Abstract
Genetic analysis of the wound response pathway in tomato indicates that systemin and its precursor protein, prosystemin, are upstream components of a defensive signaling cascade that involves the synthesis and subsequent action of the octadecatrienoic acid (18:3)–derived plant hormone jasmonic acid (JA). The suppressor of prosystemin-mediated responses2 (spr2) mutation, which was isolated previously as a suppressor of (pro)systemin-mediated signaling, impairs wound-induced JA biosynthesis and the production of a long-distance signal for the expression of defensive Proteinase inhibitor genes. Using a map-based cloning approach, we demonstrate here that Spr2 encodes a chloroplast fatty acid desaturase involved in JA biosynthesis. Loss of Spr2 function reduced the 18:3 content of leaves to <10% of wild-type levels, abolished the accumulation of hexadecatrienoic acid, and caused a corresponding increase in the level of dienoic fatty acids. The effect of spr2 on the fatty acyl content of various classes of glycerolipids indicated that the Spr2 gene product catalyzes most, if not all, ω3 fatty acid desaturation within the “prokaryotic pathway” for lipid synthesis in tomato leaves. Despite the reduced levels of trienoic fatty acids, spr2 plants exhibited normal growth, development, and reproduction. However, the mutant was compromised in defense against attack by tobacco hornworm larvae. These results indicate that jasmonate synthesis from chloroplast pools of 18:3 is required for wound- and systemin-induced defense responses and support a role for systemin in the production of a transmissible signal that is derived from the octadecanoid pathway.
INTRODUCTION
Many plants respond to insect attack and wounding by activating the expression of genes involved in herbivore deterrence, wound healing, and other defense-related processes. The synthesis and deployment of wound-induced phytochemicals is regulated by signal transduction pathways that operate both locally at the site of wounding and systemically in undamaged leaves (Green and Ryan, 1972). The occurrence of wound-induced defense responses in diverse plant species (Karban and Baldwin, 1997) suggests the existence of common mechanisms to generate, transport, perceive, and transduce wound signals into physiologically relevant changes in gene expression. Wound-inducible defensive proteinase inhibitors (PIs) in solanaceous plants provide an attractive model system in which to study the mechanism of long-distance wound signaling and its relationship to plant defense (Ryan, 2000; Walling, 2000; León et al., 2001; Gatehouse, 2002; Kessler and Baldwin, 2002; Li et al., 2002b).
A unique component of the wound response pathway in these species is the peptide signal systemin and the precursor protein prosystemin, from which it is derived (Pearce et al., 1991; McGurl et al., 1992). The tomato genome harbors a single copy of the Prosystemin (Prosys) gene that is transcribed and alternatively spliced to generate two functional forms of the protein (Li and Howe, 2001). Several lines of genetic evidence demonstrate that (pro)systemin performs an essential role in wound-induced defense responses. First, transgenic tomato plants that express antisense Prosys cDNA are deficient in wound-induced systemic expression of PIs and exhibit increased susceptibility to herbivores (McGurl et al., 1992; Orozco-Cárdenas et al., 1993). Conversely, overexpression of prosystemin from a 35S:Prosys transgene constitutively activates PI expression in unwounded plants and results in enhanced resistance to herbivores (McGurl et al., 1994; Li et al., 2002a). Finally, forward genetic analysis in tomato has shown that genes required for (pro)systemin-mediated signaling also are essential for wound-induced expression of PI and other defense-related genes (Howe et al., 1996; Howe and Ryan, 1999; Li et al., 2001; Lee and Howe, 2003).
Transcriptional activation of PI genes in response to wounding and systemin depends on the action of jasmonic acid (JA) and related pentacyclic oxylipins (collectively referred to here as jasmonates) that are derived from linolenic acid via the octadecanoid pathway (Farmer and Ryan, 1992; Ryan, 2000; Li et al., 2001). The systemin/jasmonate signaling pathway is initiated upon the binding of systemin to a 160-kD plasma membrane–bound receptor (SR160) that was identified recently as a member of the Leu-rich repeat receptor–like kinase family of proteins (Meindl et al., 1998; Scheer and Ryan, 1999, 2002). Binding of systemin to the cell surface is associated with several rapid signaling events, including increased cytosolic Ca2+ levels, membrane depolarization, inhibition of a plasma membrane proton ATPase, and activation of a mitogen-activated protein kinase activity (Felix and Boller, 1995; Moyen and Johannes, 1996; Stratmann and Ryan, 1997; Moyen et al., 1998; Schaller and Oecking, 1999).
How systemin triggers the release of linolenic acid from membrane lipids for jasmonate biosynthesis remains unknown. The ability of systemin to activate phospholipase A2 (PLA2) activity in tomato leaves suggests that systemin might effect the release of linolenic acid from the plasma membrane, analogous to the PLA2-mediated release of arachidonic acid in animal systems (Farmer and Ryan, 1992; Narváez-Vásquez et al., 1999). However, the demonstrated role of chloroplast-localized PLA1 in JA biosynthesis during flower development in Arabidopsis (Ishiguro et al., 2001) raises the possibility that systemin perception at the plasma membrane is coupled to the activation of a similar lipase in the chloroplast. In addition to systemin, oligogalacturonic acid (OGA) derived from plant cell walls activates PI expression through the jasmonate signaling pathway (Bishop et al., 1981; Doares et al., 1995). JA synthesized in response to wounding, systemin, and OGA acts in concert with ethylene (O'Donnell et al., 1996) and hydrogen peroxide (Orozco-Cárdenas et al., 2001) to positively regulate the expression of downstream target genes.
The signal transduction pathway that regulates wound-induced PI expression in damaged tomato leaves is distinct from the pathway that operates in distal undamaged tissues (Li et al., 2002b; Strassner et al., 2002). Grafting experiments using various wound-response mutants demonstrated that systemic PI expression requires jasmonate biosynthesis in wounded leaves but not in distal undamaged leaves (Li et al., 2002b). Conversely, the systemic response requires jasmonate perception in undamaged responding tissues but not in damaged leaves in which the long-distance signal is generated. These findings suggest that JA or a related compound derived from the octadecanoid pathway functions as a non-cell-autonomous wound signal. The wound-response phenotype of tomato mutants that are defective in the production or perception of systemin indicates that this polypeptide functions primarily in the systemic response (McGurl et al., 1992; Lee and Howe, 2003). Homologs of tomato systemin have been identified in potato, pepper, and nightshade but not in Arabidopsis or tobacco (Constabel et al., 1998; Ryan et al., 2002). However, recent studies have demonstrated that tobacco contains two nonhomologous peptide signals that are produced from a single precursor protein (Pearce et al., 2001). The systemin-like bioactivity of these peptides suggests that they play a role in wound-induced systemic defense responses. In contrast to systemins, OGAs are thought to mediate the JA-dependent expression of PI genes in the vicinity of the wound site (Baydoun and Fry, 1988; Ryan, 2000; Lee and Howe, 2003). Thus, OGAs and systemin appear to regulate the jasmonate pathway in ways that promote the local and systemic expression, respectively, of wound-responsive defense genes.
