Abstract
Cag pathogenicity island-containing Helicobacter pylori (type I) induces signal transduction pathways resulting in tyrosine phosphorylation of proteins adjacent to the site of bacterial adhesion on host gastric epithelial cells. Conventional block PCR-restriction fragment length polymorphism (RFLP) and real-time LightCycler (LC) PCR hybridization assays, validated by direct sequencing, were designed to test for the presence of three nucleotide sequences corresponding to tyrosine phosphorylation motifs (TPMs) A, B, and C in 84 isolates of H. pylori type I from patients in England. Overall, the PCR assays demonstrated that one or more TPMs were present in 62 strains (75%). Motif A was common (71% of strains), whereas motifs B and C were rarer (8% of strains). Strains lacking a TPM were typically vacuolating cytotoxin genotype vacA m2. Motif A was widely distributed in relation to disease severity and was more commonly (but not significantly [P = 0.071]) associated with gastric ulcer than with duodenal ulcer (86 versus 56%). The LC hybridization assay provided a rapid means of detecting all three motifs, but RFLP analysis was more specific for TPM-A. TPMs provide novel additional strain markers for defining cagA variation, including identification of RFLP types within TPM-A. The presence of a particular TPM was not of direct diagnostic value, either singly or in combination, but the higher proportion of TPM-A strains in gastric ulcer patients merits further investigation.
Helicobacter pylori is a gram-negative microaerophilic bacterium that colonizes the human stomach, where it induces chronic gastritis (18). Although not always the cause of clinically manifest disease, H. pylori infection is strongly associated with peptic ulceration, as well as with increased risk for the development of mucosa-associated lymphoid tissue lymphoma and gastric cancer (10, 11, 13). Gastric infections with H. pylori have a worldwide distribution, with current prevalence rates about 35% in England and Wales (32) and more than 75% in many developing countries (26). No single pathogenicity factor has yet been proved to be uniquely associated with the ability of H. pylori to cause gastroduodenal ulcer disease or cancer (6, 11). The presence of the cytotoxin-associated gene (cagA), encoded in the 40-kb cag pathogenicity island (cag PAI), and the presence of signal and midregion alleles of the vacuolating cytotoxin (vacA) gene are markers for type I strains of H. pylori (7). Such strains secrete VacA and exhibit increased interactions with host epithelial cells compared with type II strains, which lack the cag PAI (2). H. pylori exports the highly immunodominant 145-kDa CagA protein via a type IV secretion system encoded by the cag PAI. CagA is then translocated into the gastric epithelial cells, where it induces host cell kinases that phosphorylate tyrosine residues in CagA adjacent to the site of bacterial adhesion on the host gastric epithelial cells, which in turn activate eukaryotic signal transduction pathways and cytoskeletal plasticity (1, 4, 5, 21, 29, 28, 30). Three putative nucleotide tyrosine phosphorylation motifs (TPMs) in the CagA protein have been predicted and designated TPM-A, TPM-B, and TPM-C (21), although additional motifs (12), such as the EPIYA sequences, have also been identified in CagA (31). The precise link among cagA, the production of the CagA protein, and strain virulence is controversial (16, 20, 26). About 68% of strains isolated from dyspeptics in England have cagA present in the genome (24), and similar or higher frequencies have been reported in many other countries (16, 17, 20). Nevertheless, cagA status alone is insufficient to reliably predict either virulence or association with gastric ulcer (6, 15, 19, 25).
In the present study, conventional block PCR and real-time fluorescence PCR hybridization assays were developed for the detection of corresponding nucleotide sequence motifs for cagA TPM-A, -B, and -C and were applied to determine motif prevalence in H. pylori isolates from one locality in England and to test for any general associations with peptic ulcer disease.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Eighty-four isolates of H. pylori that had previously been reported to carry an intact cag PAI (23) were cultured from antral gastric biopsy specimens of dyspeptic patients undergoing routine upper gastrointestinal tract endoscopy (Chelmsford, mid-Essex, United Kingdom). No specific clinical selection criteria were applied for inclusion of patients. Five disease groups of isolates were defined from endoscopic investigation: the duodenal ulcer (DU) group (n = 23 strains), the gastric ulcer (GU) group (n = 14 strains), the DU-plus-GU group (n = 4 strains), the gastric neoplasia (GN) group (n = 2 strains), and the nonulcer dyspepsia (NUD) group (n = 41 strains). H. pylori isolates were cultured on Columbia agar containing 10% (vol/vol) defibrinated horse blood at 37°C under microaerobic conditions (5% O2, 5% CO2, 2% H2, 88% N2) in a Variable Atmosphere Incubator (Don Whitley Scientific, Shipley, Yorkshire, England) for 48 to 72 h. Five reference strains of H. pylori with defined TPMs were included as controls (Table 1).
