Abstract
The hormone-mediated control of plant growth and development involves both synthesis and response. Previous studies have shown that gibberellin (GA) plays an essential role in Arabidopsis seed germination. To learn how GA stimulates seed germination, we performed comprehensive analyses of GA biosynthesis and response using gas chromatography–mass spectrometry and oligonucleotide-based DNA microarray analysis. In addition, spatial correlations between GA biosynthesis and response were assessed by in situ hybridization. We identified a number of transcripts, the abundance of which is modulated upon exposure to exogenous GA. A subset of these GA-regulated genes was expressed in accordance with an increase in endogenous active GA levels, which occurs just before radicle emergence. The GA-responsive genes identified include those responsible for synthesis, transport, and signaling of other hormones, suggesting the presence of uncharacterized crosstalk between GA and other hormones. In situ hybridization analysis demonstrated that the expression of GA-responsive genes is not restricted to the predicted site of GA biosynthesis, suggesting that GA itself, or GA signals, is transmitted across different cell types during Arabidopsis seed germination.
INTRODUCTION
Gibberellin (GA) is an essential phytohormone that controls many aspects of plant development, including seed germination, leaf expansion, stem elongation, flowering, and seed development (Davies, 1995). Previous studies have suggested that these GA-mediated growth responses are regulated in part by the modulation of cellular GA concentrations and by altering the ability of cells to respond to this hormone (Richards et al., 2001). To understand the roles of GA in these developmental processes, it is necessary to study how GA concentrations are regulated and how plants respond to GA.
GAs are tetracyclic diterpenoids synthesized from geranylgeranyl diphosphate. The biosynthesis pathway that converts geranylgeranyl diphosphate to biologically active GAs, such as GA1 and GA4, has been well studied (Hedden and Kamiya, 1997). Recent studies have started to identify some regulatory factors that control this pathway (Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000). On the other hand, genetic and cell biological studies have revealed key components in the GA response pathway (Olszewski et al., 2002). However, additional GA signaling components and downstream cellular and biochemical events need to be investigated further to better understand the molecular nature of the GA response.
Seed dormancy and germination are complex physiological processes that are controlled by a range of developmental and external cues (Koornneef et al., 2002). Germination begins with water uptake by seeds and terminates with the initial elongation of the embryonic axis (Bewley, 1997). Completion of germination is visible by the emergence of the radicle. Previous studies have shown that GA plays an essential role in promoting the germination of Arabidopsis seeds. This is evident by the defects in seed germination of severe GA-deficient mutants such as ga1-3 and ga2-1 (Koornneef and van der Veen, 1980). The inhibitory effect of GA biosynthesis inhibitors, such as paclobutrazol and uniconazole, on seed germination (Nambara et al., 1991; Jacobsen and Olszewski, 1993) suggests that newly synthesized GAs are required after imbibition for radicle emergence. These lines of evidence demonstrate that the level of GA is a critical determinant for seed germination. However, it remains unclear how endogenous GA concentrations are modulated during Arabidopsis seed germination.
At least two roles for GA have been proposed in stimulating germination in small-seeded plant species such as tomato and Arabidopsis (Hooley, 1994; Debeaujon and Koornneef, 2000; Yamaguchi and Kamiya, 2002). First, GA is necessary to overcome the mechanical restraint conferred by tissues that surround the embryo, such as the aleurone and the testa. In Arabidopsis, embryos of nongerminating GA-deficient mutants can develop into dwarf plants when the surrounding tissues are removed mechanically (Silverstone et al., 1997; Telfer et al., 1997). Therefore, one of the roles of GA in the induction of radicle protrusion appears to be to weaken the tissue that surrounds the embryo. Second, GA also increases the growth potential of the embryo, as indicated by the reduced growth rate of GA-deficient embryos (Groot and Karssen, 1987). Several GA-inducible genes related to cell wall loosening have been identified in tomato seeds, including those encoding endo-β-mannanase (Nonogaki et al., 2000), xyloglucan endotransglycosylase/hydrolase (Chen et al., 2002), expansin (Chen and Bradford, 2000; Chen et al., 2001), and β-1,3-glucanase and chitinase (Wu et al., 2001), some of which are expressed specifically in the micropylar endosperm cap around the radicle.
Genetic studies have identified several GA-signaling components in Arabidopsis, some of which play a role in GA-induced seed germination, as shown by their loss-of-function mutant phenotypes (Olszewski et al., 2002). RGA-LIKE1 (RGL1) and RGL2, members of the DELLA subfamily of GRAS proteins, and SPINDLY, a Ser/Thr O-linked N-acetylglucosamine transferase, act as negative regulators of GA-dependent seed germination (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Lee et al., 2002; Wen and Chang, 2002). By contrast, SLEEPY1 (SLY1) is proposed to be a positive regulator in the GA response pathway (Steber et al., 1998). Altered GA responses in these mutants have been characterized principally by visible GA-related phenotypes. In some cases, GA-responsive gene markers also have been used to evaluate the specificity of responses modulated in GA response mutants. For example, the GA biosynthesis gene AtGA3ox1 (GA4) has been shown to be downregulated by GA activity; therefore, repression of this gene by the repressor of ga1-3 (rga) mutation is consistent with the partial activation of the GA response pathway (Silverstone et al., 2001). In germinating cereal grains, GA-induced α-amylase gene expression in aleurone cells has been a useful model system in which to study the GA response pathway (Olszewski et al., 2002). Identification of additional GA-regulated genes would help to better describe the GA response pathway and to characterize potential GA response mutants.
The importance of defining specific GA responses is emphasized by the fact that germination in GA-deficient nongerminating seeds can be rescued when the abundance of, or the sensitivity to, other hormones is modulated. Abscisic acid (ABA) plays a critical role in the maintenance of seed dormancy and inhibits germination (Leung and Giraudat, 1998). ABA biosynthesis mutants were first isolated as suppressors of the nongerminating GA-deficient mutants (Koornneef et al., 1982). Ethylene acts positively on seed germination in Arabidopsis (Kepczynski and Kepczynska, 1997). Exogenous ethylene treatment induces the germination of GA-deficient mutant seeds (Karssen et al., 1989). Brassinosteroids (BRs) also play a role in promoting germination, and exogenous BR application enhances the germination of GA-deficient nongerminating Arabidopsis mutants (Steber and McCourt, 2001). These results indicate that seed germination is determined by the net effect of multiple hormones and suggest the presence of crosstalk among actions by GA, ABA, ethylene, and BR.
To understand how GA stimulates seed germination, we performed comprehensive studies of endogenous GA levels by gas chromatography–mass spectrometry (GC-MS) and large-scale analyses of GA-regulated gene expression using DNA microarrays. Here, we show that active GA levels are increased just before radicle emergence and that a unique set of genes is upregulated or downregulated in response to exogenous GA treatment. We demonstrate that these GA-responsive genes constitute subsets of genes with different temporal patterns of expression during wild-type seed germination. The diverse nature of GA-responsive genes also is suggested by in situ hybridization analysis, which indicates that at least a fraction of GA-regulated genes are expressed predominantly in non-GA-producing cell types. Finally, we use known or deduced functions of the GA-responsive genes to predict metabolic and cellular pathways modulated by GA activity during seed germination. This investigation suggests some novel crosstalk between GA and other hormones; some of these findings are supported by the results of statistical analysis of putative cis elements in the promoter regions of GA-responsive genes.
