DOI] [PubMed] [Google Scholar]
- 26.Llewelyn C. A., Hewitt P. E., Knight R. S., Amar K., Cousens S., Mackenzie J., Will R. G. Possible transmission of variant Creutzfeldt–Jakob disease by blood transfusion. Lancet. 2004;363:417–421. doi: 10.1016/S0140-6736(04)15486-X. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann L. M., Davis W. C., Knowles D. P., Wardrop K. J., Sy M. S., Gambetti P., O'Rourke K. I. Cellular prion protein is expressed on peripheral blood mononuclear cells but not platelets of normal and scrapie-infected sheep. Haematologica. 2001;86:146–153. [PubMed] [Google Scholar]
- 28.Holada K., Simak J., Risitano A. M., Maciejewski J., Young N. S., Vostal J. G. Activated platelets of patients with paroxysmal nocturnal hemoglobinuria express cellular prion protein. Blood. 2002;100:341–343. doi: 10.1182/blood.v100.1.341. [DOI] [PubMed] [Google Scholar]
- 29.Holada K., Vostal J. G. Different levels of prion protein (PrPc) expression on hamster, mouse and human blood cells. Br. J. Haematol. 2000;110:472–480. doi: 10.1046/j.1365-2141.2000.02158.x. [DOI] [PubMed] [Google Scholar]
- 30.Thackray A. M., Ryder S. J., Bujdoso R. Modification of blood cell PrP epitope exposure during prion disease. Biochem. J. 2005;390:563–571. doi: 10.1042/BJ20050571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornemann S., Korth C., Oesch B., Riek R., Wider G., Wuthrich K., Glockshuber R. Recombinant full-length murine prion protein, mPrP(23–231): purification and spectroscopic characterization. FEBS Lett. 1997;413:277–281. doi: 10.1016/s0014-5793(97)00921-6. [DOI] [PubMed] [Google Scholar]
- 32.Foster J. D., Wilson M., Hunter N. Immunolocalisation of the prion protein (PrP) in the brains of sheep with scrapie. Vet. Rec. 1996;139:512–515. doi: 10.1136/vr.139.21.512. [DOI] [PubMed] [Google Scholar]
- 33.Feraudet C., Morel N., Simon S., Volland H., Frobert Y., Creminon C., Vilette D., Lehmann S., Grassi J. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J. Biol. Chem. 2005;280:11247–11258. doi: 10.1074/jbc.M407006200. [DOI] [PubMed] [Google Scholar]
- 34.Korth C., Stierli B., Streit P., Moser M., Schaller O., Fischer R., Schulz-Schaeffer W., Kretzschmar H., Raeber A., Braun U., et al. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature. 1997;390:74–77. doi: 10.1038/36337. [DOI] [PubMed] [Google Scholar]
- 35.Thackray A. M., Yang S., Wong E., Fitzmaurice T. J., Morgan-Warren R. J., Bujdoso R. Conformational variation between allelic variants of cell-surface ovine prion protein. Biochem. J. 2004;381:221–229. doi: 10.1042/BJ20040351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thackray A. M., Madec J. Y., Wong E., Morgan-Warren R., Brown D. R., Baron T., Bujdoso R. Detection of bovine spongiform encephalopathy, ovine scrapie prion-related protein (PrPSc) and normal PrPc by monoclonal antibodies raised to copper-refolded prion protein. Biochem. J. 2003;370:81–90. doi: 10.1042/BJ20021280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thuring C. M., van Keulen L. J., Langeveld J. P., Vromans M. E., van Zijderveld F. G., Sweeney T. Immunohistochemical distinction between preclinical bovine spongiform encephalopathy and scrapie infection in sheep. J. Comp. Pathol. 2005;132:59–69. doi: 10.1016/j.jcpa.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Haire L. F., Whyte S. M., Vasisht N., Gill A. C., Verma C., Dodson E. J., Dodson G. G., Bayley P. M. The crystal structure of the globular domain of sheep prion protein. J. Mol. Biol. 2004;336:1175–1183. doi: 10.1016/j.jmb.2003.12.059. [DOI] [PubMed] [Google Scholar]
- 39.Hornemann S., Schorn C., Wuthrich K. NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 2004;5:1159–1164. doi: 10.1038/sj.embor.7400297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez Garcia F., Zahn R., Riek R., Wuthrich K. NMR structure of the bovine prion protein. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8334–8339. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leclerc E., Peretz D., Ball H., Solforosi L., Legname G., Safar J., Serban A., Prusiner S. B., Burton D. R., Williamson R. A. Conformation of PrPC on the cell surface as probed by antibodies. J. Mol. Biol. 2003;326:475–483. doi: 10.1016/s0022-2836(02)01365-7. [DOI] [PubMed] [Google Scholar]
- 42.Halliday S., Houston F., Hunter N. Expression of PrPC on cellular components of sheep blood. J. Gen. Virol. 2005;86:1571–1579. doi: 10.1099/vir.0.80561-0. [DOI] [PubMed] [Google Scholar]
- 43.Mishra R. S., Gu Y., Bose S., Verghese S., Kalepu S., Singh N. Cell surface accumulation of a truncated transmembrane prion protein in Gerstmann-Straussler-Scheinker disease P102L. J. Biol. Chem. 2002;277:24554–24561. doi: 10.1074/jbc.M200213200. [DOI] [PubMed] [Google Scholar]
- 44.Paramithiotis E., Pinard M., Lawton T., LaBoissiere S., Leathers V. L., Zou W. Q., Estey L. A., Lamontagne J., Lehto M. T., Kondejewski L. H., et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat. Med. 2003;9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 45.Vey M., Pilkuhn S., Wille H., Nixon R., DeArmond S. J., Smart E. J., Anderson R. G., Taraboulos A., Prusiner S. B. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madore N., Smith K. L., Graham C. H., Jen A., Brady K., Hall S., Morris R. Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J. 1999;18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parizek P., Roeckl C., Weber J., Flechsig E., Aguzzi A., Raeber A. J. Similar turnover and shedding of the cellular prion protein in primary lymphoid and neuronal cells. J. Biol. Chem. 2001;276:44627–44632. doi: 10.1074/jbc.M107458200. [DOI] [PubMed] [Google Scholar]
- 48.Caughey B., Raymond G. J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 49.Moss M. L., Lambert M. H. Shedding of membrane proteins by ADAM family proteases. Essays Biochem. 2002;38:141–153. doi: 10.1042/bse0380141. [DOI] [PubMed] [Google Scholar]
- 50.Parkin E. T., Watt N. T., Turner A. J., Hooper N. M. Dual mechanisms for shedding of the cellular prion protein. J. Biol. Chem. 2004;279:11170–11178. doi: 10.1074/jbc.M312105200. [DOI] [PubMed] [Google Scholar]
- 51.Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 52.Liu T., Li R., Pan T., Liu D., Petersen R. B., Wong B. S., Gambetti P., Sy M. S. Intercellular transfer of the cellular prion protein. J. Biol. Chem. 2002;277:47671–47678. doi: 10.1074/jbc.M207458200. [DOI] [PubMed] [Google Scholar]