TABLE 1.
cagA TPM types of H. pylori strains deduced from cagA sequences
| Strain | GenBank accession no. for cagA | TPM typea |
|---|---|---|
| Reference strains | ||
| NCTC 11637 (ATCC 43504) | AF202973 | C |
| NCTC 11916 (ATCC 43526) | AF001357 | C |
| NCTC 11638 | AF282853 | A |
| J99 | AE001483 | None |
| NCTC 12455 (strain 26695) | AE000511 | A, B |
| Other strains | ||
| CAPM N62 | AF249275 | A, B |
| F32 | AF202972 | A, B |
| SS1 | AF247651 | A, B |
| 15818 | AF083352 | A |
According to the work of Odenbreit et al. (22) and J. Puls (personal communication).
Preparation of template DNA.
Genomic DNA was obtained from H. pylori by the cetyltrimethylammonium bromide extraction method described previously (33).
PCR-RFLP assays.
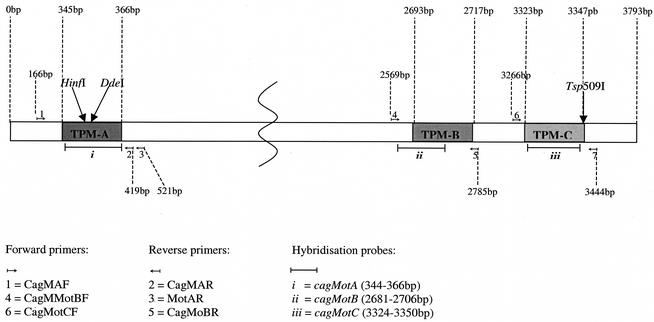
Conventional block cycler PCR-based restriction fragment length polymorphism (PCR-RFLP) assays were developed for detecting TPM-A- and TMP-C-specific sequences in cagA (Fig. 1). Primer pairs MotAF-MotAR2 and MotCF-MotCR (Table 2), for amplification of fragments containing TMP-A and TPM-C, respectively (product sizes, 356 and 179 bp, respectively), were designed by using cagA sequences of the reference strains and additional sequences in GenBank (Table 1). The fragments containing TPM-A and TPM-C were amplified by using standard protocols with the following block cycler conditions: an initial denaturation at 95°C for 5 min; 35 cycles, each consisting of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and elongation at 72°C for 1 min; and a final elongation step of 5 min at 72°C. Amplified fragments were subjected to RFLP analysis to detect the presence of motif-specific cut sites. Following incubation for 4 h at 37°C with HinfI [(G/A)NTC recognition sequence] and DdeI [(C/T)NAG recognition sequence] or at 65°C with Tsp509I [(N/A)ATT recognition sequence], fragments were separated by electrophoresis in 3.0% (wt/vol) agarose.
FIG. 1.
Schematic representation of H. pylori cagA showing locations of TPMs A, B, and C; primer and probe positions for each LC-PCR assay; and primer sites and motif-determining restriction enzyme cut sites for TPM-A and TPM-C RFLP assays. The diagram is not drawn to scale.
TABLE 2.
Oligonucleotide primers and fluorescence probes for identification of cagA TPM types in H. pylori
| TPM type | Assay | Primer or probe designationa | Nucleotide sequence (5′-3′) | Positionb | Product size (bp) | Tm (°C)c |
|---|---|---|---|---|---|---|
| A | PCR | MotAF | GAT AGG GAT AAC AGG CAA G | 166 | 356 | 54 |
| MotAR | CCT GCA AAA GAT TGT TTG GC | 521 | 56 | |||
| A | Real-time PCR | MotAF | GAT AGG GAT AAC AGG CAA G | 166 | 254 | 54 |
| MotAR2 | GTG TTG ATT TTA GAC GGA TC | 419 | 54 | |||
| CagMotA | Cy5-GAA ATT TGG GGA TCA GCG TTA C-biotin | 62 | ||||
| B | PCR and real-time PCR | MotBF | AAC CCT AGT CGG TAA TGG | 2569 | 216 | 52 |
| MotBR | GCA ACT TGA GCG TAA ATG G | 2784 | 54 | |||
| CagMotB | Cy5-AAT AAT GGA CTC AAA AAC AGC ACA GA-biotin | 68 | ||||
| C | PCR and real-time PCR | MotCF | CAA GCG GTA TCA GAA GCT A | 3266 | 179 | 54 |
| MotCR | TTA ATG CGT GTG TGG CTG TT | 3444 | 56 | |||
| CagMotC | Cy5-AGC TCA AAG ATT CTA CAA AAT ACA AT-biotin | 64 |
Probes are italicized.