RESULTS AND DISCUSSION
Global Gene Expression Profiles in Response to Exogenous GA in Imbibed ga1-3 Mutant Seeds
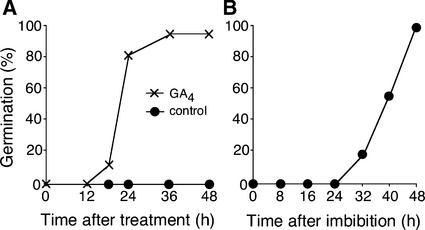
To identify GA-dependent gene expression during seed germination, we used the GA-deficient mutant ga1-3. The ga1 mutants are defective in ent-copalyl diphosphate synthase in an early step of the GA biosynthesis pathway, and ga1-3 is a null allele (Sun and Kamiya, 1994). ga1-3 seeds are nongerminating until GA is supplied exogenously. To analyze GA-regulated gene expression, ga1-3 seeds were stratified by imbibing at 4°C in the dark for 48 h and then incubated for 24 h under continuous white light before GA4 or mock (control) treatment. We included the stratification treatment because it increases tissue sensitivity to exogenous GA (Derkx and Karssen, 1993). Under these conditions, radicle emergence was observed initially at 18 h after GA4 treatment, whereas the control seeds did not germinate, as shown in Figure 1A. To monitor GA-responsive gene expression at early and late time points, RNA samples were prepared from seeds treated with or without GA4 for 1, 3, 6, and 12 h. Because GA4-treated seeds had not germinated at 12 h, transcriptional changes detected in this experiment reflect, at least in part, GA responses necessary for radicle protrusion.
Figure 1.
Germination Profiles of Seeds Used for Microarray Analysis.
Seeds were scored as germinated when radicle protrusion was visible.
(A) Germination of ga1-3 seeds. The seeds were stratified at 4°C in the dark for 48 h and then incubated at 22°C for 24 h under continuous white light before treatment with 50 μM GA4 or a mock solution (control).
(B) Germination of wild-type seeds at 22°C under continuous white light.
Genes expressed differentially between GA4-treated and nontreated samples were identified using oligonucleotide-based DNA microarrays representing ∼8200 Arabidopsis genes. To evaluate the reliability of our microarray data, we determined the expression profiles of 30 genes, which exhibited varying degrees of GA responsiveness when examined using DNA microarrays, by quantitative reverse transcription–PCR (QRT-PCR) using identical RNA materials. We confirmed that the expression profiles of all genes by QRT-PCR were very similar to those determined by microarray analysis, as presented graphically in the supplemental data online. GA-responsive genes identified in this study consist of 230 GA-upregulated and 127 GA-downregulated genes (Table 1), corresponding to 2.8 and 1.5% of the genes examined by microarray analysis, respectively. Transcripts of 36.3% of the 8200 genes were undetectable in the ga1-3 seeds. The identities of the GA-regulated genes determined in this study are listed in the supplemental data online, where the following information also is provided: known or deduced functions, duration required for the response to GA4, and the maximum induction/repression fold.
Table 1.
Number of Genes Responsive to Exogenous GA4 in ga1-3 Seeds
Initial time points at which GA responsiveness (greater than fourfold changes in transcript levels between GA-treated and control samples) was detected.
GA-upregulated genes.
GA-downregulated genes.
Regulation of the GA Biosynthesis Pathway during Wild-Type Seed Germination
In the initial set of microarray analyses, GA-deficient ga1-3 mutant seeds were exposed to exogenous GA4 to monitor GA-regulated gene expression. This experiment was designed to create a drastic difference in GA concentrations to identify genes that respond to this hormone. However, our ultimate goal is to understand how seed germination is controlled by endogenous GAs. To this end, we first analyzed time-course changes in GA levels during wild-type seed germination, and the results were compared with temporal expression patterns of GA-responsive genes in the same set of samples. These experiments were performed using wild-type seeds imbibed at 22°C under continuous white light.
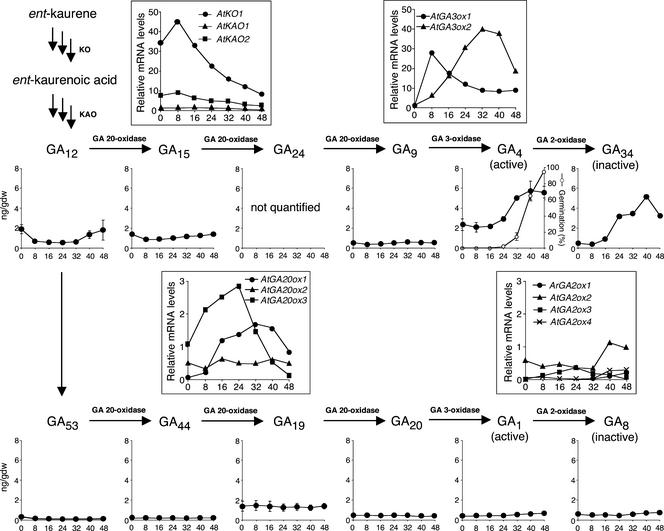
Time-course changes in endogenous contents of precursor, active, and deactivated GAs were determined using GC-MS to predict how the overall GA biosynthesis pathway is regulated during wild-type seed germination (Figure 2). To interpret these data better, we combined the expression profiles of individual GA biosynthesis genes (Figure 2), which were determined by microarray analysis (described in detail below) or QRT-PCR. Our data showed that 13-nonhydroxylated GAs (GA12, GA15, GA9, GA4, and GA34) are dominant in germinating Arabidopsis seeds compared with the corresponding 13-hydroxylated forms (GA53, GA44, GA20, GA1, and GA8). Therefore, GA4 is likely to be the major endogenous active GA in germinating seeds, as in shoots (Talón et al., 1990; Derkx et al., 1994).
Figure 2.
GA Biosynthesis and Deactivation during Wild-Type Seed Germination.
Wild-type seeds were imbibed at 22°C under continuous white light. The x axis in all graphs indicates hours after the onset of imbibition. Samples at 0 h are dry seeds. Graphs in boxes indicate relative transcript levels of GA biosynthesis genes that were determined by microarray analysis, except for those of AtGA2ox genes, which were analyzed by QRT-PCR. AtKO1 (At5g25900) encodes KO (ent-kaurene oxidase). AtKAO1 (At1g05160) and AtKAO2 (At2g32440) encode KAOs (ent-kaurenoic acid oxidase). AtGA20ox1 (At4g25420), AtGA20ox2 (A5g51810), and AtGA20ox3 (At5g07200) encode GA 20-oxidase. AtGA3ox1 (At1g15550) and AtGA3ox2 (At1g80340) encode GA 3-oxidase. AtGA2ox1 (At1g78440), AtGA2ox2 (At1g30040), AtGA2ox3 (At2g34550), and AtGA2ox4 (At1g02400) encode GA 2-oxidase. Other graphs show endogenous levels of GAs as labeled. The germination profile of wild-type seeds used for GA measurements is plotted in the GA4 graph (open circles). GA24 was detected in all samples but could not be quantified because of comigration of impurities. gdw, grams dry weight.
A dramatic increase in GA4 levels was observed between 24 and 32 h, when initial radicle protrusion was just observed (Figure 2). This finding is consistent with the premise that GA plays a key role in late stages of seed germination (Gallardo et al., 2001, 2002). In agreement with the increase in GA4 levels, several GA biosynthesis genes were upregulated after imbibition of wild-type seeds with varying temporal expression profiles (Figure 2). For example, genes encoding an ent-kaurene oxidase (AtKO1), a GA 20-oxidase (AtGA20ox3), and a GA 3-oxidase (AtGA3ox1) were upregulated within 8 h of imbibition and then downregulated at later time points. The decline in GA12 levels within 8 h appears to be correlated with an increase in AtGA20ox3 expression soon after imbibition. However, GA12 present in dry seeds would not be sufficient for the production of active GAs necessary for germination, because inhibition of ent-kaurene oxidase by uniconazole or paclobutrazol prevents germination (Nambara et al., 1991; Jacobsen and Olszewski, 1993). Two genes that encode GA 3-oxidases, AtGA3ox1 and AtGA3ox2, displayed distinct temporal expression patterns, consistent with previous RNA gel blot analysis (Yamaguchi et al., 1998). The expression profile of AtGA3ox2 is best correlated with changes in GA4 contents up to 40 h, suggesting a direct role for this gene in increasing active GA levels to cause radicle emergence.
