Abstract
ACTIN-RELATED PROTEINS 2 and 3 form the major subunits of the ARP2/3 complex, which is known as an important regulator of actin organization in diverse organisms. Here, we report that two genes, WURM and DISTORTED1, which are important for cell shape control in Arabidopsis, encode the plant ARP2 and ARP3 orthologs, respectively. Mutations in these genes result in misdirected expansion of various cell types: trichome expansion is randomized, pavement cells fail to produce lobes, hypocotyl cells curl out of the normal epidermal plane, and root hairs are sinuous. At the subcellular level, cell shape changes are linked to severe filamentous actin aggregation and compromised vacuole fusion. Because all seven subunits of the ARP2/3 complex are present in plants, our data indicate that this complex may play a pivotal role during plant cell morphogenesis.
INTRODUCTION
Observations of diverse plant cell types have shown that the cytoskeleton plays a vital role during cell morphogenesis (Kost and Chua, 2002). In general, microtubules are believed to play a role in determining and maintaining cell polarity, whereas actin microfilaments ensure the targeted delivery of vesicles that carry plasma membrane and cell wall components to the site of growth (Mathur and Hülskamp, 2002). Among other model cell types, leaf epidermal cells (trichomes) in Arabidopsis have emerged as an attractive system and have been used to dissect the role of microtubules and microfilaments during cell morphogenesis (Mathur et al., 1999; Szymanski et al., 1999; Mathur and Chua, 2000; Schwab et al., 2003). Arabidopsis trichomes are unicellular, with precisely angled branches, and develop through a well-coordinated sequence of morphogenetic events (Hülskamp et al., 1994; Szymanski et al., 2000). Thus, after branch initiation, the trichome cell undergoes rapid elongation, producing, ultimately, a perpendicular, stellate, 300- to 500-μm-tall cell. Interference with the actin cytoskeleton using different actin-interacting drugs, such as cytochalasin D, latrunculin B, phalloidin, and jasplakinolide, during the rapid expansion phase results in randomly distorted trichomes with short unextended branches (Mathur et al., 1999; Szymanski et al., 1999). At the genetic level, eight complementation groups (alien, crooked, distorted1, distorted2, gnarled, klunker, spirrig, and wurm) characterized by a common “distorted” trichome phenotype have been described (Hülskamp et al., 1994). Using a green fluorescent protein (GFP) fused to the filamentous (F) actin binding domain of a mouse talin (mTalin) gene (McKann and Craig, 1997; Kost et al., 1998), the actin cytoskeleton was found to be aberrant in these mutants, leading to the speculation that the affected genes might interact directly with the actin cytoskeleton (Mathur et al., 1999).
Arabidopsis contains eight functional actin isoforms (Meagher et al., 2000) and numerous actin-interacting proteins that exhibit a high degree of genetic redundancy (Staiger et al., 2000). Thus, there are at least 5 profilin, 9 ADF (Actin Depolymerization Factor), 3 fimbrin, 4 villin, and 11 Rho-type small GTPase genes, a capping protein, and an actin-interacting cyclase-associated protein (AtCAP1) (Barrero et al., 2002; Kost and Chua, 2002). In addition, at least eight ARP (ACTIN-RELATED PROTEIN) genes have been identified in Arabidopsis (McKinney et al., 2002). However, mutant/antisense transgenic Arabidopsis plants for profilin (Ramachandran et al., 2000; McKinney et al., 2001) and ADF (Dong et al., 2001) genes, which are likely candidates for the dis group, do not display trichome distortions. Transgenic plants for the AtRop genes display changes in actin organization (Yang, 2002) but not the dis phenotype. The different ARPs have been described and cloned from Arabidopsis (Klahre and Chua, 1999; McKinney et al., 2002), but in the absence of mutant/antisense/overexpression studies, their function in plants has remained speculative (Vantard and Blanchoin, 2002).
From other organisms, however, it is known that ARP2 and ARP3 associate with five other novel subunits to produce an ARP2/3 complex, which is acknowledged as a multifunctional modulator of the actin cytoskeleton (Machesky and Gould, 1999). The ARP2/3 complex localizes to actin filaments at cellular regions of dynamic actin assembly (Welch et al., 1997a), enhances actin nucleation (Welch, 1999), is involved in the actin polymerization–driven motility of organelles and pathogenic organisms (Welch et al., 1997b; Machesky, 1999), and affects membrane fusion and endocytotic activities (Moreau et al., 1997; Qualmann et al., 2000). In unicellular eukaryotes such as budding yeast, disruption of ARP2/3 complex subunits results in a slow-growth phenotype characterized by defects in cortical actin patches. In addition, whereas the ARPC1 homolog Arc40p was found to be unconditionally essential (Winter et al., 1999), mutants in ARPC5 (Arc16p homolog) resulted in reduced mitochondrial motility (Boldogh et al., 2001). In fission yeast, arp2, arp3, and sop2 (Arc40p homolog) all have been shown to be essential, and conditional mutants in these genes exhibit cortical actin cytoskeleton defects (Balasubramanian et al., 1996; McCollum et al., 1996; Morrell et al., 1999). In a multicellular organism such as Drosophila, loss-of-function mutations in the Arpc1 and Arp3 genes indicate that the complex is required for ring canal expansion during oogenesis (Hudson and Cooley, 2002). Thus, although the participation of the ARP2/3 complex in various actin polymerization–mediated processes such as cell crawling, intracellular motility, and phagocytosis is well documented (Bailly et al., 1999; May et al., 2000), its role in cell morphogenesis is less understood. This complex has not been described in plants.
In our attempt to understand the molecular basis of cell shape development, we aimed to molecularly characterize some of the Arabidopsis DISTORTED genes (Hülskamp et al., 1994). Here, we show that WURM (WRM) and DISTORTED1 (DIS1) encode the Arabidopsis ARP2 and ARP3 orthologs, respectively. Our cell biological data from various cell types in these mutants indicate that expansion growth in Arabidopsis requires ARP2/3 complex activity, and its loss results in inefficient fine F-actin formation while leading to enhanced F-actin aggregation and bundling. In addition, both wrm and dis1 mutants display severe defects in vacuole morphology, suggesting that membrane fusion also may be one of the processes in which ARP2/3 complex–mediated actin polymerization plays an important role.
RESULTS
WRM and DIS1 Encode for ARP2 and ARP3, Respectively
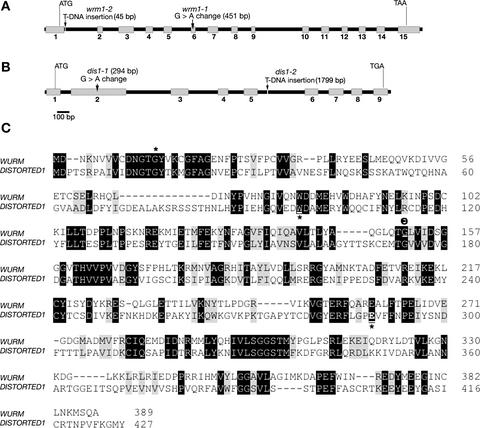
The dis1 mutant is a classic genetic marker in Arabidopsis and has been mapped to the upper arm of chromosome 1 (Feenstra, 1978). The wrm mutant was mapped to the upper arm of Arabidopsis chromosome 3, ∼20 and 30 centimorgan from markers nga162 and nga126, respectively (Schwab et al., 2003). After the sequencing and annotation of the Arabidopsis genome, we searched the defined region for actin-related genes and identified ARP2 (BAC clone moj10; gene At3g27000) and ARP3 (BAC clone f3f19; At1g13180) as putative “distorted” gene candidates. A perusal of the SALK T-DNA insertion mutant collection (Ecker single T-DNA lines) revealed the presence of two mutant lines, SALK_003448 and SALK_010045, in the At3g27000 and At1g13180 genes, respectively. Complementation tests revealed allelism between the ethyl methanesulfonate (EMS) allele wrm (wrm1-1) and T-DNA insertion line SALK_003448 (subsequently called wrm 1-2) and between the EMS allele dis1-1 and T-DNA insertion line SALK_010045 (hereafter referred to as dis1-2). Sequencing revealed single nucleotide changes (G to A) in both wrm1-1 and dis1-1 at the positions indicated in Figures 1A and 1B, respectively, and confirmed T-DNA insertions in the wrm1-2 and dis1-2 alleles (Figures 1A and 1B).
Figure 1.
Molecular Organization and Homology between WRM (Arabidopsis ARP2) and DIS1 (Arabidopsis ARP3).