Positions of primers in the cagA gene of strain 26695.
Predicted annealing temperature.
Oligonucleotide primers and fluorescently labeled probes for real-time PCR hybridization assays.
Real-time PCR assays for the LightCycler (LC) instrument (Roche Diagnostics Ltd., Lewes, East Sussex, England) were designed to detect each TPM according to principles described previously (14). The oligonucleotide primers and probes (Table 2) (for locations in cagA, see Fig. 1) were designed following multiple alignment of nine cagA sequences in GeneBase (version 1.0; Applied Maths, Kortrijk, Belgium). Bi-probes CagMotA, CagMotB, and CagMotC were designed to be exactly complementary to motifs in the respective reference strains (Table 1) and were labeled with the fluorescent dye Cy5 at the 5′ end and biotinylated at the 3′ end. Real-time PCR was performed as follows: 2 μl of DNA (10 ng/μl) was added to an 18-μl reaction mixture containing 200 μM each deoxynucleoside triphosphate, 50 mM Tris-HCl, 3 mM MgCl2, SYBR Green 1 diluted 1/10,000 (Bio/Gene Ltd.), 2 pmol of the forward primer (Table 2), 10 pmol of the reverse primer, 5 pmol of the probe, and 0.8 U of Platinum Taq DNA polymerase. TPM-containing fragments were amplified in the LC by denaturation (95°C for 10 s), followed by 50 cycles of denaturation (94°C), annealing (49 to 53°C for TPM-A, 48 to 51°C for TPM-B, and 52 to 56°C for TPM-C, all with a slow temperature transition rate of 3°C/s between the two temperatures), and extension (72°C for 10 s).
Nucleotide sequence analysis of amplicons.
After block cycler PCR amplification, cagA amplicons from 37 strains were purified and then sequenced in-house by using the CEQ Dye Terminator Cycle Sequencing Quick Start kit (Beckman Coulter Inc., High Wyckam, England). In addition, TPM-A products of four isolates were sequenced commercially. Chromatograms were checked in Chromas (version 1.42; Griffith University, Queensland, Australia), and sequences were aligned and analyzed in GeneBase.
vacA genotyping.
Vacuolating cytotoxin genotyping assays for vacA midregion (m1 and m2) alleles were performed by using the primers and PCR conditions described previously (3, 24).
Statistical analysis.
Fisher's exact test, the chi-square test, odds ratios with 95% confidence intervals (95% CI), and P values were determined (Epi-Info program, version 6). A P value below 0.05 was considered significant.
RESULTS
Detection of cagA TPM-A.
Amplification and melting conditions were optimized for the LC TPM-A assay, and the 254-bp fragment was subjected to probe hybridization melting point analysis. The predicted melting temperature (Tm) was based on a probe sequence with full complementarity to the template DNA. Tms for each reference strain are listed in Table 2. Reference strain DNA for each motif was included in analyses to correct for minor run-to-run variations in the Tms. The observed Tm for probe cagMotA in the LC-PCR assay for reference strains containing TPM-A ranged from 60.4 to 63.3°C (mean, 62.2°C). The Tms for reference strains lacking TPM-A varied between 55.6 and 60.3°C (mean, 58.0°C). A Tm of ≥62°C, corrected relative to the reference DNA samples, was used as the cutoff for a TPM-A-positive PCR product.
For identification by PCR-RFLP, the 356-bp fragment containing TPM-A, defined by the amino acid motif KFGDGRY located at site 122, was digested with HinfI to target a cut site within the asparagine residue that resulted in fragments of either 199, 87, and 70 bp (type H3) (Fig. 2A) or 199, 70, 58, and 29 bp (type H2). By contrast, HinfI restriction of PCR products lacking the TPM-A sequence resulted in DNA fragments of either 167, 102, and 87 bp (type H4) or 167, 102, 58, and 29 bp (type H5). Digestion with DdeI generated fragments of either 316 and 40 bp (RFLP type D6) or no digestion (356 bp) (type D2) (Fig. 2B) for TPM-A positive strains, while most TPM-A negative strains produced fragments of 165, 151, and 40 bp (type D1) or 191 and 165 bp (type D7) (Fig. 2B). The presence of other fragments, attributed to one or more additional mutations at the cut sites, was interpreted as absence of TPM-A.
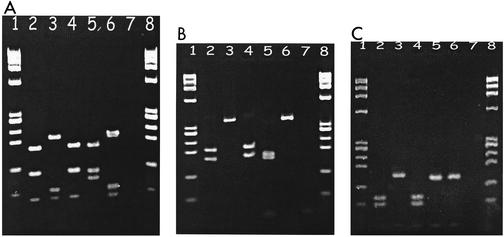
FIG. 2.