Our microarray analysis did not detect transcripts for GA 2-oxidases. However, because some 2β-hydroxylated GAs, such as GA8 and GA34, were detected in our samples, we analyzed AtGA2ox transcripts using a more sensitive QRT-PCR technique (Figure 2). Our QRT-PCR analysis confirmed that all AtGA2ox transcripts remained at low levels before radicle emergence, suggesting that altered AtGA2ox transcript abundance is not the principal mechanism for the increase in GA4 levels. This hypothesis also is supported by the observation that the curve for GA34 nearly paralleled that for GA4, suggesting that the formation of GA34 is dependent on the amount of GA4 (Figure 2). These results suggest that increased synthesis of active GAs, rather than reduced deactivation by 2β-hydroxylation, plays a major role in the increase in active GA concentrations during wild-type seed germination.
Expression Profiles of GA-Responsive Genes during Wild-Type Seed Germination
To determine whether genes regulated by exogenous GA4 are expressed in accord with the increase in active GA levels during seed germination, we performed microarray analysis using a set of wild-type seed samples that was used for GA analysis. The germination profile of wild-type seeds for this microarray analysis is shown in Figure 1B. Our data showed that transcripts of 39% of the genes were detectable in dry seeds. In our hands, approximately equal amounts of total RNA were yielded from dry seeds and seeds imbibed for up to 32 h, and similar amounts of labeled target RNAs were prepared from these samples. Thus, it appears that gross total mRNA levels in dry seeds do not change dramatically until radicle emergence. Therefore, our comparison of relative mRNA levels in this study approximately represents relative amounts on a per seed basis.
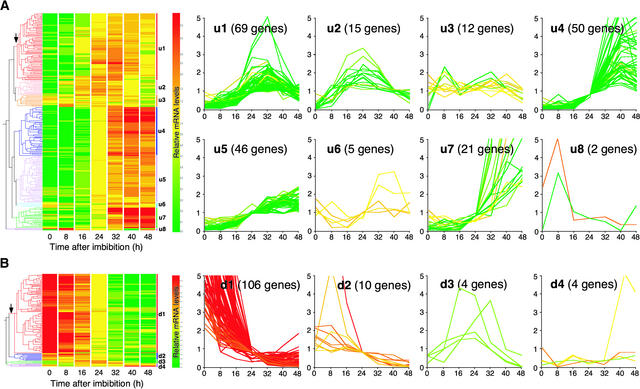
Genes responsive to GA4 treatment in ga1-3 seeds were classified further into subgroups by their expression profiles during wild-type seed germination. We used a hierarchical clustering algorithm (Eisen et al., 1998) that allowed us to group genes by constructing a hierarchical tree (Figure 3) in which genes are arranged in order based on the similarities in their expression patterns. We reasoned that if GA is the principal regulator of these genes, their expression profiles would correlate with changes in GA levels during wild-type seed germination. In addition, because GA activates seed germination, GA-inducible and GA-repressible genes would play positive and negative roles, respectively, in controlling germination. Our data showed that the majority of GA-upregulated transcripts increased in abundance at some time after imbibition relative to dry seeds, consistent with their positive roles in seed germination (Figure 3A). Likewise, most GA4-repressible genes were downregulated after imbibition of wild-type seeds, in agreement with their negative role in controlling seed germination (Figure 3B).
Figure 3.
Hierarchical Clustering of GA-Responsive Genes by Temporal Expression Patterns during Wild-Type Seed Germination.
Genes responsive to exogenous GA4 in ga1-3 seeds were used in this analysis. Hierarchical trees (at left) were generated based on similarities in expression profiles during wild-type seed germination. Branching at the level indicated by vertical arrows was used to designate eight (A) and four (B) clusters. Branches shown in different colors in the tree represent these clusters, and their names are indicated at right. The graphs depict temporal expression profiles of genes in each of the clusters (x axis, hours after imbibition; y axis, relative mRNA levels [see Methods]). The expression pattern of each gene is colored according to the relative mRNA level at 0 h. The number of genes categorized into each cluster is indicated on the graph. The identities of genes in each cluster are listed in the supplemental data online.
(A) GA-upregulated genes.
(B) GA-downregulated genes.
However, when the time-course changes were examined closely, it was noted that the expression of some of the GA-regulated genes did not correlate perfectly with endogenous GA4 levels (Figures 2 and 3). For example, GA-repressible genes in the d1 cluster were downregulated soon after imbibition of wild-type seeds, when GA4 levels were as low as in dry seeds. Transcripts for the u1, u2, and u8 clusters increased earlier than GA4 and then decreased when GA4 levels were high. This transient induction of the u1, u2, and u8 clusters cannot be explained by endogenous GA4 levels alone. Therefore, it appears that these subgroups also are regulated by other factors and that GA4 levels play only a partial role in determining the levels of these transcripts. It should be noted that GA4 was present in dry seeds at one-third of the maximum level (Figure 2). Therefore, some GA-regulated genes would become responsive to GA4 without changing GA4 concentrations if tissue responsiveness to this hormone were increased upon the onset of seed imbibition. In this context, it would be important to assess the role of GA4 present in dry seeds by comparing gene expression between GA-deficient and wild-type seeds. However, our GC-MS analysis revealed that the ga1-3 dry seeds contained active GAs that were derived from exogenous GA treatment to rescue the fertility of parental mutant plants (M. Ogawa, A. Hanada, and S. Yamaguchi, unpublished results). Thus, we found it difficult to obtain dry seeds with reduced active GA levels even in the ga1-3 background because of the necessity of active GA for normal seed set.
In contrast to the u1, u2, u8, and d1 clusters, expression of the GA-upregulated u4 and u7 clusters correlated largely with active GA levels; low levels of gene expression were seen until an increase at 16 to 24 h. Also, some GA-repressible transcripts in the d2 and d3 cluster were detectable at early time points and then downregulated nearly in agreement with the increase in GA4 levels.
Cellular Distribution of GA Biosynthesis and Response
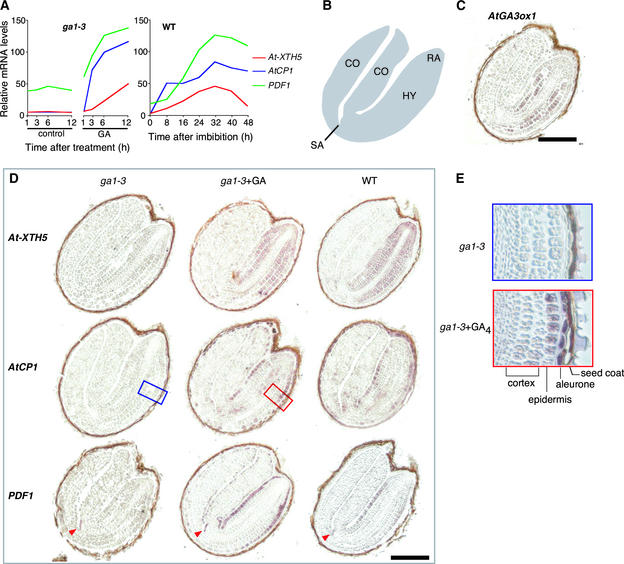
Above, we discussed the temporal correlations between GA4 levels and GA-regulated transcript abundance. However, the distribution of GA-regulated genes within a seed are largely unknown. It has been shown that two GA biosynthesis genes, AtGA3ox1 and AtGA3ox2, are expressed predominantly in the cortex and endodermis of the embryonic axis in germinating Arabidopsis seeds (Yamaguchi et al., 2001). Because these are the major genes encoding GA 3-oxidases that catalyze the final step to produce active GAs during seed germination, we postulate that active GAs are synthesized mainly in these cell types at this stage. To determine which cell types respond to GA, we examined the cellular distribution of GA-regulated transcript accumulation using in situ RNA hybridization. For this purpose, we chose GA-regulated genes whose transcript abundance deviated drastically between GA4-treated and control ga1-3 seeds (Figure 4A).