(A) and (B) Schemes of the genomic organization and mutations (arrows) in WRM (A) and DIS1 (B) in Arabidopsis. The positions of mutations are indicated relative to the A of the start codon. wrm 1-1 and dis1-1 are EMS-generated alleles, whereas wrm1-2 and dis1-2 are T-DNA insertion alleles. Gray boxes represent exons, and black lines represent introns.
(C) Amino acid sequence alignment of WRM and DIS1 using the CLUSTAL W program. Black-shaded areas indicate amino acid identity, and gray-shaded areas indicate sequence similarity. Mutations indicated by asterisks in the two genes are predicted to truncate the protein at amino acid position 14 in wrm1-2 (SALK_003448), at amino acid 97 in dis1-1, and at amino acid position 290 in dis1-2 (SALK_010045), whereas a G-to-A change in wrm1-1 changes a conserved Gly (G) at position 151 (θ) to Asp (D).
All homozygous mutant lines exhibited distorted trichomes. Subsequent Agrobacterium tumefaciens–mediated transformation of wrm1-1 with the ARP2 genomic DNA clone (∼3 kb without the 5′ upstream promoter region) and of dis1-1 with the ARP3 genomic clone (∼2.8 kb without the promoter region) under the strong 35S promoter of Cauliflower mosaic virus rescued the distorted trichome phenotype completely in both genes, proving the cloning of the two genes.
WRM and DIS1 Genes Are Integral Components of the ARP2/3 Complex
Based on the mutations, the protein truncations in wrm1-2, dis1-1, and dis1-2 would be predicted to occur after amino acids 14, 97, and 290, respectively (Figure 1C). The G-to-A change in wrm1-1 results in the alteration of a conserved Gly (position 151; Figure 1C) to Asp and may lead to a pivotal change in the ARP2 protein structure. Although WRM and DIS1 show 44.4 and 45.7% amino acid identity, respectively, with Arabidopsis ACTIN2 (McKinney et al., 2002), between themselves the two proteins display low sequence identity of 30% and an overall similarity of only 47% (Figure 1C). The occurrence of a similar distorted trichome phenotype in both mutants strongly suggested that WRM and DIS1 act in a similar pathway or may act in concert. Indeed, from other organisms, the ARP2 and ARP3 proteins encoded by WRM and DIS1 are known to function as a complex, called the ARP2/3 complex, along with five other smaller subunits designated ARPC1 to ARPC5 (Machesky and Gould, 1999; Cooper et al., 2001). Our protein homology–based search (Basic Local Alignment Search Tool [BLASTP]) of the databases revealed the other five subunits of the ARP2/3 complex to be present in Arabidopsis (Table 1). With the exception of ARPC1 (p41), all other subunits occur as single genes in Arabidopsis. Protein similarity for the different subunits in Arabidopsis with orthologs from other organisms ranges from 46% for ARPC2 to 80% for ARP2. The Arabidopsis ARPC4 displays the least identity to the ARPC4 subunit from other organisms and shares domain similarity only with a kinesin-like protein (Table 1).
Table 1.
ARP2/3 Complex Subunits in Arabidopsis and Their Amino Acid Similarity to Other Organisms
| Subunit |
Arabidopsis Gene Acc. No. MATDB |
BAC Clone | % amino-acid identity/similarity
|
|||||
|---|---|---|---|---|---|---|---|---|
| Dd | Sp | Sc | Ce | Dm | Hu | |||
| ARP2 | At3g27000 | moj10 | 63/80 | 53/73 | 57/73 | 60/79 | 61/80 | 62/80 |
| ARP3 | At1g13180 | f3f19 | 59/75 | 55/72 | 53/66 | 56/73 | 57/71 | 59/74 |
| ARPC1p41 | At2g30910 | f7f1 | 43/59 | 34/51 | 34/51 | 39/58 | 37/54 | 41/60 |
| ARPC1p41 | At2g31300 | f16d14 | 43/58 | 34/51 | 34/51 | 38/57 | 24/41 | 42/61 |
| ARPC2p34 | At1g30825 | t17h7 | 33/54 | 26/46 | 28/46 | n.a.a | 26/48 | 26/46 |
| ARPC3p20 | At1g60430 | t13d8 | 41/58 | 43/59 | 39/58 | 41/60 | 40/62 | 47/66 |
| ARPC4p20 | At4g14150 | t1j24 | n.a. | 49/72 | n.a. | 48/69 | n.a. | 57/72 |
| ARPC5p16 | At4g01710 | t15b16 | 31/51 | 25/44 | 23/45 | 32/50 | 35/55 | 32/53 |
Dd, Dictyostelium discoideum; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hu, human.
n.a., not applicable.
The occurrence of all subunits of the ARP2/3 complex in Arabidopsis suggests that the WRM and DIS1 gene products also may function together in a complex. In support of this idea, wrm1-1 dis1-1 and wrm1-2 dis1-2 double mutants show a phenotype indistinguishable from that of the single mutants.
Given the different roles played by the ARP2/3 complex in diverse organisms (Machesky and Gould, 1999), what probable role could it play during plant development? Detailed phenotypic characterization of wrm and dis1 mutants combined with a cell biological approach was used to address this question.
wrm and dis1 Mutants Exhibit Epidermal Defects during Rapid Growth
Randomly distorted trichome cells were the most consistent feature in both mutants. However, previous studies have shown both the ARP2 and ARP3 genes to be expressed ubiquitously at low levels in Arabidopsis (Klahre and Chua, 1999; McKinney et al., 2002). Consistent with this observation, the defects in the wrm and dis1 alleles were not confined solely to the occurrence of distorted trichomes (Figures 2Aa and 2Bb) but were detected in other epidermal cell types as well (Figure 2). In nearly 73 ± 4% of mutant epidermal cells, the characteristic lobed, jigsaw-puzzle shape of wild-type Arabidopsis cotyledon cells failed to develop (Figures 2C and 2D). Under light-growth conditions, mutant seedlings had 1.5 to 3 times shorter hypocotyl and cotyledon cells than the wild type. Furthermore, in nearly 74% (n = 130) of mutant seedlings, stomatal complexes did not differentiate clearly on hypocotyls (Figures 2E and 2F). Under in vitro conditions on agar-gelled MS medium (Murashige and Skoog, 1962) plates, the length and growth characteristics of root and root hair cells did not appear altered compared with those of the wild type. Homozygous mutant plants grown in soil exhibited normal fertility and seed set, suggesting that under these conditions pollen-tube growth occurred normally. Because the phenotype appeared consistently in diffusely growing, expanding cells, we challenged mutant seedlings further by growing them in the dark to induce rapid cell elongation.
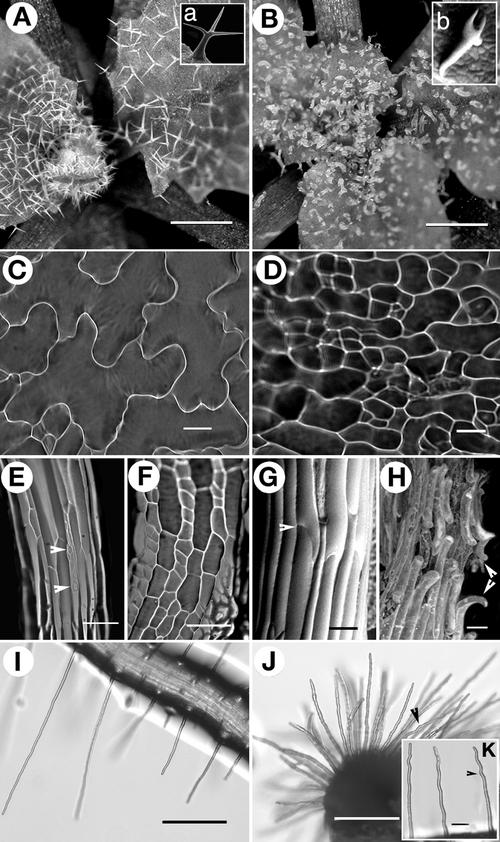
Figure 2.
Representative Images Comparing Salient Phenotypic Characters of wrm and dis1 Mutants with Those of Wild-Type Arabidopsis (ecotype Landsberg erecta).
(A) Typical two- to three-branched trichomes on wild-type leaves. Inset (a) shows a single, mature wild-type trichome.
(B) Short, nearly supine, distorted trichomes on dis1-1 mutant leaves. Inset (b) shows a single wrm1-1 trichome for comparison with inset (a).