Examples of PCR-RFLP analysis of cagA TPM regions to detect the presence of motifs A and C in H. pylori isolates from mid-Essex. (A) HinfI digestion of the 356-bp amplicon separated by agarose gel electrophoresis to detect the presence of TPM-A. Lanes: 1 and 8, X174 HaeIII ladder; 2, NCTC 11637 (TPM-A negative, RFLP type H5); 3, NCTC 11638 (TPM-A positive, RFLP type H3); 4, ATCC 43526 (TPM-A negative, RFLP type H5); 5, J99 (TPM-A negative, RFLP type H4); 6, strain 26695 (TPM-A positive, RFLP type H3); 7, negative control. (B) DdeI digestion of the 365-bp amplicon to detect the presence of TPM-A. Lanes: 1, 7, and 8, as for panel A; 2, NCTC 11637 (TPM-A negative, RFLP type D7); 3, NCTC 11638 (TPM-A positive, RFLP type D2); 4, ATCC 43526 (TPM-A negative, RFLP type D7); 5, J99 (TPM-A negative, RFLP type D1); 6, strain 26695 (TPM-A positive, RFLP type D2). (C) Tsp509I digestion of the 179-bp amplicon to detect the presence of TPM-C. Lanes: 1, 7, and 8, as for panel A; 2, NCTC 11637 (TPM-C positive, RFLP type T3); 3, NCTC 11638 (TPM-C negative, RFLP type T1); 4, ATCC 43526 (TPM-C positive, RFLP type T3); 5, J99 (TPM-C negative, RFLP type T1); 6, strain 26695 (TPM-C negative, RFLP type T1).
Overall, TPM-A was detected by at least one of the assays in 60 isolates (71%), either singly (64%) or in combination (7%) (Table 3). NCTC 11638, NCTC 12455, and 52 strains of H. pylori were TPM-A positive by all three assays. Four isolates were negative by the LC-PCR assay but positive by both PCR-RFLP assays. Product amplification failed in the LC-PCR assay of one isolate but gave a positive result in both PCR-RFLP assays, while another strain yielded no PCR product for RFLP analysis but was identified as TPM-A positive by the LC-PCR assay. There was good concordance between the HinfI and DdeI assays except that two strains were negative in the latter assay. The remaining 24 strains and 3 reference strains (NCTC 11637, NCTC 11916, and J99) were negative for TPM-A by the LC-PCR assay and by the HinfI RFLP assay. Nineteen strains for which partial DdeI digests were observed were recorded as negative for TPM-A.
TABLE 3.
Distribution of cagA TPMs in H. pylori isolates in relation to patient disease
| Gastric disease (n) | No. of H. pylori isolates with:
|
||||||
|---|---|---|---|---|---|---|---|
| No motif | Single motifs
|
Multiple motifs
|
|||||
| A | B | C | A, B | A, C | A, B, C | ||
| GU (14) | 2 | 10 | 2 | ||||
| DU (23) | 9 | 13 | 1 | ||||
| GU + DU (4) | 1 | 3 | |||||
| GN (2) | 2 | ||||||
| NUD (41) | 11 | 26 | 2 | 1 | 1 | ||
| Total (84) | 23 | 54 | 1 | 2 | 3 | 1 | |
The different RFLP profiles predicted from inspection of GenBank sequences provided a measure of the variation within the motif A region. For example, there were two different HinfI profiles (types H2 and H3) for TPM-A-positive strains and four different HinfI RFLP profiles (types H1 and H4 to 6) for TPM-A-negative strains. Likewise, there were three DdeI profiles (types D2, D4, and D6) for TPM-A-positive strains and five DdeI profiles (types D1, D3, D5, D7, and D8) for TPM-A-negative strains. Most (90%) of the TPM-A-positive strains had the same combined DdeI/HinfI profile (D2H2), which was also a feature of NCTC 11638 and strain 26695, while the remaining strains were represented by three other combined profiles. The TPM-A-negative strains, by contrast, were more diverse, with 12 combined profiles.
Sequence analysis of PCR products of selected strains was performed to assess PCR assay specificities. The presence of TPM-A in two strains that were positive by both LC-PCR and PCR-RFLP was confirmed. Furthermore, fragments of cagA from 26 strains that were TPM-A negative by the LC-PCR hybridization assay were confirmed as negative.
Detection of cagA TPM-B.