Studies in Arabidopsis present a more complex view of wound signaling than that proposed for solanaceous species. In particular, wounding of Arabidopsis activates multiple signaling pathways that regulate the expression of distinct sets of target genes. One pathway uses OGAs to induce the expression of wound-response genes at or near the site of wounding (Rojo et al., 1999). In contrast to tomato, in which JA and ethylene work synergistically to coordinate wound-induced gene expression, OGA-induced signaling in Arabidopsis occurs independently of JA and ethylene. Moreover, OGAs and ethylene appear to work together in Arabidopsis to negatively regulate JA-dependent wound responses (Titarenko et al., 1997; Kubigsteltig et al., 1999; Rojo et al., 1999). Antagonism of the JA-dependent pathway by OGA and ethylene in wounded leaves may provide a mechanism to regulate the temporal and spatial expression of distinct sets of wound-response genes (Rojo et al., 1999; León et al., 2001). Although jasmonates clearly are required for a subset of wound-induced systemic responses in both Arabidopsis and tomato, other remote responses to localized wounding in these plants are not dependent on JA (Malone, 1996; Titarenko et al., 1997; O'Donnell et al., 1998; Howe et al., 2000; LeBrasseur et al., 2002). The physiological role of JA-independent wound responses, and their relationship to plant defense, remain to be determined.
The jasmonate pathway plays a central role in the regulation of induced defense responses in species throughout the plant kingdom (Gatehouse, 2002; Kessler and Baldwin, 2002; Liechti and Farmer, 2002; Turner et al., 2002; Wasternack and Hause, 2002). In this context, the apparent differences in the function of ethylene, OGAs, and systemin in tomato versus Arabidopsis may reflect mechanisms that optimize the ability of each plant to control, through modulation of the jasmonate pathway, the temporal and spatial expression patterns of target genes in response to a range of environmental stress conditions and developmental cues. This idea is consistent with increasing evidence that jasmonates regulate different target processes in different plants. For example, genes that encode two of the major classes of systemin- and JA-induced defense proteins in tomato, ser proteinase inhibitor II (PI-II) and polyphenol oxidase, are absent in Arabidopsis (Allen, 2002; Van der Hoeven et al., 2002). It also has been noted that mutants of Arabidopsis that are defective in the jasmonate pathway are male sterile, whereas comparable mutants in tomato are male fertile and, in one case, female sterile (Li et al., 2001; Wallis and Browse, 2002). The role of systemin as a positive regulator of jasmonate signaling in tomato offers a unique opportunity to understand peptide-mediated responses in plants and to clarify the differences between the wound signaling pathways in diverse plant species.
We are using a forward genetic approach to understand the role of systemin and JA in long-distance wound signaling and to determine how these signals interact to coordinate host defense responses. Toward this goal, we conducted a genetic screen to identify mutations that suppress the constitutive wound-signaling phenotype of transgenic plants that overexpress a 35S:Prosys transgene (Howe and Ryan, 1999). Among several ethyl methanesulfonate–induced spr (suppressor of prosystemin-mediated responses) mutants identified, one (spr2) was shown to be impaired in wound-induced JA accumulation and the production of the transmissible wound signal (Li et al., 2002b). Here, we show that Spr2 encodes a chloroplast fatty acid desaturase that catalyzes the ω3 desaturation of linoleic acid (18:2) to the jasmonate precursor, linolenic acid (18:3). Our findings demonstrate that wound- and systemin-induced activation of anti-herbivore defense responses, as well as the production of the long-distance wound signal for PI expression, are dependent on chloroplastic pools of trienoic fatty acids that give rise to JA. The dependence of systemin action on Spr2 is consistent with the hypothesis that systemin functions to increase jasmonate synthesis to a level that is required for the systemic response.
RESULTS
The Wound-Response Phenotype of spr2 Plants Results from a Defect in the Octadecanoid Pathway for JA Biosynthesis
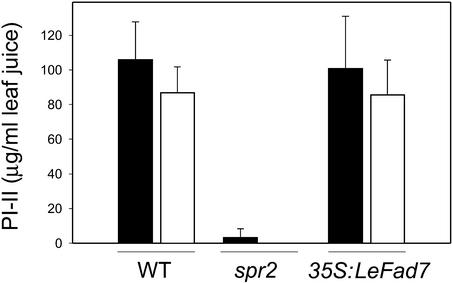
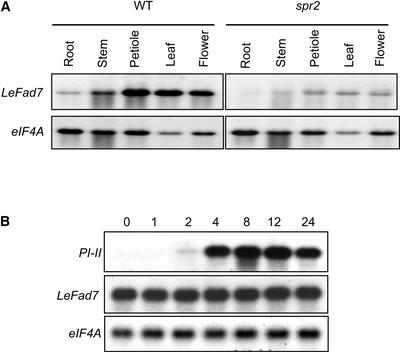
The recessive spr2 mutation was shown previously to suppress the 35S:Prosys-mediated expression of several defense-related genes, including the Ser PI genes PI-I and PI-II (Howe and Ryan, 1999). Further characterization of the mutant was performed using an spr2/spr2 homozygous line in which the 35S:Prosys transgene was removed by outcrossing, followed by three successive backcrosses to the wild-type cv Castlemart. Aside from a slight chlorotic phenotype of young leaves, the overall growth rate and morphology of spr2 homozygotes were indistinguishable from those of wild-type plants. In response to mechanical wounding, however, spr2 plants accumulated ∼5% of the wild-type levels of PI-II in the wounded leaf (local response) and no detectable PI-II in the upper, unwounded leaf (systemic response; Figure 1A). Analysis of PI-II levels in an F2 population derived from a cross between spr2 and the wild type showed that the ratio of wound-responsive wild-type to nonresponsive mutant plants was 89:34 (χ2 = 0.46 for the 3:1 hypothesis). Consistent with the PI-II protein data (Figure 1A), RNA gel blot analysis showed that PI-II was expressed very weakly in wounded spr2 leaves and was not expressed in systemic undamaged leaves (Figure 1B). The time course of this residual PI expression in the mutant was similar to that of wound-induced PI-II expression in wild-type plants, indicating that spr2 affects the amplitude but not the timing of wound-induced signaling. These results demonstrate that, in an otherwise wild-type genetic background, spr2 behaves as a single recessive mutation that impairs local and systemic PI gene expression in response to wounding.
Figure 1.
spr2 Impairs the Wound-Induced Expression of Proteinase Inhibitors.
(A) PI-II accumulation in tomato leaves in response to mechanical wounding. Sixteen-day-old wild-type (WT) and spr2 plants were wounded with a hemostat. Twenty-four hours after wounding, PI-II levels were measured in the wounded leaf (light gray bars; L, local response) and an upper, unwounded leaf (dark gray bar; S, systemic response). As controls (C), PI-II levels were measured in leaf juice expressed from pooled upper and lower leaves of unwounded plants (black bar). Values represent means ± sd from 12 plants per genotype.
(B) Time course of PI-II transcript levels in response to mechanical wounding. Sixteen-day-old seedlings of wild-type (WT) and spr2 plants were mechanically wounded at the distal end of the terminal leaflet of the lower leaf. At various times thereafter, the lower damaged leaf (L, local response) and an upper, unwounded leaf (S, systemic response) were harvested separately for RNA extraction. RNA gel blots were hybridized to a 32P-labeled PI-II cDNA or, as a loading control, a cDNA for translation initiation factor eIF4A.
The deficiency in wound-induced JA accumulation in spr2 plants (Li et al., 2002b) suggested that the mutation affects the octadecanoid pathway for JA biosynthesis. To test this hypothesis further, the responsiveness of spr2 plants to various chemical elicitors of PI expression was determined. Systemin and OGA have been shown to activate PI expression via the octadecanoid pathway (Doares et al., 1995). When supplied to young tomato plants through the cut stem, these compounds induced PI-II accumulation in wild-type but not spr2 plants (Table 1). However, spr2 plants were fully responsive to exogenous JA as well as its metabolic precursors 13(S)-hydroperoxy linolenic acid (13-HpOTrE) and 12-oxo-phytodienoic acid (12-OPDA). The mutant also expressed PI-II in response to bestatin, an amino peptidase inhibitor that activates PI gene expression via a pathway that does not require JA production (Schaller et al., 1995). These results are fully consistent with the hypothesis that spr2 abrogates wound-induced PI expression by disrupting an early step in the octadecanoid pathway.