(C) Epidermal cells in wild-type cotyledons arranged in the characteristic jigsaw-puzzle pattern with prominent lobes.
(D) Randomly shaped, less lobed, and relatively unexpanded cotyledon epidermal cells seen in both wrm and dis1 mutants.
(E) Elongated hypocotyl cells in light-grown wild-type plants show rows of stomatal complexes (arrowheads).
(F) A dis1-1 hypocotyl shows that cells are short and broad and stomatal rows often are absent in the mutants.
(G) and (H) Dark-grown wild-type hypocotyl cells are elongated and maintain firm contact with each other (arrowhead) at their ends (G), whereas contiguous hypocotyl cells developing under the same conditions in both wrm and dis1 mutants lose contact with their neighbors along the long axis and curl out of the epidermal plane (arrowheads) (H).
(I) Wild-type root hairs are straight and up to 1 mm long.
(J) and (K) Root hairs of dis1-1 representing the relatively short, stubby cells (J) and cells with varying degrees of waviness (K) (arrowheads) seen in the mutants.
Bars = 500 μm in (A) and (B), 50 μm in (C), (D), (G), (H), and (K), 100 μm in (E) and (F), and 200 μm in (I) and (J).
Rapid Cell Expansion Leads to Aberrant Behavior of Both Diffusely Growing and Tip-Growing Cells
Wild-type seedlings grown in the dark for 7 to 9 days displayed 2.5- ± 0.4-cm-long hypocotyls with a cell length (middle region of the hypocotyl) of 300 ± 40 μm and a width-to-length ratio of 1:12. Mutant hypocotyls were shorter, ∼1.6 ± 0.4 cm, with an average cell length of 198 ± 20 μm and a width-to-length ratio of 1:8. Whereas in the wild type, elongating hypocotyl cells maintained straight rows and a firm contact with their neighbors (Figure 2G), contiguous epidermal cells in the mutants broke contact at their ends, resulting in disconnected cells with curling ends (Figure 2H). However, within a given seedling population, the final length attained by different mutant epidermal cells, the degree of separation between contiguous cells, and the frequency and degree of cell end curling appeared as random parameters in which clear patterns of behavior could not be discerned.
To challenge roots hair cells to elongate rapidly, seeds were germinated on plates tilted to an angle of ∼20° from the vertical so that extending root hairs had to grow out into the air before they could touch the medium surface again. Wild-type root hairs grew straight (Figure 2I) under these conditions to lengths of 800 ± 150 μm, whereas mutant hairs remained relatively short (350 ± 200 μm) and stubby (Figure 2J) and displayed considerable waviness upon extension (Figure 2K).
We concluded that the WRM/DIS1 gene products are essential during rapid cell expansion in several cell types. The nature of the mutated genes as well as previous studies that have clearly indicated a link between cell expansion and the actin cytoskeleton (Mathur et al., 1999; Baluska et al., 2001) prompted us to investigate the actin cytoskeleton in mutant cells.
Actin Organization Is Aberrant in wrm and dis1 Mutants
Visualization of F-actin organization in all living cells of the mutants was achieved by crossing the different mutant alleles to a transgenic GFP-mTalin Arabidopsis line (Kost et al., 1998; Mathur et al., 1999). Although actin organization during the earliest stage of development in both wild-type and mutant trichomes was indistinguishable, differences became apparent after branch initiation as the cells embarked on a rapid expansion phase. Wild-type trichome cells assembled longitudinally, extending F-actin cables (Figure 3A) that persisted into the mature trichome. However, expanding mutant trichomes at the same stage of development displayed unusual localized accumulation of F-actin and a conspicuous absence of long actin cables (Figure 3B) that continued through subsequent mutant cell development (compare Figures 3C and 3F with Figures 3D and 3E). As a consequence, mature mutant trichomes characteristically displayed a randomly arrayed, polygonal cytoplasmic organization linking the brightly fluorescent F-actin foci together that clearly differed from the thin, longitudinally running F-actin cables seen in the wild type (compare Figures 3D and 3G).
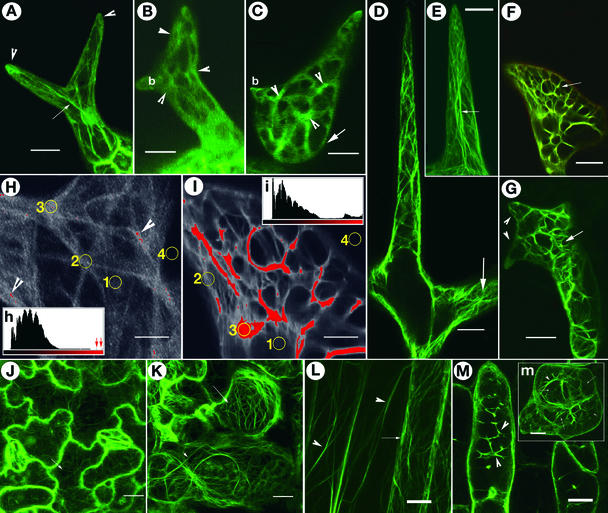
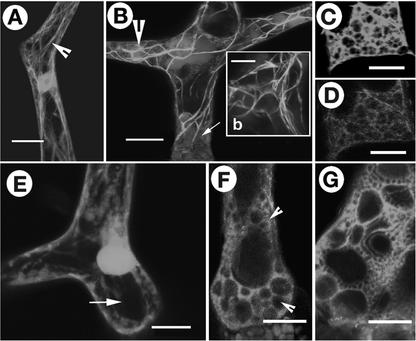
Figure 3.
F-Actin Organization in Different Cell Types in Wild-Type Arabidopsis and wrm/dis1 Mutants.
Localized increase in F-actin aggregation is a feature observed in both mutants.
(A) A wild-type trichome with extending branches organizes long F-actin cables (arrow) while maintaining a dense actin cap at the tips of branches (arrowheads).
(B) A dis1-1 trichome at approximately the same stage as that in (A) displaying abnormal accumulation of F-actin (arrowheads) and a conspicuous absence of long actin cables. Note that one branch (b) remains relatively unextended compared with the other branch.
(C) A developing wrm1-2 trichome showing increased F-actin aggregation in reticulate cytoplasmic networks containing brightly fluorescent actin foci (arrowheads). Similar cytoplasmic and F-actin organization is not observed in wild-type trichomes. Note that the lower portion of the trichome (arrow) corresponding to the stalk region is more radially expanded, whereas a small branch (b) remains unextended.
(D) Portion of a wild-type trichome showing a well-extended branch. Note that the F-actin organization consists predominantly of longitudinally oriented subcortical cables and a finer reticulum of cortical F-actin visible in the area shown by the arrow.
(E) The tip region of a wild-type trichome branch contains fine, longitudinally oriented F-actin strands (arrow) that appear to meet at the apex.
(F) An abnormally expanded tip region in a dis1-2 trichome cell representing the reticulate actin-filled cytoplasm in nonexpanding branch tips in mutant trichomes (arrow), clearly different from the wild-type situation seen in (E).
(G) A wrm1-2 trichome representing the characteristic actin organization seen in mature mutant trichomes. The abundance of intersecting, actin-rich cytoplasmic strands produces a loose polygonal mesh (arrow) that is clearly different from the longitudinally oriented cytoplasmic strands containing relatively thin, long, extended F-actin cables typical of mature wild-type trichomes (D). Note that the mutant branches (arrowheads) remain unextended as short spikes.
(H) and (I) Comparison of F-actin organization between wild-type and mutant trichome cells based on the fluorescence levels of the F-actin binding GFP-mTalin probe. Comparison of the wild-type image (H) and the dis1-2 mutant cell image (I) demonstrates the relative difference in fluorescence levels. The large areas covered with red in (I) have only small counterparts (arrowheads) in (H). Insets (h) and (i) are histograms that differentiate between the red and black levels to accentuate the relative differences in (H) and (I). Small arrows in histogram (h) indicate the minute red regions in (H). Although the images shown are of mature trichomes, the yellow circles in (H) and (I) indicate representative areas that were used to obtain the gray-scale readings reported in Table 2. Circle 1, very fine, diffuse F-actin; circle 2, defined F-actin strand; circle 3, brightest fluorescent spots in an image; circle 4, background.