Amplification and melting conditions were optimized for the TPM-B assay, and the 216-bp fragment containing TPM-B was amplified, followed by probe hybridization melting point analysis. The mean observed Tm for probe cagMotB in the LC-PCR for NCTC 12455 was 65.9°C. The mean observed Tms for reference strains lacking TPM-B varied between 52.2 and 60.9°C, and because the Tm was predicted to be in the range of 42.0 to 66.0°C, an observed Tm of ≥65.9°C was defined as the cutoff for a positive TPM-B assay. Overall, the LC-PCR assay detected TPM-B in H. pylori reference strain NCTC 12455 and in three clinical isolates (4%). A specific RFLP assay for TPM-B corresponding to the amino acid sequence motif KNSTEPIY at amino acid site 899 could not be designed due to the high degree of nucleotide sequence diversity evident from the alignment of different cagA sequences. Sequence analysis confirmed the presence of TPM-B in two of the LC-PCR-positive strains, whereas four other strains that generated equivocal Tms lacked TPM-B.
Detection of cagA TPM-C.
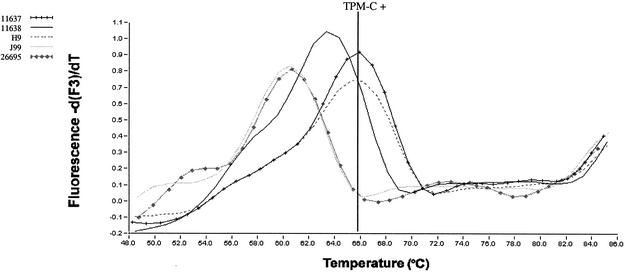
The 179-bp fragment containing TPM-C was amplified by LC-PCR, followed by probe hybridization melting point analysis. The observed Tms of probe cagMotC of TPM-C-containing control strains ranged from 64.6 to 66.3°C (mean Tm, 66.0°C). Observed Tms for reference strains lacking TPM-C were between 53.0 and 62.0°C, so a Tm of ≥66.0°C was used as the cutoff for a TPM-C-positive strain (Fig. 3). The LC-PCR hybridization assay detected TPM-C in four isolates.
FIG. 3.
Example of melting curve analysis performed in the LC instrument to detect the presence of TPM-C in H. pylori cagA. Amplification was monitored by single fluorescent acquisition (F1 channel gains set at 20) at the end of each extension. Probe hybridization melting point analysis was then performed after denaturation (94°C for 10 s) by continuous measurement of Cy5 fluorescence (F3 channel gains set at 50) over a temperature gradient of 43 to 90°C (temperature transition rate, 0.2°C/s).
For detection of TPM-C, defined by the amino acid motif KLKDSTKY located at amino acid site 1029, the Tsp509I RFLP analysis of the 175-bp amplicon was performed to cut just downstream of the tyrosine residue. The predicted fragment sizes were 82, 66, and 32 bp (type T3) (Fig. 2C) when TPM-C was present and either 139, 32, and 8 bp (type T1) (Fig. 2C) or 147 and 32 bp (type T2) when TPM-C was absent. In practice, the Tsp509I assay generated two profiles for positive strains (T3 and T4) and two profiles for negative strains (T1 and T2). Most TPM-C-positive strains had profile T3 (13 strains), although one T3 profile was interpreted as a mixture of TPM-C-positive and -negative forms. TPM-C-negative strains were either T1 (11 strains) or T2 (46 isolates).
Sequence analyses confirmed the presence of TPM-C in the two isolates that were positive by both assays and in one of the two isolates that generated an equivocal LC-PCR assay result. PCR-RFLP suggested that TPM-C was present in an additional nine isolates, but LC hybridization did not confirm this, nor did sequence analysis of five of these samples. All 70 remaining strains were negative for TPM-C by both PCR assays.
Overall, TPM-C was confirmed in five strains (6%) either singly or in combination (Table 3).
Distribution of cagA TPMs by disease group.
TPM-A was widely distributed among isolates regardless of disease symptoms, whereas TPM-B and TPM-C were rare as single motifs. No associations of TPMs with disease symptoms were evident (Table 3). Although the prevalence of TPM-A was higher in isolates from GU patients (12 of 14 [86%]) than in those from DU patients (13 of 23 [56%]), the association was not significant (P = 0.071), with an estimated odds ratio of 5.5 (95% CI, 1.0 to 30.1).
Associations between TPMs and vacA genotype.
There were 43 H. pylori vacA m1 and 37 vacA m2 strains as well as 4 strains with mixed genotypes. For the TPM-A-positive group, 26 strains were vacA m1 and 15 strains were vacA m2. Likewise, there were no associations between vacA type and the presence of TPM-B and -C. Analysis of the combined TPM-A and vacA m genotypes in relation to disease group also showed no specific associations. However, of the strains that had no TPM, only four were vacA m1 whereas the majority (83% [19 strains]) were vacA m2 or a mix of m1 and m2; of the peptic ulcer isolates, all 10 TPM-negative strains were vacA m2.