Table 1.
Proteinase Inhibitor II Accumulation in Wild-type and spr2 Plants in Response to Elicitors
| Elicitor | Wild Type | spr2 |
|---|---|---|
| Buffer | 16 ± 10 | 0 |
| Systemin | 135 ± 26 | 0 |
| OGA | 92 ± 15 | 0 |
| 13-HpOTrE | 79 ± 10 | 71 ± 9 |
| 12-OPDA | 126 ± 18 | 119 ± 21 |
| JA | 142 ± 24 | 136 ± 19 |
| Bestatin | 115 ± 9 | 100 ± 16 |
Fifteen-day-old seedlings were supplied with buffer (15 mM sodium phosphate, pH 6.5), systemin (5 pmol/plant), OGA (20 μg/plant), 13-HpOTrE (25 nmol/plant), 12-OPDA (10 nmol/plant), JA (10 nmol/plant), or bestatin (60 nmol/plant) through the cut stem. PI-II levels (μg/mL leaf juice) were measured in leaf juice expressed from leaves 24 h later. Values indicate means ± sd of at least 12 plants. The detection limit of the PI-II assay was ∼5 μg of PI-II/mL of leaf juice.
Recent studies have shown that tomato floral tissues constitutively accumulate high levels of JA, 12-OPDA, and jasmonate amino acid conjugates (Hause et al., 2000). To obtain additional evidence for a role of Spr2 in JA biosynthesis, gas chromatography–mass spectrometry was used to quantify the endogenous levels of JA in wild-type and spr2 flowers. The level of JA in wild-type and spr2 flowers of the same developmental stage was 1485 ± 290 and 265 ± 36 pmol JA/g fresh weight of tissue, respectively. This result indicates that spr2 disrupts not only wound-induced JA synthesis in leaves but also constitutive JA production in reproductive tissues.
Map-Based Cloning of Spr2
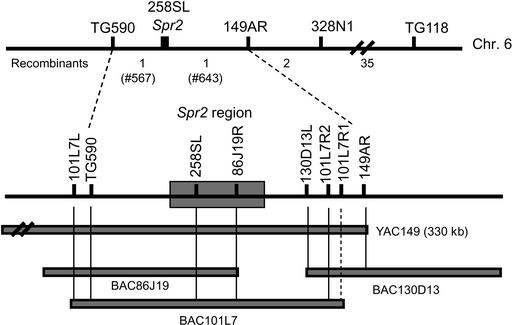
The deficiency in wound-induced PI-II protein accumulation in spr2 plants provided a facile assay to identify the Spr2 gene using a positional cloning approach. A combination of amplified fragment length polymorphism and restriction fragment length polymorphism (RFLP) markers was used to localize Spr2 to a region of ∼3 centimorgan between RFLP markers TG590 and TG118 (see Methods). A high-resolution map of the Spr2 region was constructed by scoring a large mapping population (1436 BC1 plants) for recombination events within the TG590-Spr2-TG118 interval (Figure 2). Markers showing cosegregation (258SL) or very tight linkage (TG590 and 149AR) to Spr2 were used to construct a BAC contig spanning the Spr2 locus. Mapping of sequences corresponding to the BAC ends showed that Spr2 is located physically on BAC101L7. To identify putative genes at the Spr2 locus, the genomic insert of BAC101L7 (∼120 kb) was sequenced to approximately eightfold coverage using a shotgun DNA-sequencing approach. A Basic Local Alignment Search Tool (BLAST) search (Altschul et al., 1990) of a 43-kb contiguous cosegregating genomic sequence against the tomato EST database (http://www.tigr.org) identified exact matches to genes encoding phosphoribosylanthranilate isomerase (EST275992), a putative nucleic acid binding protein (EST273694), and an ω3 fatty acid desaturase (EST474001). On the basis of homology with the well-characterized FAD7 gene from Arabidopsis (Iba et al., 1993), the desaturase-encoding gene was designated LeFad7.
Figure 2.
Map-Based Cloning of the Spr2 Gene.
The positions of molecular markers within the TG590-TG118 interval relative to the position of Spr2 on chromosome 6 are shown at top. Numbers below the line indicate the number of recombination events identified among 1436 BC1 plants analyzed. The positions of markers corresponding to the right (R) and left (L) ends of yeast artificial chromosome (YAC) and BAC clones were determined by mapping experiments using the 39 recombination events in the TG590-TG118 interval. The dotted vertical line at bottom denotes a BAC end that could not be mapped as a result of repetitive sequences. Placement of Spr2 on BAC101L7 was determined by the phenotypic data associated with recombination events 567 and 643.
LeFad7 was considered to be a strong candidate for Spr2 because (1) ω3 desaturases convert 18:2 to the JA precursor, 18:3, and (2) spr2 plants are deficient in JA biosynthesis. Three experiments were performed to test the hypothesis that Spr2 is equivalent to LeFad7. First, an RFLP marker that distinguishes LeFad7 in Lycopersicon pennellii (Fad7pen) from the tomato allele (Fad7esc) was mapped using the recombination events within the TG590-TG118 interval. The results showed that the genotype of all wound-responsive BC1 plants was Fad7pen/Fad7esc, whereas the genotype of all mutant plants was Fad7esc/Fad7esc. This finding suggested that LeFad7 and Spr2 are genetically identical. Second, comparison of the sequence of a LeFad7 cDNA obtained from spr2 plants with the wild-type cDNA revealed the existence of a single nucleotide difference between the two sequences. This polymorphism, which was confirmed in the genomic exon sequence from wild-type and spr2 plants, resulted in the transition of a G to an A at nucleotide 12 of the open reading frame. This mutation changed the codon TGG, which specifies Trp, to the stop codon TGA, resulting in premature termination of the protein. This mutation is predicted to abolish the function of LeFad7 and thus could account for the JA deficiency in spr2 plants.
To confirm this possibility, experiments were performed to determine whether the wild-type LeFad7 gene would complement the spr2 mutant phenotype. For this purpose, a 2.05-kb genomic fragment containing the wild-type LeFad7 gene was placed under the control of the 35S promoter of Cauliflower mosaic virus in the T-DNA vector pBI121. This construct was introduced into the spr2 genetic background using Agrobacterium tumefaciens–mediated transformation. DNA and RNA gel blot analyses identified six independent kanamycin-resistant transformants (T0) that expressed one or more copies of the 35S:LeFad7 transgene (data not shown). All six of these lines showed normal levels of local and systemic PI-II accumulation in response to mechanical wounding (Figure 3). This result demonstrates that 35S:LeFad7 complements the wound-signaling defect in spr2 plants, proving that Spr2 and LeFad7 are equivalent.
Figure 3.
Functional Complementation of spr2 with 35S:LeFad7.
Wound-induced PI-II accumulation in leaves from wild-type (WT), spr2, and 35S:LeFad7-expressing spr2 plants. Plants of each genotype (at the five- to six-leaf stage) were wounded once across the midvein of each leaflet on the two lower leaves. Wounding of the same leaflets was repeated 3 h later. One day after the wound treatment, PI-II levels were determined both in the wounded leaflets (black bars) and in leaflets from the upper, unwounded leaves (white bars). Values represent means ± sd from six independent transgenic T0 lines. PI-II levels in unwounded control plants were below the detection limit of the assay (∼5 μg of PI-II/mL of leaf juice).