Because the GFP-mTalin fusion protein used to visualize the actin organization exhibits high specificity for binding to F-actin (Kost et al., 2000, and references therein), the observation of brightly fluorescent F-actin label in cytoplasmic strands of the mutant suggested the presence of densely bundled F-actin. To confirm this possibility, the actin organization in wild-type and mutant trichomes was subjected to image analysis for regional differences in fluorescence levels that were taken to reflect the degree of F-actin aggregation at each spot (Figures 3H and 3I, Tables 2 and 3). Based on gray-scale values, regional fluorescence was categorized into three classes (Tables 2 and 3) corresponding to areas containing very fine F-actin (which appear diffuse gray in compressed confocal image stacks; e.g., circle 1 in Figures 3H and 3I), a second class comprising regions of thin but clearly defined F-actin strands (e.g., circle 2 in Figures 3H and 3I), and class three, comprising regions with brightly fluorescent F-actin aggregates (e.g., circle 3 in Figures 3H and 3I).
Table 2.
Region- and Cell Type–Specific F-Actin Fluorescence Values in Developing Trichome Cells of Arabidopsis Wild Type (Landsberg erecta) and the dis1-1 Mutant
| Region | Stage of trichome development
|
|||||
|---|---|---|---|---|---|---|
| Young
|
Middle
|
Mature
|
||||
| Wild Type | dis1-1 | Wild Type | dis1-1 | Wild Type | dis1-1 | |
| Diffuse fluorescence | 40 ± 7 | 42 ± 7 | 43 ± 11 | 41 ± 9 | 36 ± 5 | 36 ± 5 |
| Well-defined F-actin strand | – a | – | 68 ± 11 | 68 ± 15 | 89 ± 10 | 98 ± 33 |
| Bright fluorescence | – | – | – | 189 ± 18 | – | 210 ± 52 |
Average fluorescence values and standard deviations are based on gray-scale readings from five separate regions in 15 cells each of the wild type and the dis1-1 mutant. The three regions correspond to areas marked 1, 2, and 3, respectively, in Figures 3H and 3I. The stages of trichome development are defined as follows: young, when it is not possible to distinguish F-actin strands within diffuse fluorescence; middle, when extending F-actin strands are visible (Figures 3A to 3C); mature, bright fluorescence (Figures 3D to 3G).
Regions/stages that are absent and cannot be compared.
Table 3.
Region- and Cell Type–Specific F-Actin Fluorescence Values in Developing Hypocotyl Cells of Arabidopsis Wild Type (Landsberg erecta) and the dis1-1 Mutant
| Region | Stage of hypocotyl cell development
|
||||
|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
||
| (Wild Type) | (dis1-1) | (dis1-1) | (dis1-1) | (dis1-1) | |
| Cell end | 63 ± 9 | 65 ± 17 | 89 ± 32 | 123 ± 38 | 128 ± 40 |
| Cell middle | 51 ± 12 | 55 ± 18 | 58 ± 33 | 55 ± 19 | 53 ± 18 |
Average fluorescence values and standard deviations are based on gray-scale readings from five separate regions in 15 cells each of the wild type and the dis1-1 mutant. Stages 1 to 4 of expanding hypocotyl cells refer to the labels defined in the legend to Figure 4.
The fluorescence values for diffuse F-actin (class 1) and well-defined F-actin strands (class 2) were similar between wild-type and mutant cells. However, a nearly threefold increase in fluorescence levels was observed in the bright actin foci seen in mutant cells over that of the F-actin strands in wild-type trichomes (Table 2). Spot values comparable to the brightly fluorescent actin foci (class 3) seen in mutant cells were not obtained in wild-type trichomes. Within individual trichomes themselves, the ratio between the lowest and the highest fluorescence intensity was 1:2.5 for the wild type and 1:6 for the mutants. Additionally, a visual assessment of the compressed image stacks obtained under identical confocal settings and normalized to an equal gray-scale value for the background gave the impression that mutant cells displayed larger areas with bright fluorescence (compare Figure 3A with Figure 3C and Figure 3D with Figures 3F and 3G). Because comparable readings for the bright fluorescence (class 3) were not obtained in wild-type trichomes, the threshold-exclusion function in the MetaMorph image-analysis program was used to pseudocolor and overlay cellular areas displaying gray-scale values beyond a specified threshold limit (Figures 3H and 3I). A threshold limit of 150 gray-scale units was found to cover the maximally bright areas in mutant cell images (Figure 3I) and was applied to wild-type images. Figures 3H and 3I show the relative red overlaid images. The red areas seen in the resulting histograms (Figures 3H and 3I) show that compared with wild-type cells at the same developmental stage, a much greater volume of mutant cells is occupied by F-actin aggregates.
We surmised that during growth, mutant trichomes progressively accumulate an excess of bundled/aggregated F-actin in random locations for which comparisons are not found in the wild type.
Abnormal actin accumulation and thicker F-actin bundles also were observed in expanding cotyledon cells (compare Figures 3J and 3K) and hypocotyl cells (compare Figures 3L and 3M) in both mutants, but a clear pattern of actin distribution could not be deciphered. This finding prompted the question of whether increased F-actin accumulation and bundling was the primary cause of cell deformation or was a consequence of cell shape change. This notion was tested in dark-grown hypocotyl cells that in the mutants displayed cell separation and curling.
Actin Organization Defects Precede Cell Deformation
Technical limitations precluded the repeated visualization of the same dark-grown hypocotyl cell during a time-course study. Therefore, the following sequence of events was threaded together through the simultaneous visualization of cell shape and actin organization in 270 different hypocotyl cells from 27 dark-grown dis1-1 mutant seedlings carrying the GFP-mTalin transgene. Controls consisted of 30 hypocotyl cells from 10 wild-type seedlings (GFP-mTalin transgenic plants) grown and observed under identical conditions. Based on a purely morphological criterion and after staining with propidium iodide to provide contrast and highlight intercellular gaps, the hypocotyl cells were placed into one of four sequential stages (Figure 4A): (1) unaffected; (2) bulged; (3) less expanding and separated; and (4) curled. Figures 4B to 4E and Table 3 show representative images/fluorescence values of F-actin in cells corresponding to the different expansion stages. Cells of the dis-1 mutant at stage 1 elongated like wild-type cells and maintained a fine cortical F-actin mesh (Figure 4B) and the predominantly longitudinally oriented subcortical F-actin organization (Figure 4B, arrow).
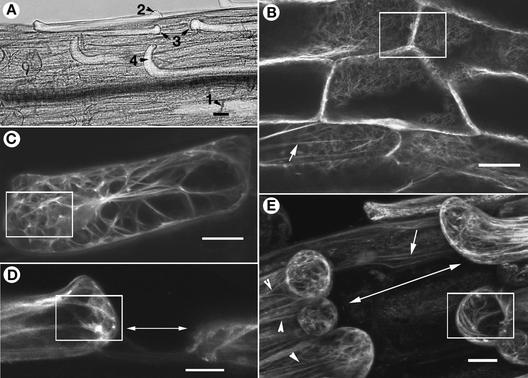
Figure 4.
Defects in F-Actin Organization Precede Morphological Changes in Mutant Cells.
Elongating mutant hypocotyl epidermal cells of the dis1-1 mutant were categorized into different sequential stages of expansion that lead ultimately to contact breakage between contiguous cells. This categorization formed the basis for construing the sequence of changes in F-actin organization that occur in these cells. Hypocotyl cells in GFP-mTalin transgenic plants (Landsberg erecta ecotype) were used as wild-type controls. Bars = 40 μm.
(A) Portion of the hypocotyl in a dis1-1 seedling carrying a GFP-mTalin transgene challenged to elongate rapidly by growing for 8 days in the dark shows cells (labeled 1 to 4) in different stages of expansion. (1) Normally expanding cells in firm contact with their neighbors along the long axis. (2) A bulging cell end signifying the initiation of abnormal expansion; such cells later would break contact with their neighbors along their end walls. (3) Cells with gaps between them resulting from different elongation rates. (4) Cells that have broken contact with their neighbors along the long axis and whose ends have curled out of the epidermal plane.
(B) The actin cytoskeleton at cell ends in control wild-type hypocotyl cells showing the cortical organization (box) and a subcortical, longitudinally oriented F-actin cable (arrow). Nonresponding and stage-1 mutant hypocotyl cells exhibit a similar organization.
(C) The complete F-actin organization of a stage-2 mutant cell depicted by compression of a stack of 30 confocal images shows increased actin accumulation at one end of the cell (box), whereas the opposite side remains relatively clear.
(D) A single median-longitudinal confocal section through two contiguous epidermal cells (stage 3) that have broken contact with each other along the longitudinal growth direction (double-headed arrow). The boxed area shows brightly fluorescent spots indicative of dense F-actin aggregation at the cell end.