Figure 4.
Cellular Localization of GA-Regulated Transcripts
(A) Expression profiles of GA-upregulated genes used for in situ hybridization experiments. At left, profiles of ga1-3 seeds treated with GA4 or mock solution (control); at right, profiles of wild-type (WT) seeds imbibed under continuous white light. Samples at 0 h are dry seeds.
(B) Scheme of an Arabidopsis mature embryo. CO, cotyledon; HY, hypocotyl; RA, radicle; SA, shoot apical meristem.
(C) In situ hybridization analysis of the GA biosynthesis gene AtGA3ox1 in wild-type seeds imbibed for 24 h. This result using embryos without seed coats has been described previously (Yamaguchi et al., 2001). Bar = 0.1 mm.
(D) In situ hybridization analysis of GA-upregulated genes. Digoxigenin-labeled antisense probes were hybridized with longitudinal sections of ga1-3 seeds treated with or without GA4 for 6 h and wild-type seeds (WT) imbibed for 24 h. AGI gene codes are as follows: At-XTH5 (At5g13870), AtCP1 (At4g36880), and PDF1 (At2g42840). The boxed areas are magnified in (E). The arrowhead indicates the PDF1 transcript detected in the shoot apical meristem. Bar = 0.1 mm.
(E) Magnified views highlighting AtCP1 transcript accumulation in the aleurone and the epidermis after GA4 treatment.
Figure 4 shows that each of these GA-inducible genes displayed a unique spatial expression pattern in wild-type seeds and that each expression pattern in wild-type seeds was similar to that in GA4-treated ga1-3 seeds. Weak or no hybridization signals in the ga1-3 seeds without GA4 treatment confirm the responsiveness of these genes to GA4. At-XTH5 transcripts were detected mainly in the embryonic axis (Figure 4D), including cell types exhibiting AtGA3ox1 expression (Figures 4B and 4C). However, unlike AtGA3ox1, At-XTH5 expression appeared abundant in the radicle. Upregulation of AtCP1 (encoding a putative Cys proteinase) by GA4 was most evident in the epidermis and provasculature of the embryonic axis and the aleurone layer (Figure 4D), where AtGA3ox transcripts were not detected (Figure 4C). GA4-induced transcript accumulation of the PROTODERMAL FACTOR1 (PDF1) gene (Abe et al., 1999) was observed mainly in the epidermis of the embryonic axis (Figure 4D). Interestingly, PDF1 expression in the L1 layer of the shoot apical meristem also was detectable in the ga1-3 seeds without GA4 treatment, indicating that the response of this gene to GA4 varies in different cell types.
These data from in situ hybridization studies provide an important insight into GA response at the cellular level. We showed that at least some GA-inducible transcripts exist primarily in the predicted non-GA-producing cell types in wild-type seeds. These observations suggest that GA4 itself or its secondary signal moves across different cell types in Arabidopsis seeds. Indeed, in germinating cereal grains, active GAs synthesized in the embryonic epithelium have been shown to cause the induction of α-amylase in the aleurone (Kaneko et al., 2002). Our results suggest a similar embryo–aleurone interaction in Arabidopsis, provided that the expression of AtGA3ox genes is restricted to the embryo (Yamaguchi et al., 2001), whereas AtCP1 expression responded to exogenous GA4 in the aleurone (Figure 4E). This finding is consistent with the hypothesis that GAs produced in the radicle trigger the weakening of endosperm tissues during tomato seed germination (Groot and Karssen, 1987).
We assume that GA4 treatment of seeds results in the exposure of this hormone to all cells. Therefore, it is interesting that, for all GA-inducible genes tested, cell type–specific expression patterns in wild-type seeds are largely identical to those in GA4-treated ga1-3 seeds. These results suggest that the cell type specificity of these genes in wild-type seeds is not attributable to GA4 localization to those cells but is determined by other cell type–specific factor(s).
Statistical Prediction of cis Elements for GA Responsiveness
The identification of a number of GA-regulated genes offered an opportunity to search for sequences common in their promoter regions. We first examined whether the GA-regulated genes selected in this study contain known GA-responsive cis elements. Analysis of α-amylase gene promoters in barley aleurone cells has identified a sequence, TAACAAA/G (GA-responsive element; GARE), that is necessary and sufficient for GA responsiveness (Skriver et al., 1991). A GA-inducible transcription factor, HvGAMYB, binds specifically to GARE to activate transcription (Gubler et al., 1995). We surveyed the GARE motif in the 500 bp immediately upstream from the ATG of Arabidopsis genes. The occurrence of GARE in GA-inducible, GA-repressible, and GA-nonresponsive genes was 20, 18, and 12%, respectively. Any more specific subgroup (e.g., GA upregulated within 6 h or the clusters shown in Figure 3) did not contain GARE at a much higher frequency. These results suggest that GARE does not serve as a major cis element for GA-induced gene expression in Arabidopsis seeds. An 8-bp motif (CAACTGTC) responsible for the GA induction of LEAFY in Arabidopsis (Blázquez and Weigel, 2000) was found in only 1 gene among 230 GA-inducible genes identified in this study (Table 1), suggesting that this sequence does not serve as a common cis element for GA responsiveness in Arabidopsis seeds.
Second, to identify candidates for uncharacterized cis elements for GA responsiveness, we searched for DNA sequences that are found more frequently in the promoter regions (500 bp upstream from the initial ATG) of GA-responsive genes relative to all annotated Arabidopsis gene promoters using GeneSpring software (see Methods). This investigation revealed that the sequence ACGTGTC is present in 24 genes in the GA-downregulated d1 cluster (106 genes; Figure 3B) and that the frequency of the occurrence of this sequence in the d1 cluster is significantly greater than that in all annotated Arabidopsis genes (false-positive probability: P < 2.5 × 10−4). Curiously, this motif is identical to an ABA-responsive element (ABRE) for ABA-induced gene expression (Busk and Pages, 1998).
GA–ABA Interactions
Our data showed that many GA-downregulated genes contain ABRE in their putative promoter regions. There are at least three possible mechanisms by which GA downregulates ABA-upregulated genes: (1) GA reduces ABA levels by affecting ABA biosynthesis; (2) GA negatively regulates the ABA response pathway; and (3) GA and ABA signals are targeted independently to distinct cis-regulatory sequences of a single gene.
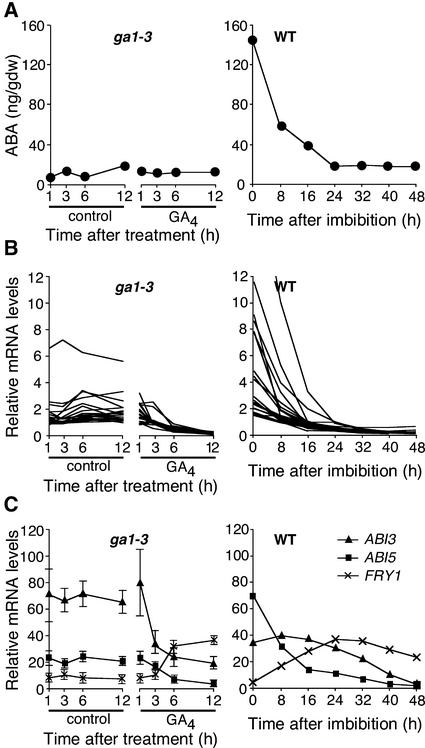
In germinating lettuce seeds, exogenous GA3 treatment has been shown to reduce endogenous ABA levels when the seeds are incubated in the dark after far-red light treatment that inhibits germination (Toyomasu et al., 1994). To determine whether the downregulation of ABRE-containing genes by GA4 results from a reduction in ABA content, we performed ABA measurements using the same set of samples used for microarray analyses. Figure 5A (left graph) shows that endogenous ABA levels did not change dramatically between GA-treated and nontreated ga1-3 seeds, whereas ABRE-containing genes were downregulated by GA4 (Figure 5B, left graph). These results suggest that the downregulation of ABRE-containing genes by exogenous GA4 is not caused by a decrease in endogenous ABA levels in the experimental conditions used in this study. Further investigation is required to determine whether GA decreases endogenous ABA levels in Arabidopsis seeds under conditions similar to those used by Toyomasu et al. (1994).