Spr2/LeFad7 Encodes a Chloroplast ω3 Fatty Acid Desaturase
ω3 fatty acid desaturases catalyze the conversion of dienoic fatty acids to trienoic fatty acids both in the chloroplast via the so-called prokaryotic pathway and in the endoplasmic reticulum (ER) via the eukaryotic pathway (Browse and Somerville, 1991; Wallis and Browse, 2002). Although plastid- and ER-localized ω3 desaturases share significant sequence similarity, they form distinct phylogenetic groups (Figure 4A). The Spr2/LeFad7 gene (henceforth referred to as LeFad7) is predicted to encode a 435–amino acid protein (LeFAD7) that clearly belongs to the chloroplast group of desaturases. The deduced amino acid sequence of LeFAD7 is most similar to those of chloroplast ω3 desaturases from other solanaceous plants, including potato (94% identity; Martín et al., 1999), tobacco (82% identity; Hamada et al., 1996), and pepper (81% identity; Kwon et al., 2000). Similar to other chloroplast ω3 desaturases, the N terminus of LeFAD7 has characteristic features of a chloroplast-targeting sequence (Keegstra et al., 1989). A plastidic location for the protein was further supported by computer analyses using the ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) programs.
Figure 4.
Spr2 Encodes a Chloroplast ω3 Fatty Acid Desaturase.
(A) Phylogenetic relationship of the Spr2 gene product, LeFAD7, to other plant ω3 fatty acid desaturases. Shown is an unrooted neighbor-joining phylogenic tree constructed in PAUP4.0* from the predicted amino acid sequences of representative desaturases. The phylogenic groups on the left and right sides of the tree correspond to ER-localized and chloroplast-localized ω3 desaturases, respectively. Sequences were from Arabidopsis (AtFAD3, AtFAD7, and AtFAD8), potato (StFAD7), rice (OsFAD3), wheat (TaFAD3 and TaFAD7), maize (ZmFAD7 and ZmFAD8), perilla (PfFAD7), soybean (GmFAD3 and GmFAD7), tobacco (NtFAD3 and NtFAD7), sesame (SiFAD7), and tomato (LeFAD7 and putative LeFAD3).
(B) Gene structure of Spr2/LeFad7 compared with homologous FAD7 and FAD8 genes from Arabidopsis. Intron and exon sequences are indicated by horizontal lines and closed boxes, respectively, and are drawn to scale. Numbers above and below each gene denote the number of nucleotides in each exon and intron, respectively. Only the translated portion of the first and last exon of each gene is shown, together with the start (ATG) and stop (TAG or TGA) codons within the respective exons.
Comparison of the genomic and cDNA sequences revealed that the LeFad7 gene contains seven exons and six introns (Figure 4B). The coding capacity of individual exons, as well as the positions of intron sequences within the gene, indicated that LeFad7 is homologous with the well-characterized FAD7 and FAD8 genes that encode chloroplast-localized ω3 desaturase isozymes in Arabidopsis (Figure 4B) (Iba et al., 1993; Gibson et al., 1994). A notable difference between the tomato and Arabidopsis genes was the presence of an additional intron at the 5′ end of AtFAD7 and AtFAD8. Genomic DNA hybridization experiments using the LeFad7 cDNA as a probe suggested that this gene exists as a single copy in the tomato genome (data not shown). Consistent with this notion, all 42 tomato ESTs annotated as chloroplast ω3 fatty acid desaturases constitute a single tentative consensus sequence (TC99514; release of April 25, 2002) that exactly matches the sequence of LeFad7. A survey of the tomato EST database also revealed the existence of another gene (TC99736) that encodes a putative ER-localized ω3 fatty acid desaturase, designated LeFAD3 (Figure 4A).
LeFad7 Is Expressed Constitutively in Various Tissues
RNA gel blot analysis was used to determine the expression pattern of LeFad7 in various tissues of tomato. In wild-type plants, LeFad7 transcripts were relatively abundant in leaf, petiole, and flower, with somewhat lower expression observed in stems and roots (Figure 5A). This broad expression pattern is consistent with the fact that the ESTs corresponding to LeFad7 were identified in cDNA libraries constructed from leaf, fruit, flower, ovary, pollen, trichome, germinating seed, and root (http://www.tigr.org/). The level of LeFad7 mRNA accumulation in tissues from spr2 plants was significantly less than that in the corresponding wild-type tissues (Figure 5A). This observation is consistent with the well-documented existence of RNA surveillance mechanisms that target mRNAs containing premature termination codons for rapid degradation (Hilleren and Parker, 1999).
Figure 5.
LeFad7 Is Expressed Constitutively in Various Tissues.
(A) Blots containing total RNA from root, stem, petiole, and leaf tissue from 16-day-old wild-type (WT) and spr2 plants were probed with the LeFad7 cDNA. Flower RNA was obtained from mature unopened or newly opened flowers harvested from 7- to 8-week-old plants. Blots also were hybridized to a probe for eIF4A as a loading control.
(B) Expression of LeFad7 and PI-II in response to wounding. Sixteen-day-old wild-type seedlings were wounded on both lower and upper leaves, and total RNA was extracted from the wounded tissue at the times indicated (h) thereafter. RNA was subjected to RNA gel blot analysis as described for (A).
In leaf tissue of several plants, transcripts that encode chloroplast ω3-desaturases have been shown to accumulate in response to wounding (Hamada et al., 1996; Nishiuchi et al., 1997; Martín et al., 1999; Kwon et al., 2000). To determine whether LeFad7 expression is wound inducible, we mechanically wounded wild-type plants and measured the levels of LeFad7 transcript by RNA gel blot analysis at various times thereafter (Figure 5B). This experiment showed that wounding does not significantly affect the steady state level of LeFad7 mRNA in tomato leaves. Hybridization of the same RNA gel blots to a probe for PI-II confirmed that the treatment was effective in activating wound-induced gene expression.
spr2 Impairs the Desaturation of Dienoic Fatty Acids in Chloroplast Lipids
To determine the effect of spr2 on the biosynthesis of polyunsaturated fatty acids, the levels of various C16 and C18 fatty acids in total lipids of wild-type and spr2 leaves were assessed by gas chromatography (Figure 6A). The major fatty acid components of the total lipids in wild-type tomato leaves were 18:3, 18:2, hexadecatrienoic acid (16:3), and palmitic acid (16:0), consistent with previous studies on tomato (Whitaker, 1986; Conconi et al., 1996; Ouariti et al., 1997). In spr2 leaves, 18:3 constituted ∼5% of the total fatty acid content, whereas the accumulation of 16:3 was undetectable (Figure 6A). The decreased content of trienoic fatty acids in spr2 leaves was accompanied by a corresponding increase in the level of dienoic fatty acids (i.e., 16:2 and 18:2). With the exception of a small but significant increase in the proportion of oleic acid (18:1) in spr2 leaves, the levels of other C16 and C18 fatty acids in the mutant were comparable to wild-type levels. To determine whether the spr2 mutation was responsible for the altered fatty acid composition of spr2 leaves, the total fatty acyl content in leaves of spr2 transgenic lines expressing the 35S:LeFad7 transgene was determined (Figure 6A). The results showed that 35S:LeFad7 expression in the spr2 genetic background restored the normal profile of fatty acids in total leaf lipids in each of the transgenic lines tested.
Figure 6.
Fatty Acid Composition in Lipids from Wild-Type and spr2 Plants.
(A) Lipids were extracted from young leaves of 4-week-old wild-type (black bars), spr2 (light gray bars), and 35S:LeFad7-expressing spr2 (dark gray bars) plants. Fatty acids obtained by lipid saponification were quantified by gas chromatography as their methyl ester derivatives. The y axis shows the mole percent of each fatty acid species indicated on the abscissa. Values represent means ± sd from three plants per genotype.