(E) Mutant hypocotyl cells in the final stage (stage 4) of development. After breaking contact, the gaps have further widened between cells (double-headed arrow) and many cell ends have curled out. Note the presence of thick F-actin bundles in the boxed area, the occurrence of long F-actin cables (arrowheads), and the nearly normal F-actin organization in an underlying hypocotyl cell (arrow).
Fluorescence measurements at different intracellular locations were used subsequently to substantiate our microscopic observations on regional changes occurring in hypocotyl cells. Wild-type cells maintained a nearly uniform average in their middle regions, although a slight increase in F-actin density was suggested by fluorescence measurements at the cell ends (Table 3). By contrast, as shown by confocal imaging and fluorescence measurements, increased aggregation of F-actin–rich cytoplasm was observed at one (Figure 4C) or both ends of all stage-2 mutant cells (Table 3). The unequal aggregation of F-actin often was accompanied by localized bulging and detachment of the cell end(s) from the underlying layer of cells. Further increases in end-localized F-actin accumulation and apparent bundling (Table 3) produced cells that did not elongate properly. As a result, stage-3 cells displayed different elongation rates and broke contact with contiguous cells along their long axes (Figure 4D). Having broken contact, the ends of affected cells usually curled out of the epidermal plane (stage 4; Figure 4E). Although cell curling was most apparent in the uppermost epidermal layer, actin bundling extended to underlying cells as well. In all cell types examined, regions free from localized actin accumulation and bundling maintained normal growth characteristics and morphology, whereas cell shape defects always were preceded by aberrant cytoplasmic and F-actin organization.
The overall altered F-actin organization in various rapidly expanding cells caused by an apparent increase in F-actin aggregation (Figures 3 and 4) suggested that the mutants might be impaired in the efficiency with which fine actin filaments could be generated. To provide specific proof for this notion, we used latrunculin B (Lat-B), a well-characterized G-actin agonist known to interfere with actin polymerization (Morton et al., 2000; Baluska et al., 2001; Vidali et al., 2001).
Lat-B–Induced Inhibition of Actin Polymerization Phenocopies wrm/dis1 Mutants
A GFP-mTalin line (Landsberg erecta background) that allows F-actin visualization (Kost et al., 1998) was used as the wild type with respect to trichomes in these experiments. At high concentrations, Lat-B treatment has been shown to rapidly inhibit actin polymerization and result in distorted trichomes (Mathur et al., 1999). Here, we used low concentrations of Lat-B (1 to 10 nM) that did not inhibit cytoplasmic streaming to follow the gradual changes in F-actin organization in wild-type trichomes. Within 2 h of Lat-B application (5 nM), an increase in cytoplasmic granularity was observed and thick, brightly fluorescent actin bundles became visible in trichomes (Figures 5A and 5B). At this stage, the bundled F-actin cytoskeleton in the wild-type Lat-B–treated trichomes (Figure 5B) closely resembled that observed in mutant trichomes (Figure 3G). The F-actin bundles disappeared completely within 6 h and were replaced by a diffuse green fluorescence, suggesting a complete disruption of F-actin organization in the treated cells. Consistent with these observations, trichomes that developed over the next 24 h on medium containing low concentrations of Lat-B were short and distorted. Increased actin bundling before the complete disappearance of F-actin also was observed in cotyledon, hypocotyl, and leaf cells, all of which generally remained short and unexpanded in Lat-B–treated seedlings (data not shown).
Figure 5.
The Intracellular Organization in wrm/dis1 Trichomes Resembles That Achieved upon Lat-B Treatment of Wild-Type Trichome Cells.
(A) Actin cytoskeleton in an untreated mature wild-type trichome (carrying a GFP-mTalin transgene for F-actin visualization) serving as a control in the Lat-B treatment experiments displays the normal long F-actin cables (arrowhead).
(B) A mature wild-type trichome treated with 5 nM Lat-B for 2 h displaying a general increase in cytoplasmic granularity (arrow) and thick F-actin bundling. Inset (b) shows a magnified view of the region indicated by the arrowhead.
(C) and (D) Basal portion of a trichome cell treated with Lat-B for 2 h and then stained simultaneously with FDA (C) and FM4-64 (D). FDA is excluded from the miniature vacuoles, whereas FM4-64 staining labels vacuolar membranes specifically.
(E) An FDA-stained wild-type trichome shows a large central vacuole (arrow).
(F) An FDA-stained wrm1-2 trichome at a developmental stage comparable to that of the wild-type trichome in (E) shows many small vacuoles (arrowheads) of varying size in the vicinity of larger vacuoles.
(G) The base of an FDA-stained trichome that developed on medium containing 5 nM Lat-B showing numerous apparently unfused vacuoles of varying size (cf. with [E] for the absence of a large central vacuole and with [F] for the presence of small vacuoles).
Bars = 80 μm in (A), (B), and (E) to (G) and 40 μm in inset (b), (C), and (D).
Whereas the Lat-B treatment clearly showed an effect on actin organization and trichome morphology, the initial increase in cytoplasmic granularity was not clearly understood. This was investigated further in Lat-B–treated trichomes.
Vacuole Morphology Is Altered in Lat-B–Treated and Mutant Trichomes
Under a light microscope, Lat-B–treated trichomes were found to have pockets containing numerous small, membrane-bound compartments. Because wild-type trichomes possess a single large vacuole whose expansion correlates closely with trichome cell growth and morphogenesis (Hülskamp et al., 1994), we attempted to identify these membranous compartments. Fluorescein diacetate (FDA), which stains the cytoplasm but is excluded from the vacuole (Fricker et al., 2001), and FM4-64, which specifically stains vacuolar membranes (Vida and Emr, 1995; Kutsuna and Hasezawa, 2002), were used. Addition of these dyes along with Lat-B to wild-type trichomes showed an increasing accumulation of small bodies (Figures 5C and 5D) near existing large vacuoles and suggested that they could be unfused miniature vacuoles.
Staining of mutant seedlings with FDA revealed that in contrast to wild-type trichomes, in which a single large vacuole is seen (Figure 5E), mutant trichomes possess numerous, randomly distributed miniature vacuoles that accumulate around larger vacuoles (Figure 5F). Although large vacuoles were never completely absent, in most mutant trichomes small vacuoles were observed moving around in the general cytoplasmic flow and accumulating in randomly located small pockets near or well separated from a large vacuole. Areas with extensive actin bundles were conspicuously devoid of large vacuoles.
The presence of small unfused vacuoles in Lat-B–treated cells (Figure 5G) and the existence of similar miniature vacuoles in the mutants (Figure 5F) suggested that vacuolar-membrane fusion could be impaired in wrm/dis1 mutants and also may contribute to the mutant phenotype.
DISCUSSION
Cell Shape Determination Involves the Actin Cytoskeleton
The cytoskeleton plays an important role in cell shape determination in higher plants. This has been demonstrated clearly in drug-based experiments involving microtubules and actin microfilaments using diverse cell types (Mathur and Hülskamp, 2002). Clear molecular links to the microtubule cytoskeleton have been established with the cloning of genes such as ZWICHEL, which encodes a kinesin-like protein, mutation of which leads to short, less-branched trichomes (Oppenheimer et al., 1997); MOR1, which encodes a homolog of the TOG-XMAP215 family, with the mutant characterized by rapid alterations in microtubule arrays in response to temperature shifts (Whittington et al., 2001); FRA2/ERH3/BOT1/FTR, which encode a katanin p60-like protein, with mutants exhibiting pleiotropic phenotypes, including altered morphology of root and leaf trichome cells (Bichet et al., 2001; Burk et al., 2001; Webb et al., 2002); and members of the PILZ group, which encode different tubulin-folding cofactors, mutation of which results in phenotypes ranging from mere cell shape changes to embryo lethality (Kirik et al., 2002; Mayer and Jürgens, 2002).
Molecular evidence for actin cytoskeleton involvement in the shape development of single cells also is beginning to accumulate. Thus, the demonstration of Arabidopsis ACTIN2-linked defects in the deformed root hairs1 mutants (Ringli et al., 2002), observations of altered cell expansion and flowering time in profilin (Ramachandran et al., 2000) and ADF (Dong et al., 2001) transgenic plants, and the cloning of the BRK1 (BRICK1) gene from maize mutations that display epidermal cell shape alterations (Frank and Smith, 2002) all add to our understanding of the behavior of the actin cytoskeleton in plant development. The BRK1 gene is especially relevant to this study, because the HSPC300 gene product of its human ortholog has been shown to interact with the ARP2/3 complex (Eden et al., 2002; Smith, 2003). Thus, our cloning of WRM/DIS1 genes not only reveals their molecular identities but also suggests that the ARP2/3 complex, hitherto unrecognized in plants, may play a pivotal role in cellular morphogenesis, perhaps in interaction with the novel BRK1 gene.