Figure 5.
Endogenous ABA Levels and Expression of ABRE-Related Genes.
For all panels, the x axis of the graphs at left indicates time (hours) after GA4 or mock treatment (control) of ga1-3 seeds. Graphs at right depict time courses in wild-type (WT) seeds imbibed under continuous white light. Samples at 0 h are dry seeds. gdw, grams dry weight.
(A) ABA levels.
(B) Expression profiles of GA4-repressible ABRE-containing genes in the d1 cluster (see Figure 3B). The identities of these genes are listed in the supplemental data online.
(C) ABI3 (At3g24650), ABI5 (At2g36270), and FRY1 (At5g63980) transcript levels. In the graph at left, means and standard errors are plotted from triplicate measurements from DNA microarray and QRT-PCR analysis.
Transcriptional regulation through ABREs has been studied in detail (Shen and Ho, 1995). A basic domain/Leu zipper transcription factor, ABI5, binds to ABRE (Kim et al., 2002), and another transcription factor, ABI3, binds to a sequence called a Sph/RY element (Suzuki et al., 1997; Ezcurra et al., 2000), which often is found adjacent to ABREs. Because both abi3 and abi5 confer ABA insensitivity and they behave as recessive mutations, ABI3 and ABI5 are positive regulators of the ABA response (Finkelstein et al., 2002). Recently, FIERY1 (FRY1), an inositol polyphosphate 1-phosphatase, was shown to function as a negative regulator of ABRE-containing ABA-inducible genes (Xiong et al., 2001). Although ABI3, ABI5, and FRY1 genes were not classified as being GA responsive, our microarray data indicated that transcript levels of these ABRE-related genes were modulated slightly by GA4 in ga1-3 seeds. QRT-PCR was used to confirm that GA4 treatment of ga1-3 seeds significantly decreased ABI3 and ABI5 mRNA levels and increased FRY3 transcript abundance (Figure 5C, left graph). These observations suggest that ABI3, ABI5, and FRY1 may partially mediate the downregulation of ABRE-containing genes by GA4.
Many GA-downregulated ABRE-containing genes belong to the d1 cluster (Figure 3B); the mRNA levels of these genes decline soon after imbibition of wild-type seeds, before the increase in GA4 (Figures 2 and 5B). On the other hand, ABA contents decreased drastically immediately after imbibition (Figure 5A, right graph). Thus, the temporal profile of ABA is analogous to that of the d1 cluster. These results suggest that ABA contributes primarily to the initial downregulation of the ABRE-containing genes, which then are downregulated further to nearly undetectable levels by active GAs that increase at later times during wild-type seed germination. An early decrease in the ABA level and a late increase in the active GA level after seed imbibition also have been reported in germinating barley embryos (Jacobsen et al., 2002).
Cellular Events and Metabolic Pathways Modulated by GA Activity
One of the objectives of this study was to predict GA-dependent alterations in metabolic and cellular processes to specify the roles of GAs during seed germination. To this end, we examined whether any set of genes involved in particular metabolic or cellular pathways is regulated coordinately by GA activity.
Cell Elongation and Cell Division
Among GA-upregulated genes identified in this study, there are some classes of genes implicated in cell elongation, as shown in Table 2. Aquaporin, a class of intrinsic membrane proteins, facilitates water uptake into symplast to increase turgor for cell expansion (Quigley et al., 2002; Aharon et al., 2003). Xyloglucan endotransglycosylase/hydrolase (XTH) loosens the cell wall by hydrolyzing cell wall components (Campbell and Braam, 1999; Rose et al., 2002), whereas expansin does it by disrupting hydrogen bonding (Cosgrove, 2000; Lee et al., 2001). Although the roles of pectin methylesterase (PME) have not been elucidated fully, this enzyme also may be involved in cell elongation by modifying cell wall pectin (Pilling et al., 2000; Micheli, 2001). These cell wall–loosening activities may be associated with weakening of the tissue surrounding the embryo and/or the facilitation of embryo growth. GA-dependent expression of cell wall–related enzymes during germination already has been reported for other species (Chen and Bradford, 2000; Nonogaki et al., 2000; Ren and Kermode, 2000; Chen et al., 2001, 2002; Wu et al., 2001). Our large-scale expression analysis demonstrated that the majority of expansin and PME gene family members were upregulated by GA4 (Table 2), suggesting that these cell wall–modifying activities are dependent in large part on GA during Arabidopsis seed germination.
Table 2.
GA-Responsive Genes Implicated in Cell Elongation
| Protein Namea | Totalb | Microarrayc | Germinationd | GA Upe | GA Downf |
|---|---|---|---|---|---|
| Expansin | 26 | 12 | 10 | 7 | 0 |
| XTH | 33 | 24 | 12 | 5 | 1 |
| PME | 67 | 32 | 9 | 7 | 0 |
| Aquaporin | 38 | 24 | 18 | 5 | 1 |
PME, pectin methylesterase; XTH, xyloglucan endotransglycosylase/hydrolase.
Number of genes identified in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000).
Number of genes represented on the microarray.
Number of genes whose mRNAs were detectable within 32 h after imbibition of wild-type seeds.
Number of GA-upregulated genes.
Number of GA-downregulated genes.
Most cells in mature dry seeds are in G1-phase (Deltour, 1985; Georgieva et al., 1994). Our microarray analysis indicated that classes of cell cycle–related genes, notably those involved in the transition from G1- to S-phase, were upregulated after GA4 treatment. These include genes that encode D-type cyclin (At1g70210) and the DNA replication licensing factor MCM protein (At1g44900 and At2g16440) (Springer et al., 2000; Vandepoele et al., 2002). GA also upregulated a gene that encodes replication protein A (RPA; At2g24490). The GA-induced expression of OsRPA1 (which encodes an RPA) in internodes of deepwater rice has been reported (van der Knaap et al., 1997). Because the expression of the majority of genes implicated in the transition from G2- to M-phase was not GA dependent, we speculate that the GA signal for cell division is targeted mainly to G1 cells to initiate DNA replication.
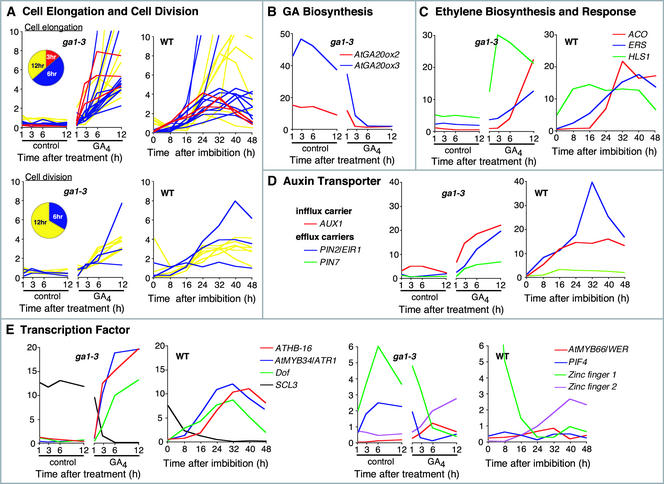
Figure 6A shows time-course expression profiles of genes implicated in cell elongation or cell division in wild-type and GA4-treated or control ga1-3 seeds. Our data show that transcriptional changes for both cell elongation and cell division took place before radicle appearance. However, cell elongation–related genes tended to be upregulated earlier than genes related to cell division (Figure 6A).