(B) Lipids were extracted from roots of 18-day-old wild-type (black bars) and spr2 (white bars) plants and analyzed as described for (A). Values represent means ± sd from six plants per genotype.
To assess the contribution of LeFAD7 to fatty acid desaturation in nonphotosynthetic tissues, we compared the fatty acyl content of total lipids from wild-type and spr2 roots. The major lipid-derived fatty acids in wild-type roots were 18:2 (48.4%), 16:0 (26.1%), and 18:3 (15.0%) (Figure 6B). This profile is essentially identical to that reported by Ouariti et al. (1997). The fatty acid composition of spr2 roots was very similar to that of wild-type roots, with only minor differences (<5% deviation from wild-type composition) observed in the relative proportions of 18:2 and 18:3. These results indicate that LeFAD7 functions primarily in the desaturation of lipids in leaves and other photosynthetic tissues.
To gain additional insight into the biochemical function of LeFAD7 in lipid biosynthesis, we separated individual glycerolipids from wild-type and spr2 leaves and analyzed them for fatty acid composition (Table 2). The results showed that all major lipids synthesized in spr2 leaves contained reduced levels of 18:3 and a corresponding increase in the proportion of 18:2. It was apparent, however, that spr2 had a greater effect on the desaturation of lipids that are synthesized in the chloroplast (monogalactosyldiacylglycerol [MGDG], digalactosyldiacylglycerol [DGDG], sulfoquinovosyldiacylglycerol [SQDG], and phosphatidylglycerol [PG]) than it does on lipids such as phosphatidylcholine (PC), phosphatidylinositol (PI), and phosphatidylethanolamine (PE) that accumulate primarily in extrachloroplastic membranes. For example, the proportion of 18:3 in chloroplast galactolipids (MGDG and DGDG) from the mutant ranged from 5 to 8% of that found in the corresponding lipids from wild-type plants, whereas the proportion of 18:3 in extrachloroplast lipids (PC, PI, and PE) ranged from 28 to 55% of wild-type levels (Table 2). These analyses also showed that spr2 abolished the accumulation of 16:3, which in wild-type plants is found predominantly in the sn-2 position of the chloroplast-specific lipid MGDG (Jamieson and Reid, 1971; Browse and Somerville, 1991). These results provide direct biochemical evidence for a role of LeFAD7 in the desaturation of dienoic fatty acids in the chloroplast. The spr2 mutation appeared to have little or no effect on the level of stearic acid (16:0 and 18:0) in any of the lipid classes examined. Consistent with the analysis of total fatty acids (Figure 6A), however, the amount of 18:1 in some lipids (e.g., MGDG) was increased slightly in spr2 leaves relative to wild-type levels.
Table 2.
Fatty Acid Composition of Lipids from Wild-Type and spr2 Leaves
| Fatty acid compositiona
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lipid | Genotype | 16:0 | 16:1 | 16:2 | 16:3 | 18:0 | 18:1 | 18:2 | 18:3 |
| MGDG | Wild type | 6.7 | 1.2 | – b | 15.8 | 0.8 | 0.7 | 3.7 | 71.1 |
| spr2 | 6.0 | 2.1 | 0.3 | – | 1.1 | 3.2 | 83.4 | 3.8 | |
| DGDG | Wild type | 31.2 | 0.8 | 0.2 | 1.0 | 1.5 | 1.6 | 3.4 | 60.1 |
| spr2 | 26.5 | 1.0 | 0.1 | – | 1.4 | 2.4 | 63.9 | 4.6 | |
| SQDG | Wild type | 70.7 | – | 0.4 | 2.3 | 0.5 | 4.2 | 7.9 | 14.1 |
| spr2 | 63.7 | 0.8 | 0.3 | – | 1.3 | 3.9 | 28.4 | 1.4 | |
| PG | Wild type | 26.7 | 32.4 | – | 1.0 | – | 4.8 | 21.1 | 14.0 |
| spr2 | 25.0 | 30.0 | 0.3 | – | 0.9 | 8.6 | 33.7 | 0.8 | |
| PC | Wild type | 35.7 | 0.8 | 0.2 | 0.6 | 1.3 | 7.0 | 40.9 | 13.6 |
| spr2 | 33.7 | 0.8 | 0.2 | – | 1.3 | 6.9 | 53.3 | 3.8 | |
| PI | Wild type | 58.2 | 0.4 | 1.4 | 2.0 | 3.0 | 8.7 | 21.9 | 4.5 |
| spr2 | 57.2 | – | 0.7 | – | 2.5 | 6.7 | 30.4 | 2.5 | |
| PE | Wild type | 42.5 | 0.7 | 0.6 | 1.0 | 1.0 | 3.6 | 41.7 | 8.9 |
| spr2 | 42.4 | 0.7 | 0.3 | – | 1.1 | 3.7 | 48.0 | 3.7 | |
Values represent means of triplicate samples and are presented as mole percent. Standard deviations between triplicates was <2% of the indicated values.
Present at trace levels (<0.1% of total fatty acid).
Loss of LeFad7 Function Results in Increased Susceptibility to Insect Attack
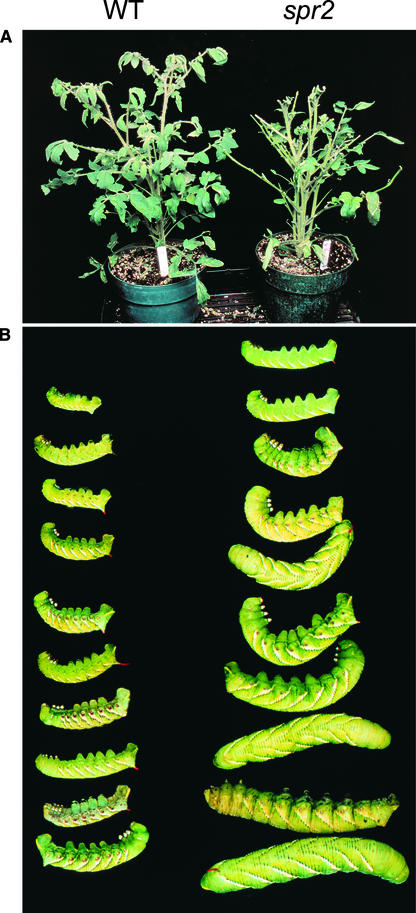
The inability of spr2 plants to express significant levels of defensive PIs in response to mechanical wounding (Figure 1) suggested that the mutant might be compromised in resistance to herbivorous insects. To test this possibility, 3-week-old (experiment 1) and 6-week-old (experiment 2) wild-type and spr2 plants were challenged with tobacco hornworm larvae. After termination of the feeding trial, PI-II protein accumulation in damaged leaf tissue was measured, as was the weight gain of larvae reared on the two host genotypes. In contrast to high levels of PI-II accumulation in herbivore-damaged wild-type leaves, no PI-II accumulation was detected in leaves from damaged spr2 plants (Table 3). The average weight of larvae reared on spr2 plants was 2.3-fold (experiment 1) and 3.1-fold (experiment 2) greater than that of larvae reared on wild-type plants. Furthermore, it was apparent that larvae consumed more foliage from the mutant plants than from the wild-type plants (Figure 7). These data indicate that spr2 compromises the plant's defense response against herbivory. As a result, foliage from spr2 plants is a better food source for hornworm larvae.
Table 3.