The ARP2/3 Complex Exists in Higher Plants
The seven-subunit ARP2/3 complex is an acknowledged multifunctional modulator of actin organization in diverse organisms (Machesky et al., 1997; Machesky and Gould, 1999; Welch, 1999). Using amino acid sequence comparisons, we show that all components of the complex are present in Arabidopsis. Structure-function relationship studies of the ARP2/3 complex have suggested that ARP2 and ARP3 come together through the concerted movement of several subunits to form a nucleation site for the growth of a new actin filament (Robinson et al., 2001; Volkmann et al., 2001). Functional dissection of the complex in yeast supports this view and clearly indicates that loss of either of the two major subunits (ARP2 or ARP3) severely compromises the actin-nucleating activity of the complex (Winter et al., 1999). Even in the absence of protein data, the similar phenotypes displayed by the wrm and dis1 alleles and their double mutants strongly suggest that the two proteins exist and function in a complex. We conclude that the defects observed in wrm and dis1 are attributable to a loss of ARP2/3 complex activity.
The ARP2/3 Complex Plays a Role in Organizing the Fine Cortical Actin Cytoskeleton in Rapidly Expanding Plant Cells
These observations of wrm/dis1 mutants present compelling evidence for the need to organize actin efficiently during rapid growth. An inability to do so results in spectacularly aberrant cell shapes. Under in vitro conditions, the ARP2/3 complex is known to dramatically enhance the slow spontaneous rate of actin nucleation (Mullins et al., 1998a). In vivo, the role of the ARP2/3 complex in organizing actin is most apparent in regions of rapid actin activity, such as the leading edge of lamellipodia (Welch et al., 1997a), and stems from its ability to initiate fresh actin polymerization on the sides of an existing actin filament (Mullins et al., 1998b). Daughter actin filaments initiated by the ARP2/3 complex arise at nearly 70° angles and result in a dendritic actin array (Svitkina and Borisy, 1999). Such actin organization is believed to enhance the efficiency of membrane protrusion by distributing force generated by actin polymerization over a large area. Because actin monomers also can nucleate and initiate polymerization spontaneously, the loss of a process-enhancing complex such as the ARP2/3 complex may become apparent only during rapid cell growth or movement.
Our observations of wrm and dis1 mutants validate the characteristics of the ARP2/3 complex described above and show that mutant cells that undergo rapid expansion via diffuse growth (i.e., growth spread over a large cellular area) exhibited the most apparent morphological defects. In these cells, changes in actin organization were invariably linked to and preceded cell shape alterations. As cell expansion progressed, there was a concomitant decrease in fine cortical F-actin, and the mature cells finally displayed massive F-actin bundles. Similar disorganized, dysfunctional cortical actin defects that severely compromise cell growth and viability have been reported for ARP2/3 complex mutants in both fission and budding yeast (McCollum et al., 1996; Winter et al., 1999). However, unlike these single-celled organisms, Arabidopsis is a complex multicellular organism in which cell-to-cell contact and coordinated growth rates are absolute requirements for the development of normal form and function. Thus, although the loss of ARP2/3 activity in yeast frequently is lethal, mutants in the same genes in Arabidopsis present new insights into the effects of actin dysfunction in plants.
Proper Actin Organization Is Essential for Cell-to-Cell Contact and Coordinated Targeting of Cell-Building Components
In multicellular plants, cell-to-cell contact integrates cellular processes and ensures growth symmetry. The important role played by the F-actin cytoskeleton in this process was illustrated through our experiments with dark-grown hypocotyls. When challenged to elongate rapidly, many mutant hypocotyl cells with impaired actin organization went out of phase with other cells, resulting in contact breakage and cell curling. Interestingly, contact broke transverse to the axis of cell elongation at the cell ends in regions that displayed maximally aberrant actin organization. Could this be an indication that cell wall–reinforcing material was not delivered correctly to the cell ends? Although a clear explanation must await further experimentation, the recent phenotypic analysis of quasimodo1 (qua1) mutants in Arabidopsis presents a similar cellular phenotype of curling epidermal cells. QUA1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis (Bouton et al., 2002). The qua1 mutants are deficient in the synthesis of pectic polysaccharides, resulting in weakened cross walls. The similar phenotype suggests the following mechanistic reasoning for the wrm/dis1 mutants.
It has been speculated that exocytic vesicles that carry cell-building material are delivered into a cortical vesicle incorporation zone from the ends of fine actin filaments (Ketelaar and Emons, 2001). Consistent with this view, fine actin filaments often are found associated with regions of active growth or cell repair (Foissner et al., 1996; Miller et al., 1999). Fine actin filament generation appears to be impaired in wrm/dis1 mutants because cells frequently are found congested with dense actin bundles. The delivery of vesicles into the growth zone thus may be aberrant in the absence of fine actin, whereas bundled actin probably can act as a direct impediment and also restrict entry of the vesicles into the proper cortical regions. Although not demonstrated clearly for subcellular organelles in plants to date, the results of Boldogh et al. (2001) showing decreased mitochondrial motility in yeast ARPC-5 mutants support such speculation and suggest that the transport of vesicles that carry membrane and cell wall material to the cell cortex also may be compromised in a similar manner in the wrm/dis1 mutants.
Other Actin Polymerization–Mediated Processes Also May Contribute to Cell Shape Changes in the Mutants
That wrm/dis1 mutants suffer from compromised actin polymerization activity resulting from impaired functioning of the ARP2/3 complex was suggested by our cell biological and molecular observations. The experiments with Lat-B, a specific inhibitor of actin polymerization that phenocopied the mutants, further reinforced this fact and also drew attention to yet another phenomenon involving fine F-actin that may contribute to cell shape distortion.
It is widely believed that in contrast to actin polymerization–driven tip growth (Vidali et al., 2001), cell turgor plays an important role during diffuse growth (Kost and Chua, 2002) such as that exhibited by trichomes. Treatment with Lat-B resulted in an accumulation of miniature vacuoles and finally produced distorted trichomes. Trichomes in both wrm and dis1 mutants also displayed a similar accumulation of miniature vacuoles. The ARP2/3 complex has been shown to localize to the vacuolar membrane, and observations with mutants in different subunits of the ARP2/3 complex in budding yeast have shown the existence of similar unfused vacuoles (Eitzen et al., 2002). In plants, the vacuole forms an integral component of the cell-expansion machinery. In trichomes especially, it may provide the turgor support necessary for perpendicular growth and to maintain an erect morphology. An inability of small vacuoles to coalesce into a central large vacuole may result in the loss of turgor support and lead to the recumbent trichome cells seen in wrm/dis1 mutants, whereas randomly located pockets of accumulated vacuoles may further contribute to cell distortions. Although the present experiments do not provide direct evidence, we speculate that the observations of small, apparently unfused vacuoles could be extrapolated to other membranous compartments as well. Thus, loss of ARP2/3 complex activity in the wrm/dis1 mutants generally may impair membrane fusion capability, the loss of which becomes apparent as shape distortions during rapid expansion growth.
Our observations clearly indicate that the loss of ARP2/3 complex activity in Arabidopsis leads to severe defects in F-actin organization concomitant with impaired vacuole-organizing capability, both of which contribute to the development of an aberrant cell morphology. Thus, the ARP2/3 complex is strongly implicated in cell shape development in plants.
METHODS
Drug Treatments and Microscopy
Latrunculin-B (Lat-B; Molecular Probes, Eugene, OR) was dissolved in 30% DMSO and used at 1 to 10 nM. Seedlings of an Arabidopsis thaliana transgenic line carrying a GFP-mTalin gene under the control of a 35S promoter of Cauliflower mosaic virus (Kost et al., 1998) were treated as described previously (Mathur et al., 1999). Drug-treated plants, untreated controls, and mutants carrying GFP-mTalin for visualizing actin organization were examined using a confocal laser-scanning microscope (TCS-SP2; Leica, Wetzlar, Germany) as described (Mathur et al., 2002). For fluorescein diacetate staining, seedlings were dipped into a 0.05% (w/v) fluorescein diacetate solution in deionized water (Fricker et al., 2001), prepared just before use, and observed after 15 to 30 min. A styryl fluorescent dye, FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide (Molecular Probes)], was dissolved in 30% DMSO and used at a final concentration of 1 μM (Vida and Emr, 1995) by adding it directly to wet seedlings. Confocal microscopic observations of FM4-64 staining were made between 1 and 3 h after the addition of the dye using 540-/620-nm excitation/emission settings. Scanning electron micrographs and agarose impressions of epidermal surfaces were made as described (Mathur and Koncz, 1997; Mathur et al., 1999), and the images were captured using frame-grabbing DISKUS software (Technisches Büro, Königswinter, Germany). Images were processed using Adobe Photoshop (6.0) software (Mountain View, CA).