Figure 6.
GA-Regulated Cellular and Metabolic Pathways during Seed Germination.
For each panel, the graph(s) at left indicate the ga1-3 seeds treated with 50 μM GA4 (GA) or mock solution (control). The x axis indicates time (hours) after treatment. Graph(s) at right depict temporal profiles of GA-regulated transcripts after imbibition of wild-type seeds under continuous white light. Samples at 0 h are dry seeds. For all graphs, the y axis shows relative transcript levels (see Methods).
(A) Genes implicated in cell elongation (top) and cell division (bottom). The pie chart indicates the ratio of genes whose response to GA4 was detectable within 3 (red), 6 (blue), or 12 h (yellow). This color code also is used to depict the relative transcript levels of the corresponding genes in the graph.
(B) GA biosynthesis genes. Gene identities are described in the legend to Figure 2.
(C) Ethylene biosynthesis and response genes. AGI gene codes are as follows: ACO (At2g19590), ERS1 (At2g40940), and HLS1 (At4g37580).
(D) Genes implicated in auxin transport. AGI gene codes are as follows: AUX1 (At2g38120), PIN2/EIR1 (At5g57090), and PIN7 (At1g23080).
(E) Genes encoding (putative) transcription regulators whose expression was regulated by GA4 within 6 h. For clarity, expression profiles are displayed in two separate sets of graphs. AGI gene codes are as follows: ATHB-16 (homeodomain-Leu zipper protein; At4g40060), AtMYB34/ATR1 (At5g60890), Dof (Dof zinc finger protein; At2g28510), SCL3 (SCARECROW-LIKE3; At1g50420), AtMYB66/WER (At5g14750), PIF4 (At2g43010), Zinc finger1 (At1g27730), and Zinc finger2 (At2g28200).
GA Biosynthesis
It has been demonstrated that some genes that encode GA 20-oxidases and GA 3-oxidases are negatively regulated by GA activity through feedback inhibition, whereas GA upregulates genes that encode GA-deactivating GA 2-oxidases by a positive feedforward loop (Olszewski et al., 2002). These homeostatic mechanisms would maintain the concentration of active GAs within a limited range. Our microarray analysis showed that AtGA20ox2 and AtGA20ox3, which encode GA 20-oxidases, were downregulated by exogenous GA4 in ga1-3 seeds (Figure 6B). GA4 downregulation of AtKO1, AtGA20ox1, and AtGA3ox1 also was evident, as determined by QRT-PCR (see supplemental data online) (these genes are not included in the list of GA-regulated genes because their GA responsiveness was less than fourfold; see Methods). Genes that encode GA 2-oxidases were upregulated by exogenous GA3 treatment in floral stems (Thomas et al., 1999). However, we found no evidence for the feedforward upregulation of any AtGA2ox genes in imbibed seeds by QRT-PCR (data not shown). These observations are consistent with the idea that the synthesis of active GAs, rather than their deactivation, plays an important regulatory role in determining active GA levels during Arabidopsis seed germination.
Ethylene and Auxin
Ethylene acts positively on seed germination in Arabidopsis (Kepczynski and Kepczynska, 1997). Our microarray data indicate that some ethylene-related genes are affected by exogenous GA4 treatment in imbibed ga1-3 seeds. A gene that encodes a 1-aminocyclopropane-1-carboxylic acid oxidase (ACO), which catalyzes ethylene formation, was upregulated by exogenous GA4 (Figure 6C). This finding suggests that GA4 may increase ethylene production in imbibed Arabidopsis seeds. Transcripts of ethylene-inducible genes, such as HOOKLESS1 (HLS1) (Lehman et al., 1996), and a member of the ethylene receptor gene family, ERS1 (Hua et al., 1998), also were upregulated by GA4 treatment (Figure 6C), which supports the idea that GA activates ethylene synthesis and/or response.
The roles of auxin in Arabidopsis seed germination have been unclear; however, our microarray analysis indicated that the expression of several auxin-related genes was modulated by exogenous GA4 treatment. Polar auxin transport is controlled by auxin influx and efflux carriers, which pump auxin into and out of cells, respectively (Muday and DeLong, 2001). Figure 6D indicates that several genes that encode (putative) auxin carrier proteins are upregulated in response to exposure to GA4 at relatively early time points (Figure 6D). Two genes that encode CYP79B2 (At4g39950) and CYP79B3 (At2g22330) were determined to be upregulated by exogenous GA4. Recent work has demonstrated that these CYPs are necessary not only for indole glucosinolate synthesis but also for the formation of indoleacetic acid through indole-3-acetaldoxime (Zhao et al., 2002). Thus, GA activity may be targeted to enhancing auxin biosynthesis as well. Although direct evidence for this idea remains to be found, our results suggest that GA activity results in substantial changes in auxin level and transport during Arabidopsis seed germination.
Transcriptional Regulators
Figure 6E summarizes the expression profiles of GA-responsive genes that encode (putative) transcription factors, of which response to exogenous GA4 was observed within 6 h. The roles of several of these transcription factors have been studied previously. PIF4 is a basic helix-loop-helix transcription factor that interacts physically with the photoreceptor phytochrome B (phyB) and is likely to be a negative regulator of the phyB signaling pathway during seedling development (Huq and Quail, 2002). We found that PIF4 transcript levels were decreased upon exposure of ga1-3 seeds to GA4. Repression of a negative regulator of the phyB response pathway by GA4 is intriguing because both GA and phyB act positively on seed germination in Arabidopsis (Shinomura et al., 1996). However, the role of PIF4 in red light–stimulated seed germination has yet to be clarified. A transcription factor, AtMyb34/ATR1, functions as a positive regulator of the ASA1 gene (Bender and Fink, 1998), which encodes the α-subunit of anthranilate synthase, catalyzing the first committed step in Trp biosynthesis (Niyogi and Fink, 1992). The rapid and drastic induction of ASA1 by GA4 (Figure 6E) suggests that GA modulates Trp biosynthesis during Arabidopsis seed germination.
Conclusions and General Discussion
We performed detailed analysis of the GA biosynthesis pathway and large-scale expression studies of GA-responsive genes during Arabidopsis seed germination. We demonstrated that the major active GA, GA4, increases after seed imbibition and that this increase occurs just before radicle protrusion (Figure 2). These results reinforce the importance of de novo GA biosynthesis for the control of radicle protrusion in Arabidopsis seeds. Combining the expression profiles of GA biosynthesis genes, we speculate that an increase in the synthesis of active GAs, but not a decrease in GA deactivation, contributes principally to increasing GA4 levels during Arabidopsis seed germination.
This study also revealed part of the complex mechanism by which GA activates seed germination in Arabidopsis. This complexity is attributable to at least three aspects of the diversity found in the GA response pathway. First, as expected, the GA signal is likely to be targeted to a number of metabolic and cellular pathways, some of which were discussed here (Figure 6). Although several GA-responsive genes had been described previously, this study has substantially improved our knowledge of the molecular basis of the GA response. Second, this study revealed a variety of temporal expression patterns of GA-responsive genes during wild-type seed germination (Figure 3). Therefore, it appears that subsets of GA-regulated genes might be controlled differently by other factors. Third, our study has started to reveal the diversity of cell types responding to GA in Arabidopsis seeds. Our results indicate that expression of at least some GA-responsive genes is not restricted to the major site of active GA biosynthesis (Figure 4). Therefore, active GA itself, or its signals, should move between different cell types. This may account in part for the diversity of temporal expression patterns of GA-regulated genes during wild-type seed germination. Identification of the spatial expression patterns of GA-responsive genes on a larger scale may provide a better view of how GA signals are transmitted to a series of different cell types and also should allow us to classify GA-regulated genes further into subgroups based on their cellular locations. Such classification of GA-regulated genes may help to define more specifically the GA response pathway and to predict cellular and metabolic events that occur in individual cell types.