DISCUSSION
Novel block PCR-RFLP and rapid real-time PCR hybridization assays with three fluorescently labeled bi-probes were developed to test for the presence in the H. pylori genome of nucleotide sequence motifs corresponding to deduced CagA amino acid TPMs (A, B, and C) (21). Because these motifs are associated with tyrosine phosphorylation of CagA protein, they were investigated to determine if they might provide markers of more-severe disease outcome as a result of increased H. pylori-host epithelial cell interactions. Polymorphisms within cagA may affect the biological function of the protein and might explain the lack of a consistent correlation between cagA and disease severity previously noted (6, 23, 24). Initial identification of TPMs was based on a small number of diverse isolates of H. pylori, so the purpose of the present study has been to investigate TPM variation and frequency in a larger sample of isolates from peptic ulcer as well as NUD patients.
TPM-A was a common feature (71%) of H. pylori type I strains isolated from patients in England (mid-Essex), irrespective of associations with chronic clinical disease such as the presence of gastric and/or duodenal ulcers and of gastric cancer. By contrast, TPM-B and TPM-C were less common; they were detected in only 3 and 6% of strains, respectively, and mostly in combination with TPM-A. H. pylori that contained TPM-A showed some diversity with respect to RFLP type and vacA midregion genotype, and overall the most common combined genotype was TPM-A vacA-m1 RFLP-H2D2. Interestingly, the strains with no detectable motif were usually vacA m2, which is a genotype that interacts least with the host gastric mucosa (2).
Other data on TPM frequencies are for H. pylori type I isolates from 15 gastritis and 18 gastric cancer patients in Costa Rica (20). Nucleotide sequencing and deduced amino acid sequences showed relative frequencies that were significantly higher than ours, with 100% for TPM-A and 58% for TPM-B, whereas TPM-C was not detected in any strain. The reasons for such differences from our results are unclear, particularly for motif B, but they could be due to the patient populations examined or the methodology used; for instance, TPM-B strains were defined as containing the amino acid sequence motif KNS(T/G)EPY at position 899. Neither study found an association between the number of TPMs present and the severity of disease. However, our data suggest a trend, albeit not statistically significant, indicating that a higher proportion of GU-associated strains had TPM-A—an observation that may merit further study on a larger isolate set. Analysis of other cagA sequences also revealed that only a minority contained TPM-C (27), suggesting that that motif does not play a role in the pathogenesis of H. pylori, unlike TPM-A and -B, which are more frequently associated with severe disease. However, these observations are contradicted by other reports (22, 27) showing that TPM-C and not TPM-A is required for phosphorylation and that lack of TPM-C results in lower phosphorylation activity in strains containing either TPM-A alone or both TPM-A and TPM-B. Recently, an additional TPM, characterized by the peptide sequence EPIYA, has been identified and shown to occur as multiple repeats located upstream of the TPM-C site at the 3′ end of CagA (7, 31). This TPM is identical to the R1 repeat sequence characterized previously (34). Interestingly, strains with CagA containing three EPIYA elements at the C-terminal end were isolated predominantly from patients with gastric cancer and severe gastric atrophy (type C), whereas isolates with two EPIYA repeats (type A) were evenly distributed among patients with different gastric disease symptoms. Further analysis of the C-terminal region showed that both type A and type C strains lacked TPM-C. However, all type C strains were TPM-B positive, whereas approximately one-third of the type A strains lacked TPM-B. These findings suggested that the presence of TPM-B may also enhance the pathogenicity of H. pylori, whereas the presence of TPM-A in those isolates could not be verified. Our English isolates, like those from Costa Rica (20), lacked the TPM-B-TPM-C motif combination, which may be due to the fact that these two phosphorylation sites are located in close proximity at the COOH terminus of the CagA protein. The remaining 23 isolates in our study had no motifs detectable in the PCR assays and in that respect resembled strain J99 (a DU isolate from the United States), whose genome has been sequenced and reported to contain one R1 repeat 50 bp upstream of the nonfunctional TPM-C.