Measurement of Fluorescence Levels
Indirect quantification of fluorescence levels in different regions of developing cells was performed using the image-analysis software MetaMorph version 4.5r4 (Universal Imaging, West Chester, PA). Comparative images were captured using the same confocal microscope settings and normalized for background gray level using Adobe Photoshop (6.0) software before analysis. Gray-scale values were obtained for five separate sites per image using a fixed-size circular-marquee tool (112 pixels) for spot values (Figures 3H and 3I) or a larger rectangle (5850 pixels) for area values (e.g., for end/middle regions of hypocotyl cells) and calculated as average gray-level values ± sd. Fifteen separate images each of wild-type and mutant trichomes and wild-type and mutant hypocotyls were analyzed. For pseudocolor representation of regions displaying different gray-scale values, the threshold-exclusion function of the MetaMorph program was used and the overlaid image was saved in RGB format. Subsequently, the histogram function of Adobe Photoshop (6.0) software was used to provide the relative color saturation in the image under the red channel (Figures 3H and 3I).
Molecular and Genetic Techniques and Transgenic Lines
cDNA clones of WRM (AtARP2; 1270 bp) and DIS1 (AtARP3; 1290 bp) were amplified by PCR (primers used were as follows: for WRM, 5′-ATGGACAACAAAAACGTCGTCGTTTGC-3′ [forward] and 5′-GCGCAGCTTGGCTCATTTTATTCAAAC-3′ [reverse]; for DIS1, 5′-CTAGTCGACATGGATCCGACTTCTCGACC-3′ [forward] and 5′-AGGTACCAGCTTCAATACATTCCCTTGAAC-3′ [reverse]) from a cDNA library generated from 25-day-old, soil-grown Arabidopsis plants (ecotype Landsberg erecta) using standard procedures (Sambrook and Russell, 2001). An ∼3-kb WRM genomic fragment was obtained by PCR from Arabidopsis genomic DNA (5′-CTATAGAAATCGGAGAAGATGGACAA-3′ and 5′-ATGCGTTTGAAAGTTATTATGCTTGG-3′), whereas an ∼2.8-kb fragment was excised by PCR from Arabidopsis BAC clone f3f19 for the DIS1 gene (5′-GGCTTAGTTTCTCTAATGGATCCGACTT-3′ [forward] and 5′-GTTCCCTTACATAACTTGACGAAAGTATT-3′ [reverse]). The cDNA clones were placed under the control of a 35S promoter of Cauliflower mosaic virus or a trichome-specific GLABRA2 promoter (Szymanski et al., 1998) in pCAMBIA-1300 binary vector. Genomic clones were blunt end ligated into the HindIII sites of pCAMBIA-1300 vector. For genomic complementation, Agrobacterium tumefaciens (strain GV3101)–mediated transformation of wrm-1 or dis1-1 was performed with the genomic DNA constructs (without the promoter regions) under the control of the 35S promoter of Cauliflower mosaic virus in the pCAMBIA-gARP2 or pCAMBIA-gARP3 vector, respectively, using the floral dip method (Clough and Bent, 1998). T1 plants were screened for phenotypic rescue on MS medium (Murashige and Skoog, 1962) selection plates containing 25 μg/mL hygromycin B.
Information regarding T-DNA insertion lines in the At3g27000 (WRM/ARP2; SALK_003448) and At1g13180 (DIS1/ARP3; SALK_010045) genes was obtained from the SALK T-DNA insertion mutant collection (http://signal.salk.edu/cgi-bin/tdnaexpress), and seeds were ordered from the Nottingham Arabidopsis Stock Center (NASC; http://nasc.nott.ac.uk/). Mutant lines SALK_003448 and SALK_010045 were confirmed for T-DNA insertion using the suggested sequencing primers on genomic mutant DNA (Salk Institute Genomic Analysis Laboratory). The integrity of all clones was confirmed by sequencing. DNA and protein sequence homology searches were performed using the respective Basic Local Alignment Search Tool (BLAST) programs (Altschul et al., 1997). To test for allelism, the F1 progeny resulting from reciprocal crosses between homozygous lines of the two T-DNA insertion mutants and the ethyl methanesulfonate–generated wrm (wrm1-1 allele) and dis1-1 mutants (Hülskamp et al., 1994) were scored.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the genes mentioned in this article are as follows: At3g27000/WRM/Arabidopsis ARP2, AB026649; complete coding sequence, AF095912, gi-3818623 (Klahre and Chua, 1999); At1g13180/DIS1/Arabidopsis ARP3, AC007357; pCAMBIA-1300 vector sequence, AF234296; and GL2 promoter, L32873.
Acknowledgments
We thank the ABRC/NASC Stock Center for seeds and BAC clones, N.-H. Chua for the GFP-mTalin transgenic line, R. Jefferson for the pCAMBIA-1300 vector, D. Tautz for allowing access to sequencing and confocal microscopy facilities, Anshudeep Mathur for help with computer and imaging work, and Botanical Institute III members for their comments on the manuscript. The study was supported by a Volkswagon Stiftung grant to M.H.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011676.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, M., Macaluso, F., Cammer, M., Chan, A., Segall, J.E., and Condeelis, J.S. (1999). Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 145, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, M.K., Feoktistova, A., McKollum, D., and Gould, K.L. (1996). Fission yeast Sop2p: A novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 15, 6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Baluska, F., Busti, E., Dolfini, S., Gavazzi, G., and Volkmann, D. (2001). Lilliputian mutant of maize lacks cell elongation and shows defects in organization of actin cytoskeleton. Dev. Biol. 236, 478–491. [DOI] [PubMed] [Google Scholar]
- Barrero, R.A., Umeda, M., Yamamura, S., and Uchimiya, H. (2002). Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division. Plant Cell 14, 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Hofte, H. (2001). BOTERO1 is required for normal orientation of microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25,137–148. [DOI] [PubMed] [Google Scholar]
- Boldogh, I.R., Yang, H.C., Nowakowski, W.D., Karmon, S.L., Hays, L.G., Yates, J.R., 3rd, and Pon, L.A. (2001). Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc. Natl. Acad. Sci. USA 98, 3162–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M., Talbotec, J., Granier, F., Lahaye, M., Hofte, H., and Truong, H.-N. (2002). QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cooper, J.A., Wear, M.A., and Weaver, A.M. (2001). Arp2/3 complex: Advances on the inner workings of a molecular machine. Cell 107, 703–705. [DOI] [PubMed] [Google Scholar]
- Dong, C.H., Xia, G., Hong, Y., Ramachandran, S., Kost, B., and Chua, N.H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, S., Rohtagi, R., Podtelejnikov, A.V., Mann, M., and Kirschner, M.W. (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793. [DOI] [PubMed] [Google Scholar]
- Eitzen, G., Wang, L., Thorngren, N., and Wickner, W. (2002). Remodeling of organelle bound actin is required for yeast vacuole fusion. J. Cell Biol. 158, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra, W.J. (1978). Contiguity of linkage groups 1 and 4, as revealed by linkage relationships of two newly isolated markers dis-1 and dis-2. Arabidopsis Inf. Serv. 15, 35–38. [Google Scholar]
- Foissner, I., Lichtscheidl, I.K., and Wasteneys, G.O. (1996). Actin-based vesicle dynamics and exocytosis during wound wall formation in Characean internodal cells. Cell Motil. Cytoskeleton 35, 35–48. [DOI] [PubMed] [Google Scholar]
- Frank, M.J., and Smith, L.G. (2002). A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr. Biol. 12, 849–853. [DOI] [PubMed] [Google Scholar]
- Fricker, M., Parsons, A., Tlalka, M., Blancaflor, E., Gilroy, S., Meyer, A., and Plieth, C. (2001). Fluorescent probes for living plant cells. In Plant Cell Biology, C. Hawes and B. Satiat-Jeunemaitre, eds (New York: Oxford University Press), pp. 35–84.