Finally, although we focused on GA-related genes, this study has provided expression profiles of a number of other transcripts detectable during germination and early seedling growth (see supplemental data online). These data sets, together with other related transcriptome (Girke et al., 2000; Ruuska et al., 2002) and proteome (Gallardo et al., 2001, 2002) analyses, should contribute to our understanding of the molecular basis of how seeds germinate.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Landsberg erecta was used as the wild type in this study. The ga1-3 seeds were obtained originally from Maarten Koornneef (Wageningen University, The Netherlands). To propagate seeds for germination experiments, plants were grown on soil under continuous light at 22°C. To rescue the growth of the ga1-3 mutant, GA4 solution (2 μM) was sprayed repeatedly as required. Harvested mature seeds were stored at room temperature at 30% RH for at least 3 months. The quality of each batch of seeds was assessed by monitoring the germination profile (radicle emergence) under continuous light at 22°C, and seed batches showing similar germination abilities were used throughout the study. For germination, the seeds were washed with 0.02% Triton X-100 solution, rinsed with water, and incubated on wet 3MM filter paper (Whatman, Maidstone, UK).
Gibberellin and Abscisic Acid Analysis
One gram of dry seeds was used for each gibberellin (GA) and abscisic acid (ABA) measurement. Quantitative analysis of GA and ABA was performed using 2H-labeled GAs and ABA as internal standards, as described previously (Gawronska et al., 1995). GA measurements were performed three times using different seed batches. Where data are available, the means and standard errors from triplicate experiments are indicated in Figure 2. For GA9, GA15, GA20, GA34, GA44, and GA53, the mean from two measurements (with similar results) is plotted on the graph. ABA levels were determined twice using independent plant materials with similar results.
Target Complementary RNA Synthesis and Hybridization to Affymetrix GeneChips
For microarray analysis, total RNA was extracted from dry or imbibed seeds (from 40 mg of dry seeds) using the RNAqueous RNA Isolation Kit with Plant RNA Isolation Aid (Ambion, Austin, TX). Double-stranded cDNA was synthesized from 5 μg of total RNA using the SuperScript Choice cDNA Synthesis Kit (Invitrogen, Carlsbad, CA) with an oligo(dT)24 primer containing a T7 polymerase promoter site at the 3′ end. Biotin-labeled complementary RNA (cRNA) was synthesized by T7 RNA polymerase using the double-stranded cDNA as a template (BioArray High-Yield RNA Transcript Labeling Kit; Enzo Diagnostics, Farmingdale, NY) and purified with the use of the RNeasy RNA Purification Kit (Qiagen, Valencia, CA). The biotin-labeled cRNA was fragmented and hybridized to a GeneChip microarray (Affymetrix, Santa Clara, CA) for 16 h at 42°C. After hybridization, the arrays were washed and stained with biotinylated anti-streptavidin antibody (Vector Laboratories, Burlingame, CA) and a phycoerythrin-streptavidin conjugate (Molecular Probes, Eugene, OR) according to the manufacturer's protocol. Signals were scanned using a confocal microscope scanner (Gene Array Scanner; Hewlett-Packard, Palo Alto, CA) at 570 nm.
Data Analysis
Signal values for individual genes were obtained using statistical algorithms on Microarray Suite software version 5.0 (Affymetrix). The sum of signal values from all probe sets was used for normalization across different samples. Genes were classified as GA responsive if the signal values deviated either positively or negatively fourfold or more between GA4-treated and control samples reproducibly at least at one time point in duplicated experiments using independent plant materials. The presence or absence of a reliable hybridization signal for each gene was judged by the detection call on Microarray Suite. Genes for which transcripts were determined to be undetectable (absence) in GA4-treated samples were eliminated from the list of GA-upregulated genes. Similarly, genes for which transcripts were determined to be undetectable (absence) in control samples were eliminated from the list of GA-downregulated genes. When the transcript was undetectable in only the GA-treated or the control sample, we gave the background signal intensity (determined by Microarray Suite software) to the undetectable transcript. If the signal intensity from the other sample was greater by fourfold or more relative to the background value, this gene was regarded as being GA regulated. To ensure the GA responsiveness of selected genes, we inspected the time-course curve for each gene visually in both GA4-treated and control samples from duplicate experiments. This inspection confirmed that the expression profiles of most of the selected genes were reproducible, except for those of 11 genes (3%) that we eliminated from the list because their erratic expression patterns were inconsistent between the duplicate experiments.
Signal values from one of the duplicated experiments were used for further data analysis using a GeneSpring software package (version 5.0; Silicon Genetics, Redwood, CA). Relative transcript levels in Figures 2, 4, 5, and 6 (except Figures 5B and 6A) and the supplemental data online were determined by dividing each signal value for each gene by the 50th percentile of all signal values for normalization across samples. Thus, values in these graphs reflect the strength of the original hybridization signals. In addition to the normalization procedure described above, the data in Figures 3, 5B, and 6A were normalized across genes by dividing the value by the median of different samples. In this case, the data do not reflect absolute transcript levels; rather, they reflect the degree of fluctuation among the set of samples. For example, relatively low levels of these values in the u5 cluster transcripts (Figure 3A) do not mean that their absolute levels are low but indicate that they deviate only within a narrow range among samples.
Hierarchical clustering analysis (Eisen et al., 1998) (Figure 3) and statistical prediction of promoter cis elements were performed using GeneSpring Software version 5.0 (Silicon Genetics). In this analysis, a particular sequence commonly present in the 5′ upstream region (500 bp) of selected genes (e.g., GA-upregulated genes) was searched. Statistical analysis then was performed to determine how specifically this particular sequence occurs in the selected gene group relative to the corresponding upstream regions of all annotated genes in Arabidopsis. False-positive probabilities were calculated based on the expected frequency of occurrence of a given sequence if the distributions of bases are random.
Throughout the data sets, genes are identified by the AGI gene code (e.g., At1g80340) supplied by the Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de/). Correspondence between AGI gene code and the Affymetrix gene number was determined based on data from the Salk Institution Genomic Analysis Laboratory (SIGnAL; http://signal.salk.edu/smission.html) and Sheen Lab (http://genetics.mgh.harvard.edu/sheenweb/search_affy.html) World Wide Web sites.
Quantitative Reverse Transcription–PCR
Total RNA (2 μg) was treated with RQ1 RNase-free DNase (Promega, Madison, WI) to eliminate genomic DNA contamination. First-strand cDNA was synthesized with random hexamers using a SuperScript first-strand synthesis system according to the manufacturer's instructions (Invitrogen). Quantitative real-time PCR with Taq-Man technology (Holland et al., 1991) or the SYBR Green I dye method was performed using the first-strand cDNA as a template on a sequence detector system (model 7700; Applied Biosystems, Foster, CA). The mean value of three replicates was normalized using an 18S rRNA as the internal control. Nucleotide sequences of gene-specific primers and Taq-Man probes are listed in the supplemental data online.
In Situ Hybridization
In situ hybridization experiments were performed as described previously (Yamaguchi et al., 2001). Digoxigenin-labeled antisense cRNA probes were synthesized using PCR-amplified cDNA fragments as templates. The following primer sets were used: for At-XTH5, 5′-GCTCCTTTTGTCGCGTCCTACAG-3′ and 5′-CTAGATTAAATTGTAATAAGAAG-3′; for AtCP1, 5′-GACAACTTAAGATTCATCG-3′ and 5′-CGCAAGTTCCTTGGTCTTTG-3′; and for PDF1, 5′-GAGAGGGATGGTGAGTTTTGCCGT-3′ and 5′-CTCCAGTAATCGCATGTTCCAGTG-3′.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Peter McCourt (University of Toronto, Canada), Eiji Nambara, Ian S. Curtis, and Damian P. O'Neill (RIKEN) for valuable comments on the manuscript. We also thank Yukihisa Shimada and Hideki Goda (RIKEN) for help with microarray analyses.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011650.