A total of six different PCR-based assays for detecting TPMs were developed and tested in this study. These comprised three assays for TPM-A (PCR-RFLP with HinfI, PCR-RFLP with DdeI, and LC probe hybridization), one LC probe hybridization assay for TPM-B, and two assays for TPM-C (PCR-RFLP with Tsp509I and LC probe hybridization). Direct DNA sequencing was used as the gold standard to check the specificity of the assays for selected samples. Real-time PCR assays, including those based on LC technology, are used increasingly in bacterial diagnostics because of their speed, convenience, and easy applicability to clinical specimens (9). Previously, LC-PCR was used to detect mutations in H. pylori 23S ribosomal DNA associated with clarithromycin resistance in pure cultures (14) and directly in gastric biopsy specimens without culture (8). The LC assays for TPM-A, -B, and -C have not yet been fully evaluated for direct use on gastric biopsy specimens, but with purified DNA they offer the advantage of simplicity (single-tube reaction) and speed, giving a result within 1 h as opposed to 24 h by conventional RFLP analysis.
Because of the high degree of interstrain sequence diversity within cagA, particularly within the TPM-B region, no single PCR assay format was applicable to all motifs. For TPM-A, the PCR-RFLP (HinfI) assay was the most reliable, with 100% specificity and 98.8% sensitivity, and was easy to perform and interpret. Close agreement was observed between the results of that assay and those of the real-time PCR assay. In the case of TPM-B, it was possible to develop an LC assay only because of sequence diversity, attributable to substitutions as well as insertions and deletions within the region of the motif. The LC assay showed good concordance with direct sequencing, although it generated a number of equivocal results that complicated interpretation. Likewise, for TPM-C, LC-PCR and sequencing results showed good correlation in most (seven of nine) cases, although only sequencing could define TPM-C status accurately for the two isolates that had generated equivocal melting peaks by the LC assay. In contrast, PCR-RFLP was less reliable and misidentified TPM-C in nine isolates. It was not possible to identify a restriction endonuclease specific for the TPM-C sequence, but examination of cagA sequences in GenBank suggested that the Tsp509I recognition sequence located immediately downstream of TPM-C was characteristic of that motif. However, the Tsp509I restriction sequence, while rare in strains lacking TPM-C, is occasionally present in such strains, so the PCR-RFLP assay had reduced specificity. The advantage of LC-PCR assays is that they might be applied directly to DNA extracted from gastric biopsy specimens and thus, if used in combination with real-time sequencing of short (25- to 50-bp) motifs, could provide a strategy for rapid screening of isolates.
In conclusion, our study shows that while TPM-A was common in patients in England, there was no evidence of a direct association between the presence of that TPM or other, rarer TPMs (B and C) and the occurrence of peptic ulcer disease. The presence and disease associations of these TPMs and the EPIYA motif repeats need to be investigated in a wider selection of strains in relation to the degree of inflammation in the host gastric tissue as well as to the level of tyrosine phosphorylation of CagA protein in infected epithelial cells.
Acknowledgments
We thank Neville Verlander (PHLS Statistics Unit) for performing the statistical analysis, J. Puls for helpful comments, and Louise Teare (Chelmsford PHL) and S. Saverymuttu (Broomfield Hospital, Chelmsford, United Kingdom) for isolates.
REFERENCES
- 1.Asahi, M., T. Azuma, Y. Ito, H. Suto, Y. Nagai, M. Tsubokawa, Y. Tohyama, S. Maeda, M. Omata, T. Suzuki, and C. Sasakawa. 2000. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 191:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton, J. 1998. H. pylori virulence factors. Br. Med. Bull. 54:105-120. [DOI] [PubMed] [Google Scholar]
- 3.Atherton, J. C., T. L. Cover, R. J. Twells, C. J. Hawkey, and M. J. Blaser. 1999. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J. Clin. Microbiol. 37:2979-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backert, S., E. Ziska, V. Brinkmann, U. Zimny-Arndt, A. Fauconnier, P. R. Jungblut, M. Naumann, and T. F. Meyer. 2000. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2:155-164. [DOI] [PubMed] [Google Scholar]
- 5.Backert, S., S. Moese, M. Selbach, V. Brinkmann, and T. F. Meyer. 2001. Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol. Microbiol. 42:631-644. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1999. Allelic variation in Helicobacter pylori: progress but no panacea. Gut 45:477-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Censini, R., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chisholm, S. A., R. J. Owen, E. L. Teare, and S. Saverymuttu. 2001. PCR-based diagnosis of Helicobacter pylori infection and real-time PCR determination of clarithromycin resistance directly from human gastric biopsy samples. J. Clin. Microbiol. 39:1217-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cockerill, F. R., III, and T. F. Smith. 2002. Rapid-cycle real-time PCR: a revolution for clinical microbiology. ASM News 68:77-83. [Google Scholar]