- Hudson, A.M., and Cooley, L. (2002). A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J. Cell Biol. 156, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp, M., Misera, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Ketelaar, T., and Emons, A.M.C. (2001). The cytoskeleton in plant cell growth: Lessons from root hairs. New Phytol. 152, 409–418. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Grini, P.E., Mathur, J., Klinkhammer, I., Adler, K., Bechtold, N., Herzog, M., Bonneville, J.-M., and Hülskamp, M. (2002). The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the α/β-tubulin monomer balance. Plant Cell 14, 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre, U., and Chua, N.H. (1999). The Arabidopsis actin-related protein 2 (AtARP2) promoter directs expression in xylem precursor cells and pollen. Plant Mol. Biol. 41, 65–73. [DOI] [PubMed] [Google Scholar]
- Kost, B., and Chua, N.-H. (2002). The plant cytoskeleton: Vacuoles and cell walls make the difference. Cell 108, 9–12. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., Mathur, J., Dong, C.H., and Chua, N.-H. (2000). Non-invasive F-actin visualization in living plant cells using a GFP-mouse talin fusion protein. In Actin: A Dynamic Framework for Multiple Plant Cell Functions, C.J. Staiger, F. Baluska, D. Volkmann, and P.W. Barlow, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 637–659.
- Kutsuna, N., and Hasezawa, S. (2002). Dynamic organization of vacuolar and microtubule structures during cell cycle progression in synchronized tobacco BY-2 cells. Plant Cell Physiol. 43, 965–973. [DOI] [PubMed] [Google Scholar]
- Machesky, L. (1999). Rocket-based motility: A universal mechanism? Nat. Cell Biol. 1, E29–E31. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., and Gould, K.L. (1999). The Arp2/3 complex: A multifunctional actin organizer. Curr. Opin. Cell Biol. 11, 117–121. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., Reeves, E., Wientjes, F., Mattheyse, F.J., Grogan, A., Totty, N.F., Burlingame, A.L., Hsuan, J.J., and Segal, A.W. (1997). Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodia protrusion and is composed of evolutionarily conserved proteins. Biochem. J. 328, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, M., and Chua, N.-H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., and Hülskamp, M. (2002). Microtubules and microfilaments in cell morphogenesis in higher plants. Curr. Biol. 12, R669–R676. [DOI] [PubMed] [Google Scholar]
- Mathur, J., and Koncz, C. (1997). Method for preparation of epidermal imprints using agarose. Biotechniques 22, 280–282. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., and Hülskamp, M. (2002). Simultaneous visualization of peroxisome and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128, 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Spielhofer, P., Kost, B., and Chua, N.H. (1999). The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126, 5559–5568. [DOI] [PubMed] [Google Scholar]
- May, R.C., Caron, E., Hall, A., and Machesky, L.M. (2000). Involvement of the Arp2/3 complex in phagocytosis mediated by FcγR or CR3. Nat. Cell Biol. 2, 246–248. [DOI] [PubMed] [Google Scholar]
- Mayer, U., and Jürgens, G. (2002). Microtubule cytoskeleton: A track record. Curr. Opin. Plant Biol. 5, 494–501. [DOI] [PubMed] [Google Scholar]
- McCollum, D., Feoktistova, A., Morphew, M., Balasubramanian, M., and Gould, K.L. (1996). The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 15, 6438–6446. [PMC free article] [PubMed] [Google Scholar]
- McKann, R.O., and Craig, S.W. (1997). The I/LWEQ module: A conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. USA 94, 5679–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney, E.C., Kandasamy, M.K., and Meagher, R.B. (2001). Small changes in the regulation of one Arabidopsis profilin isovariant, PRF1, alter seedling development. Plant Cell 13, 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney, E.C., Kandasamy, M.K., and Meagher, R.B. (2002). Arabidopsis contains ancient classes of differentially expressed actin-related protein genes. Plant Physiol. 128, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., McKinney, E.C., and Kandasamy, M.K. (2000). The significance of diversity in the plant actin gene family. In Actin: A Dynamic Framework for Multiple Plant Cell Functions, C.J. Staiger, F. Baluska, D. Volkmann, and P.W. Barlow, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 3–27.
- Miller, D.D., deRuitjer, N.C.A., Bisseling, T., and Emons, A.M. (1999). The role of actin in root hair morphogenesis: Studies with lipochito-oligosaccharides as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 17, 141–154. [Google Scholar]
- Moreau, V., Galan, J.-M., DeVilliers, G., Haguenauer-Tsapis, R., and Winsor, B. (1997). The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol. Biol. Cell 8, 1361–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell, J.L., Morphew, M., and Gould, K.L. (1999). A mutant of Arp2p causes partial disassembly of the Arp2/3 complex and loss of cortical actin function in fission yeast. Mol. Biol. Cell 10, 4201–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, W.M., Ayscough, K.R., and McLaughlin, P.J. (2000). Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat. Cell Biol. 2, 376–378. [DOI] [PubMed] [Google Scholar]
- Mullins, R.D., Heuser, J.A., and Pollard, T.D. (1998. a). The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95, 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, R.D., Kelleher, J.F., Xu, J., and Pollard, T.D. (1998. b). Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments. Mol. Biol. Cell 9, 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Oppenheimer, D.G., Pollock, M.A., Vacik, J., Szymanski, D.B., Ericson, B., Feldmann, K., and Marks, M.D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann, B., Kessels, M.M., and Kelly, R.B. (2000). Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 150, F111–F116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, S., Christensen, H., Ishimaru, Y., Dong, C.H., Wen, C.M., Cleary, A.L., and Chua, N.-H. (2000). Profiling plays a critical role in cell elongation, cell shape maintenance and flowering in Arabidopsis. Plant Physiol. 124, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli, C., Baumberger, N., Diet, A., Frey, B., and Keller, B. (2002). ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 129, 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, R.C., Turbedsky, K., Kaiser, D.A., Marchand, J.-B., Higgs, H., Choe, S., and Pollard, T.D. (2001). Crystal structure of Arp2/3 complex. Science 294, 1679–1684. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schwab, B., Mathur, J., Saedler, R., Schwarz, H., Frey, B., Scheidegger, C., and Hülskamp, M. (2003). Regulation of cell expansion by the DISTORTED genes in Arabidopsis thaliana: Actin controls the spatial organization of microtubules. Mol. Gen. Genomics 10.1007/s00438-003-0843-1. [DOI] [PubMed]
- Smith, L.G. (2003). Cytoskeletal control of plant cell shape: Getting the fine points. Curr. Opin. Plant Biol. 6, 63–73. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Baluska, F., Volkmann, D., and Barlow, P.W., eds (2000). Actin: A Dynamic Framework for Multiple Plant Cell Functions. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Svitkina, T.M., and Borisy, G.G. (1999). Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Lloyd, A.M., and Marks, M.D. (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 5, 214–219. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Marks, D.M., and Wick, S.M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11, 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantard, M., and Blanchoin, L. (2002). Actin polymerization processes in plant cells. Curr. Opin. Plant Biol. 5, 502–506. [DOI] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali, L., McKenna, S.T., and Hepler, P.K. (2001). Actin polymerization is essential for pollen tube growth. Mol. Biol. Cell 12, 2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann, N., Amann, K.J., Stoilova-McPhie, S., Egile, C., Winter, D.C., Hazelwood, L., Heuser, J.E., Li, R., Pollard, T.D., and Hanein, D. (2001). Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science 293, 2456–2459. [DOI] [PubMed] [Google Scholar]
- Webb, M., Jouannic, S., Foreman, J., Linstead, P., and Dolan, L. (2002). Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3, a katanin-p60 protein. Development 129, 123–131. [DOI] [PubMed] [Google Scholar]
- Welch, M.D. (1999). The world according to Arp: Regulation of actin nucleation by the Arp2/3 complex. Trends Cell Biol. 9, 423–427. [DOI] [PubMed] [Google Scholar]
- Welch, M.D., DePace, A.H., Verma, S., Iwamatsu, A., and Mitchison, T.J. (1997. a). The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 138, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, M.D., Iwamatsu, A., and Mitchison, T.J. (1997. b). Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385, 265–269. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Winter, D.C., Choe, E.Y., and Li, R. (1999). Genetic dissection of the budding yeast Arp2/3 complex: A comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA 96, 7288–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2002). Small GTPases: Versatile signaling switches in plants. Plant Cell 14 (suppl.), S375.–S388. [DOI] [PMC free article] [PubMed] [Google Scholar]