Tobacco Hornworm Feeding Assay on Wild-type and spr2 Plants
| Experimenta | Genotype | PI-II (μg/mL) | Larval Weight (g)b |
|---|---|---|---|
| 1 | Wild type | 157 ± 15 | 0.53 ± 0.19 (n = 25) |
| spr2 | 0 | 1.21 ± 0.25 (n = 21) | |
| 2 | Wild type | 303 ± 25 | 1.82 ± 0.27 (n = 14) |
| spr2 | 0 | 5.58 ± 0.67 (n = 16) |
In experiment 1, 30 newly hatched larvae were placed randomly on the leaves of 30 3-week-old tomato plants of each genotype. Larvae were allowed to move freely between plants of the same genotype. In experiment 2, six newly hatched larvae were placed on leaves of each of four separately potted 6-week-old plants of each genotype. Experiments 1 and 2 were terminated 10 and 12 days after the start of the feeding trial, respectively. At that time, PI-II levels in damaged leaf tissue were measured, as was the weight of larvae recovered from the plants. Data represent means ± sd. The detection limit of the PI-II assay was ∼5 μg PI-II/mL leaf juice.
In both experiments, the weight of larvae grown on wild-type and mutant plants was significantly different at P < 0.001 (Student's t test).
Figure 7.
Challenge of Wild-Type and spr2 Plants with Tobacco Hornworm Larvae.
Six newly hatched hornworm larvae (∼5 mg each) were placed on each of four plants (6 weeks old) of each genotype. Larvae were allowed to feed for 12 days and remained on the same plant for the duration of the trial as described for experiment 2 in Table 3. WT, wild type.
(A) Representative wild-type and spr2 plants at the end of the feeding trial.
(B) Hornworm larvae recovered at the end of the trial from representative wild-type and spr2 plants.
DISCUSSION
Spr2 Encodes a Chloroplast ω3 Fatty Acid Desaturase
In the present study, we used a positional cloning approach to demonstrate that the Spr2 gene of tomato encodes a fatty acid desaturase (designated LeFAD7) that is required for jasmonate biosynthesis. Several observations lead us to conclude that LeFAD7 functions within the chloroplast to catalyze the conversion of 16:2 and 18:2 to 16:3 and 18:3, respectively. First, the deduced amino acid sequence of LeFAD7 showed a high degree of similarity to several well-characterized chloroplast ω3 desaturases and was phylogenetically distinct from the group of ER-localized ω3 desaturases. The structure of the LeFad7 gene also indicated that it is homologous with the FAD7 and FAD8 genes of Arabidopsis that encode chloroplast ω3 desaturase isozymes. Second, the spr2 mutation, which abolishes the function of LeFAD7, causes a massive decrease in 16:3 and 18:3 and a corresponding increase in the levels of 16:2 and 18:2 in leaf lipids. Particularly noteworthy was the absence in spr2 leaves of detectable levels of 16:3, which is synthesized exclusively in the chloroplast (Browse and Somerville, 1991). Third, the 18:2 and 18.3 content of spr2 roots differed only slightly from that of wild-type roots (Figure 6B). Because plastids are a minor component of the fatty acid content in roots, this observation supports the idea that LeFAD7 is a chloroplast desaturase. Finally, a role for LeFAD7 in the synthesis of trienoic fatty acids in the chloroplast is consistent with the fact that the octadecanoid pathway for JA biosynthesis is initiated in this cellular compartment (Weber, 2002).
Much of what is known about trienoic fatty acid biosynthesis in plants has come from the characterization of Arabidopsis mutants that are deficient in one or more of three ω3 desaturases (FAD3, FAD7, and FAD8) encoded by the Arabidopsis genome (Wallis and Browse, 2002). The FAD3 gene product is an ER-localized enzyme that desaturates 18:2, whereas the FAD7 gene product desaturates both 16:2 and 18:2 in the chloroplast. FAD8 is an isozyme of FAD7 that is expressed predominantly at low temperature (McConn et al., 1994). Our results strongly suggest that LeFAD7 is functionally homologous with the Arabidopsis FAD7 gene product. In this context, it is of interest to compare tomato and Arabidopsis with respect to the effects that mutations in these genes have on the trienoic fatty acid content of leaves. Both species are considered 16:3 plants owing to the relatively high abundance of the 16:3 acyl group in chloroplast-specific galactolipids (Jamieson and Reid, 1971). At standard laboratory temperatures, the fatty acyl content of leaves from Arabidopsis fad7 plants contains ∼2% 16:3 and 19% 18:3, whereas fad7 fad8 double mutants contain no 16:3 and ∼17% 18:3 (McConn et al., 1994). By comparison, leaves of the tomato spr2 mutant contain no detectable 16:3 and only ∼5% 18:3. This finding indicates that the contribution of LeFAD7 to the production of trienoic fatty acids in tomato leaves is greater than the combined contribution of FAD7 and FAD8 in Arabidopsis leaves. Thus, unlike in Arabidopsis, there appears to be relatively little genetic or biochemical redundancy in the pathway for trienoic fatty acid synthesis in tomato leaves.
Plant cells use both a chloroplast-localized prokaryotic pathway and an ER-localized eukaryotic pathway for the synthesis of glycerolipids and the associated production of trienoic fatty acids. Metabolic coordination between these pathways is achieved, at least in part, by lipid transport between chloroplast and extrachloroplast membranes (Roughan et al., 1980; Browse and Somerville, 1991). The effect of spr2 on the fatty acyl content of various lipids that constitute chloroplast and extrachloroplast membranes is consistent with this idea. For example, transport of a portion of the 18:3-containing lipids synthesized in the ER to the chloroplast may account for the residual level of 18:3 in chloroplast-specific lipids of spr2 leaves. Such a mechanism was proposed previously to account for the 18:3 content of chloroplast lipids in Arabidopsis mutants that are impaired in chloroplast ω3 desaturase activity (Somerville and Browse, 1991; McConn et al., 1994). By comparison with Arabidopsis, the relatively low level of 18:3 in chloroplast lipids from spr2 leaves suggests that the contribution of the eukaryotic pathway to chloroplast lipid synthesis in tomato is less than that in Arabidopsis. It also is possible that tomato contains a second chloroplast ω3 desaturase that is analogous to FAD8 of Arabidopsis. However, genomic DNA hybridization experiments and surveys of the tomato EST database failed to provide evidence for the existence of such a gene. The inability to detect 16:3 in lipids from spr2 leaves also contradicts this possibility. Another implication of the results presented in Table 2 is that LeFAD7 produces trienoic fatty acids for both chloroplast and extrachloroplast membranes. The reduced 18:3 content of extrachloroplast lipids (i.e., PI, PE, and PC) in spr2 leaves supports previous work in Arabidopsis (Browse et al., 1986), suggesting that there is substantial flux of 18:3 between the chloroplast and extrachloroplast membranes.
Role of Trienoic Fatty Acids in Systemin- and Wound-Induced Signaling
The results presented here confirm the previous proposal that spr2 abrogates wound- and systemin-induced gene expression by blocking JA biosynthesis (Li et al., 2002b). This conclusion is supported by the deficiency in JA production in spr2 leaves and flowers as well as by the ability of the mutant to respond to exogenous JA. Our conclusion that LeFad7 function is essential for wound-induced PI expression is consistent with results obtained using potato transgenic lines in which the expression of a chloroplast ω3 desaturase was reduced by antisense technology (Martín et al., 1999). In the case of tomato, it is noteworthy that the small amount of 18:3 in spr2 chloroplast lipids (∼7% of the combined 18:3 levels in MGDG, DGDG, sulfolipid [SL], and PG in wild-type leaves) was correlated with the capacity of spr2 leaves to accumulate low levels (∼7% of wild-type levels) of JA in response to wounding (Li et al., 2002b). This residual amount of JA likely accounts for the low level of PI expression in wounded spr2 leaves. Because LeFad7 appears to be a single-copy gene, a putative function for the ER-resident ω3 FAD3 in generating 18:3 that is used for the production of low levels of JA in leaves, as well as the synthesis of 18:3 in roots, can be proposed.