Footnotes
Online version contains Web-only data.
References
- Abe, M., Takahashi, T., and Komeda, Y. (1999). Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol. 40, 571–580. [DOI] [PubMed] [Google Scholar]
- Aharon, R., Shahak, Y., Wininger, S., Bendov, R., Kapulnik, Y., and Galili, G. (2003). Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 15, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bender, J., and Fink, G.R. (1998). A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 5655–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. [DOI] [PubMed] [Google Scholar]
- Busk, P.K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Campbell, P., and Braam, J. (1999). Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 4, 361–366. [DOI] [PubMed] [Google Scholar]
- Chen, F., and Bradford, K.J. (2000). Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 124, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Dahal, P., and Bradford, K.J. (2001). Two tomato expansin genes show divergent expression and localization in embryos during seed development and germination. Plant Physiol. 127, 928–936. [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Nonogaki, H., and Bradford, K.J. (2002). A gibberellin-regulated xyloglucan endotransglycosylase gene is expressed in the endosperm cap during tomato seed germination. J. Exp. Bot. 53, 215–223. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000). Loosening of plant cell walls by expansins. Nature 407, 321–326. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltour, R. (1985). Nuclear activation during early germination of the higher plant embryo. J. Cell Sci. 75, 43–83. [DOI] [PubMed] [Google Scholar]
- Derkx, M.P.M., and Karssen, C.M. (1993). Effects of light and temperature on seed dormancy and gibberellin-stimulated germination of Arabidopsis thaliana: Studies with gibberellin-deficient and -insensitive mutants. Physiol. Plant. 89, 360–368. [Google Scholar]
- Derkx, M.P.M., Vermeer, E., and Karssen, C.M. (1994). Gibberellins in seeds of Arabidopsis thaliana: Biological activities, identification and effects of light and chilling on endogenous levels. Plant Growth Regul. 15, 223–234. [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerstrom, M., and Rask, L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24, 57–66. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S.L., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15.–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo, K., Job, C., Groot, S.P.C., Puype, M., Demol, H., Vandekerckhove, J., and Job, D. (2001). Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo, K., Job, C., Groot, S.P.C., Puype, M., Demol, H., Vandekerckhove, J., and Job, D. (2002). Proteomics of Arabidopsis seed germination: A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 129, 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronska, H., Yang, Y.-Y., Furukawa, K., Kendrick, R.E., Takahashi, N., and Kamiya, Y. (1995). Effects of low irradiance stress on gibberellin levels in pea seedlings. Plant Cell Physiol. 36, 1361–1367. [Google Scholar]
- Georgieva, E.I., Lopez-Rodas, G., Hittmair, A., Feichtinger, H., Brosch, G., and Loidl, P. (1994). Maize embryo germination. Planta 192, 118–124. [Google Scholar]
- Girke, T., Todd, J., Ruuska, S., White, J., Benning, C., and Ohlrogge, J. (2000). Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124, 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot, S.P.C., and Karssen, C.M. (1987). Gibberellins regulate seed germination in tomato by endosperm weakening: A study with gibberellin-deficient mutants. Planta 171, 525–531. [DOI] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P., and Kamiya, Y. (1997). Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 431–460. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5, 523–530. [DOI] [PubMed] [Google Scholar]
- Holland, P.M., Abramson, R.D., Watson, R., and Gelfand, D.H. (1991). Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus. Proc. Natl. Acad. Sci. USA 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley, R. (1994). Gibberellins: Perception, transduction and responses. Plant Mol. Biol. 26, 1529–1555. [DOI] [PubMed] [Google Scholar]
- Hua, J., Sakai, H., Nourizadeh, S., Chen, Q.G., Bleecker, A.B., Ecker, J.R., and Meyerowitz, E.M. (1998). EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10, 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, J.V., Pearce, D.W., Poole, A.T., Pharis, R.P., and Mander, L.N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant. 115, 428–441. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, M., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., and Matsuoka, M. (2002). The α-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 128, 1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen, C., Zagórski, S., Kepczynski, J., and Groot, S. (1989). Key role for endogenous gibberellins in the control of seed germination. Ann. Bot. 63, 71–80. [Google Scholar]
- Kepczynski, J., and Kepczynska, E. (1997). Ethylene in seed dormancy and germination. Physiol. Plant. 101, 720–726. [Google Scholar]
- Kim, S.Y., Ma, J., Perret, P., Li, Z., and Thomas, T.L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130, 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58, 257–263. [DOI] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Choi, D., and Kende, H. (2001). Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 4, 527–532. [DOI] [PubMed] [Google Scholar]
- Lehman, A., Black, R., and Ecker, J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85, 183–194. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Micheli, F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419. [DOI] [PubMed] [Google Scholar]
- Muday, G.K., and DeLong, A. (2001). Polar auxin transport: Controlling where and how much. Trends Plant Sci. 6, 535–542. [DOI] [PubMed] [Google Scholar]
- Nambara, E., Akazawa, T., and McCourt, P. (1991). Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol. 97, 736–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., and Fink, G.R. (1992). Two anthranilate synthase genes in Arabidopsis: Defense-related regulation of the tryptophan pathway. Plant Cell 4, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki, H., Gee, O.H., and Bradford, K.J. (2000). A germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol. 123, 1235–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.-p., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.), S61.–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling, J., Willmitzer, L., and Fisahn, J. (2000). Expression of a Petunia inflata pectin methyl esterase in Solanum tuberosum L. en-hances stem elongation and modifies cation distribution. Planta 210, 391–399. [DOI] [PubMed] [Google Scholar]
- Quigley, F., Rosenberg, J.M., Shachar-Hill, Y., and Bohnert, H.J. (2002). From genome to function: The Arabidopsis aquaporins. Genome Biol. 3, 0001.1–0001.17. [DOI] [PMC free article] [PubMed]
- Ren, C., and Kermode, A.R. (2000). An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol. 124, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. [DOI] [PubMed] [Google Scholar]
- Rose, J.K., Braam, J., Fry, S.C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Ruuska, S.A., Girke, T., Benning, C., and Ohlrogge, J.B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14, 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., and Ho, T.H.D. (1995). Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.-S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.-p. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Mak, P.Y., Martinez, E.C., and Sun, T.-p. (1997). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver, K., Olsen, F.L., Rogers, J.C., and Mundy, J. (1991). cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 88, 7266–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, P.S., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G1 phase and is required maternally for early Arabidopsis development. Development 127, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S.E., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125, 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.-p., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., and McCarty, D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talón, M., Koornneef, M., and Zeevaart, J.A.D. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. USA 87, 7983–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96, 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu, T., Yamane, H., Murofushi, N., and Inoue, Y. (1994). Effects of exogenously applied gibberellin and red light on the endogenous levels of abscisic acid in photoblastic lettuce seeds. Plant Cell Physiol. 35, 127–129. [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap, E., Jagoueix, S., and Kende, H. (1997). Expression of an ortholog of replication protein A1 (RPA1) is induced by gibberellin in deepwater rice. Proc. Natl. Acad. Sci. USA 94, 9979–9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C.-K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.-T., Leubner-Metzger, G., Meins, F., Jr., and Bradford, K.J. (2001). Class I β-1,3-glucanase and chitinase are expressed in the micropylar endosperm of tomato seeds prior to radicle emergence. Plant Physiol. 126, 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Lee, B.-h., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.-K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., and Kamiya, Y. (2000). Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 41, 251–257. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., and Kamiya, Y. (2002). Gibberellins and light-stimulated seed germination. J. Plant Growth Regul. 20, 369–376. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Kamiya, Y., and Sun, T.-p. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28, 443–453. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G.S., Kamiya, Y., and Sun, T.-p. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Hull, A.K., Gupta, N.R., Goss, K.A., Alonso, J., Ecker, J.R., Normanly, J., Chory, J., and Celenza, J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.