- 10.Danesh, J. 1999. Helicobacter pylori infection and gastric cancer: systematic review of the epidemiological studies. Aliment. Pharmacol. Ther. 13:851-856. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, D. J., Jr., and D. G. Evans. 2001. Helicobacter pylori CagA: analysis of sequence diversity in relation to phosphorylation motifs and implications for the role of CagA as a virulence factor. Helicobacter 6:187-198. [DOI] [PubMed] [Google Scholar]
- 13.Forman, D. 1998. Is there significant variation in the risk of gastric cancer associated with Helicobacter pylori infection? Aliment. Pharmacol. Ther. 12:3-7. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, J. R., N. A. Saunders, B. Burke, and R. J. Owen. 1999. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J. Clin. Microbiol. 37:3746-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, D. Y., R. M. Genta, D. P. Graham, and J. E. Crabtree. 1996. Serum CagA antibodies in asymptomatic subjects and patients with peptic ulcer: lack of correlation of IgG antibody in patients with peptic ulcer or asymptomatic Helicobacter pylori gastritis. J. Clin. Pathol. 49:829-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenks, P. J., F. Megraud, and A. Labigne. 1998. Clinical outcome after infection with Helicobacter pylori does not appear to be reliably predicted by the presence of any of the genes of the cag pathogenicity island. Gut 43:752-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda, S., H. Yoshida, T. Ikenoue, K. Ogura, K. Kani, N. Kato, Y. Shiratori, and M. Omata. 1999. Structure of cag pathogenicity island in Japanese Helicobacter pylori isolates. Gut 44:336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell, H. M., S. L. Hazell, Y. Y. Li, and P. J. Hu. 1996. Serological response to specific Helicobacter pylori antigens: antibody against CagA antigen is not predictive of gastric cancer in a developing country. Am. J. Gastroenterol. 91:1785-1788. [PubMed] [Google Scholar]
- 20.Occhialini, A., A. Marais, M. Urdaci, R. Sierra, N. Munoz, A. Covacci, and F. Megraud. 2001. Composition and gene expression of the cag pathogenicity island in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 69:1902-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 22.Odenbreit, S., B. Gebert, J. Puls, W. Fischer, and R. Haas. 2001. Interaction of Helicobacter pylori with professional phagocytes: role of the cag pathogenicity island and translocation, phosphorylation and processing of CagA. Cell. Microbiol. 3:21-31. [DOI] [PubMed] [Google Scholar]
- 23.Owen, R. J., T. M. Peters, R. Varea, E. L. Teare, and S. Saverymuttu. 2001. Molecular epidemiology of Helicobacter pylori in England: prevalence of cag pathogenicity island markers and IS605 presence in relation to patient age and severity of gastric disease. FEMS Immunol. Microbiol. 30:65-71. [DOI] [PubMed] [Google Scholar]
- 24.Owen, R. J., J. Xerry, T. M. Peters, and E. L. Teare. 2002. Surveillance and clinical relevance of vacA genotypes of Helicobacter pylori infecting dyspetics in mid-Essex. Commun. Dis. Public Health 5:106-111. [PubMed] [Google Scholar]
- 25.Peters, T. M., R. J. Owen, E. Slater, R. Varea, E. L. Teare, and S. Saverymuttu. 2001. Genetic diversity in the Helicobacter pylori cag pathogenicity island and effect on expression of anti-CagA serum antibody in UK patients with dyspepsia. J. Clin. Pathol. 54:219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pounder, R. E., and D. Ng. 1995. The prevalence of Helicobacter pylori infection in different countries. Aliment. Pharmacol. Ther. 9:33-39. [PubMed] [Google Scholar]
- 27.Puls, J., W. Fischer, and R. Haas. 2002. Activation of Helicobacter pylori CagA by tyrosine phosphorylation is essential for dephosphorylation of host cell proteins in gastric epithelial cells. Mol. Microbiol. 43:961-969. [DOI] [PubMed] [Google Scholar]
- 28.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slater, E., R. Owen, M. Williams, and R. Pounder. 1999. Conservation of the cag pathogenicity island of Helicobacter pylori and association with vacuolating cytotoxin alleles and IS605. Gastroenterology 117:1308-1315. [DOI] [PubMed] [Google Scholar]
- 30.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA 97:1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein, M., F. Bagnoli, R. Halenbeck, R. Rappuoli, W. J. Fantl, and A. Covacci. 2002. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 43:971-980. [DOI] [PubMed] [Google Scholar]
- 32.Vyse, A. J., N. J. Gay, L. M. Hesketh, N. J. Andrews, B. Marshall, H. I. Thomas, P. Morgan-Capner, and E. Miller. 2002. The burden of Helicobacter pylori infection in England and Wales. Epidemiol. Infect. 128:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 34.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]