The broad effect of spr2 on the trienoic fatty acid content of lipids desaturated in the chloroplast (i.e., MGDG, DGDG, SL, and PG) indicates that LeFAD7, like the FAD7 enzyme in Arabidopsis (Browse et al., 1986), is not specific for the nature of the lipid head group or the length of the fatty acid. Given this feature of the enzyme, our results do not provide insight into the identity of the particular class of chloroplast lipid that provides the 18:3 substrate for JA synthesis. However, constitutive expression of LeFad7 transcript in leaf tissue supports the idea that wound- and systemin-mediated defense gene expression does not require the de novo synthesis of 18:3 but rather involves new synthesis of JA from existing pools of lipid-esterified 18:3 or an 18:3-derived intermediate in the octadecanoid pathway. These data are in general agreement with previous work showing that wound-induced JA accumulation in tomato leaves is correlated with the release of 18:3 and 18:2, most likely from chloroplast lipids (Conconi et al., 1996). The preponderance of 18:2 in chloroplast lipids from spr2 leaves indicates that oxylipins derived from this polyunsaturate (Blechert et al., 1995) are not active as signals for PI gene expression in response to wounding and systemin.
The spr2 mutation was identified initially on the basis of its ability to suppress the systemin-mediated expression of defensive genes (Howe and Ryan, 1999). Identification of the Spr2 gene product as a chloroplast ω3 desaturase provides strong evidence that systemin signaling involves the synthesis of JA from a pool of 18:3 located in chloroplast lipids rather than extrachloroplast lipids. This interpretation is supported by the observation that spr2 affects the 18:3 content of chloroplast lipids much more than that of extrachloroplast lipids. The precise mechanism by which systemin rapidly induces the massive accumulation of JA (Doares et al., 1995) remains to be determined. However, the involvement of the chloroplast in systemin-mediated defense responses implies the existence of a signaling cascade that couples the activation of the systemin receptor (i.e., SR160) at the plasma membrane to the initiation of JA biosynthesis in the plastid. Wound-inducible mitogen-activated protein kinase and phospholipase activities have been implicated in this aspect of systemin signaling (Stratmann and Ryan, 1997; Narváez-Vásquez et al., 1999). Further characterization of mutants that are impaired in systemin perception (Lee and Howe, 2003) may provide additional insight into the signaling pathway that links systemin action to the activation of the jasmonate pathway.
Molecular cloning of Spr2 provides new insight into the role of JA biosynthesis in wound-induced expression of systemic defense responses. Of relevance to this issue are grafting studies showing that spr2 impairs production of the transmissible wound signal for PI expression (Li et al., 2002b). Identification of Spr2 as a chloroplast ω3 desaturase provides solid support for the hypothesis that jasmonate synthesis is required for the production of the systemic wound signal in wounded leaves. It also is noteworthy that spr2 does not affect the ability of grafted scions to perceive the long-distance signal that emanates from wounded leaves (Li et al., 2002b). Because spr2 impairs JA synthesis by blocking 18:3 production, it can be inferred that de novo synthesis of JA (or of related compounds derived from trienoic fatty acids) is not required in cells that perceive and respond to the systemic signal. This interpretation is in agreement with the work of Strassner et al. (2002), who used gene expression profiling and direct measurements of jasmonates to study wound-induced gene expression in tomato plants. The capacity of spr2 plants to perceive the systemic wound signal, together with the inability of the mutant to respond to exogenous systemin or 35S:Prosys (Howe and Ryan, 1999), strongly suggests that jasmonate rather than systemin is the transmissible wound signal for PI gene expression. Recent studies of the systemin-insensitive mutant spr1 indicate that systemin may function at or near the site of wounding (i.e., in rootstock tissues) to activate JA synthesis to a level that is required for the systemic response (Lee and Howe, 2003). It is possible that systemin and jasmonate are both mobile components of the signaling pathway and that positive feedback interactions between them serve to propagate the long-distance signal (Ryan and Moura, 2002).
Because spr2 impairs the production of trienoic fatty acids, we cannot exclude the possibility that some defense-related phenotypes of the mutant are attributed to a deficiency in 16:3/18:3-derived oxylipins other than JA. For example, the cyclopentenone 12-OPDA has been shown to exhibit biological activity in the absence of prior conversion to JA (Blechert et al., 1995, 1999; Stintzi et al., 2001). Increasing evidence also indicates that C6 aldehydes produced from the hydroperoxide lyase branch of oxylipin metabolism play an important role in plant defense signaling (Bate and Rothstein, 1998; Vancanneyt et al., 2001). Exogenous trans-2-hexenal, a C6 oxylipin produced in tomato by the sequential action of 13-lipoxygenase and hydroperoxide lyase, was shown recently to trigger local and systemic emission of monoterpenes and sesquiterpenes that are implicated in the attraction of natural predators of herbivores (Farag and Paré, 2002). The spr2 mutant should provide a valuable tool with which to assess the function of the complete repertoire of oxylipins that are produced endogenously from trienoic fatty acids. Based on the trienoic fatty acid deficiency in spr2 leaves, it also can be predicted that the mutant is impaired in the synthesis of 16:3-derived oxylipins such as dinor-OPDA (Weber et al., 1997). Nevertheless, the ability of spr2 plants to respond to exogenous JA and its C18 precursors (Table 1) is consistent with previous studies indicating that JA is the active signal for the expression of PI genes in tomato leaves (Farmer and Ryan, 1992; Conconi et al., 1996; Wasternack et al., 1998; Strassner et al., 2002).
Role of Trienoic Fatty Acids in the Growth and Development of Tomato
The spr2 mutant provides a useful resource to investigate the broader role of trienoic fatty acids in plant growth and development. The rationale underlying this approach has been illustrated by the elegant use of comparable Arabidopsis mutants to study the role of polyunsaturates in temperature tolerance and photosynthesis (Browse and Somerville, 1991; Wallis and Browse, 2002). The slightly chlorotic phenotype of young spr2 leaves is similar to that of ω3 desaturase mutants of Arabidopsis (Wallis and Browse, 2002) and clearly suggests a role for LeFAD7 in chloroplast function. With regard to temperature tolerance, it is interesting that tomato suffers from chilling injury when exposed to temperatures of ∼10°C or less and has little capacity to adapt to cold temperatures (Jaglo et al., 2001). In many plants, including Arabidopsis, the capacity to synthesize polyunsaturated fatty acids is an important component of low-temperature fitness (Hugly and Somerville, 1992; Miquel et al., 1993; Routaboul et al., 2000). It has been proposed that the cold-inducible FAD8 gene of Arabidopsis may provide increased ω3 desaturase activity for optimal growth at low temperatures (Gibson et al., 1994). The apparent absence of a FAD8 homolog in tomato, which is of tropical origin, may contribute to the cold-sensitive trait and the inability of the plant to accumulate increased levels of trienoic fatty acids in response to low temperatures (Novitskaya et al., 2000).
The spr2 mutant also may provide insight into the role of JA and related oxylipins in development. It will be interesting to determine whether the various developmental roles fulfilled by jasmonates in Arabidopsis also apply to tomato, a solanaceous species that diverged from the lineage leading to Arabidopsis as many as 150 million years ago (Yang et al., 1999). It is well established that the biosynthesis and perception of JA is essential for male reproductive development in Arabidopsis (McConn and Browse, 1996; Wallis and Browse, 2002). Despite the 18:3 deficiency in spr2 plants, however, this mutant displayed normal fertility, as determined by seed set of self-pollinated spr2 plants and by outcrossing of spr2 pollen to a wild-type female parent. It is possible that the residual amount of JA (18% of wild-type levels) in spr2 flowers is sufficient to promote fertility. However, this interpretation is not consistent with the observation that the PLA1-deficient dad1 mutant of Arabidopsis exhibits ∼22% of wild-type levels of JA in flowers but is male sterile (Ishiguro et al., 2001). It also is possible that male fertility in tomato involves JA derivatives such as JA amino acid conjugates or 12-OPDA that accumulate to high levels in tomato flowers (Hause et al., 2000). Measurement of the endogenous levels of these and other oxylipins in spr2 tissues may help address this issue. Of course, our results also leave open the possibility that JA and its derivatives are not required strictly for male fertility in tomato. This notion is consistent with recent studies showing that the sterility of a JA-insensitive mutant of tomato results mainly from a defect in female, not male, reproductive development (Li et al., 2001).
METHODS
Plant Material and Growth Conditions
Tomato (Lycopersicon esculentum) cv Castlemart was used as the wild-type parent for all experiments involving comparison of wild-type and mutant plants. Tomato seedlings were grown in Jiffy peat pots (Hummert International, Earth City, MO) in a growth chamber maintained under 17 h of light (200 μE·m−2·s−1) at 28°C and 7 h of dark at 18°C. Seeds of Lycopersicon pennellii (LA716) and the introgression lines (Eshed and Zamir, 1994) were obtained from the C.M. Rick Tomato Genetic Resource Center (University of California at Davis).
The original spr2 mutant (line 572A; Howe and Ryan, 1999), which was identified in the 35S:Prosys genetic background, was backcrossed to cv Castlemart to generate an F2 population segregating for both 35S:Prosys and spr2. To identify spr2 homozygotes that lack the transgene, F2 plants that exhibit a wound-nonresponsive phenotype were identified. Genomic DNA from these plants was subjected to PCR screening using a primer set (5′-GCGGATCCGTGGAGATGACAAAGAGACTCC-3′ and 5′-GCGGATCCGAAGTTACTTTTCTAACGGGAGAC-3′) that specifically amplifies a 200-bp fragment corresponding to 35S:Prosys and a 1-kb fragment corresponding to the endogenous Prosys gene. DNA gel blot analysis was used to confirm the absence of the transgene in selected spr2 homozygotes. One such line was used as the female parent for three successive backcrosses to the wild type.
Wound- and Elicitor-Response Assays
The wound response of tomato plants was determined using a radial immunodiffusion assay for the detection of PI-II accumulation in leaf tissue (Ryan, 1967; Trautman et al., 1971). Briefly, 16- to 18-day-old seedlings containing two expanded leaves and a third emerging leaf were wounded with a hemostat across the midrib of all leaflets (typically three) on the lower leaf. Three hours later, the same leaflets were wounded again, proximal to the petiole. Wounded plants were incubated for 24 h under standard growth conditions, after which the wounded leaf (local response) and an upper, unwounded leaf (systemic response) were harvested separately for determination of PI-II protein levels. Feeding of elicitors was performed as described by Lee and Howe (2003). Systemin and oligogalacturonic acid were obtained from C.A. Ryan (Washington State University). 12-Oxo-phytodienoic acid and 13(S)-hydroperoxy linolenic acid were purchased from Cayman Chemical (Ann Arbor, MI). Jasmonic acid (JA) and bestatin were purchased from Sigma. All compounds were diluted from stock solutions into sodium phosphate buffer (15 mM sodium phosphate, pH 6.5) before use.
Map-Based Cloning of Spr2
The general strategy for mapping Spr2 was similar to that used to map the tomato Def1 gene (Li et al., 2002a). A BC1 mapping population was constructed from a cross between spr2/spr2 (L. esculentum) and L. pennellii (LA716), followed by backcrossing of a resulting F1 plant to spr2/spr2 (L. esculentum). Use of a BC1 population ensured that all segregating progeny contained at least one copy of the L. esculentum PI-II gene, the product of which was used to generate the antibody used for the radial immunodiffusion assay of PI-II. The wound-response phenotype of individual plants in the BC1 population was assessed quantitatively by comparison with the wound response of wild-type and spr2 plants grown together with the BC1 plants. Plants showing an unambiguous phenotype were repotted and grown in the greenhouse for several weeks, at which time leaf material was harvested for DNA extraction as described by McCouch et al. (1988).
Using the BC1 population described above, bulked segregant analysis was used in combination with amplified fragment length polymorphism (AFLP) analysis to identify molecular markers linked to Spr2. Equal amounts of DNA from 10 randomly selected wound-responsive (i.e., wild-type) and 10 mutant BC1 plants were pooled to construct a wild-type bulk (B+) and a mutant bulk (B−), respectively. The procedures of Vos et al. (1995) and Thomas et al. (1995) were used to identify AFLPs that distinguished the B+ and wild-type parental (L. pennellii) DNA from the B− and mutant parental (spr2/spr2) DNA. Among 64 primer combinations used to screen the bulks for AFLPs, one combination (E-AAG/M-CAG) generated a product of ∼200 bp that was present in DNA from both the wild-type parent (Spr2pen/spr2esc) and the B+ but absent in DNA from the mutant parent (spr2esc/spr2esc) and the B− (data not shown). The DNA fragment corresponding to this AFLP marker was designated AF1.
The polymorphic band corresponding to AF1 was excised from the polyacrylamide gel and cloned into the pGEM-T Easy vector (Promega) as described previously (Li et al., 2002a). Linkage of an AF1-derived restriction fragment length polymorphism (RFLP) marker to Spr2 was confirmed using the 20 BC1 individuals used to construct the bulks; all wound-responsive wild-type plants were heterozygous for the marker, whereas all mutant plants were homozygous for the L. esculentum RFLP pattern (data not shown). The absence of recombination events in this population of 20 BC1 plants demonstrated tight linkage between AF1 and Spr2. Tomato introgression lines (ILs) harboring defined chromosome segments of L. pennellii in an L. esculentum background (Eshed and Zamir, 1994) were used to map AF1. Probing of survey blots containing L. esculentum and L. pennellii DNA showed that restriction enzyme DraI generated a readily detectable RFLP using a 32P-labeled AF1 probe. This RFLP allowed us to map AF1 to the IL6-1 region located between RFLP markers TG612 and TG352 on chromosome 6, which is defined by IL LA3500.
Analysis of linkage between Spr2 and known RFLP markers on IL6-1 (Tanksley et al., 1992) demonstrated that Spr2 is located between TG590 and TG118. A high-resolution genetic map of the Spr2 region was constructed by scoring 1436 BC1 plants for recombination events within the TG590-Spr2-TG118 interval. Results obtained from these experiments placed Spr2 ∼2.6 map units (38 recombination events) from TG118 and 0.07 map units (one recombination event) from TG590. Three yeast artificial chromosome (YAC) end-derived markers, 258SL, 149AR, and 328N1, which were mapped previously in the TG590-TG118 interval (Ling et al., 2002), also were mapped relative to Spr2 (Figure 2). The mapping data showed that 258SL cosegregated with Spr2 in all 1436 BC1 plants, whereas 149AR and 328N1 were located one and three recombination events, respectively, distant of Spr2.
Markers TG590, 258SL, and 149AR were used individually to screen a tomato BAC library (Budiman et al., 2000; Ling et al., 2002). Restriction enzyme digests were used to “fingerprint” the genomic inserts of 25 BAC clones, prepared using the procedure of Budiman et al. (2000). Thermal asymmetric interlaced (TAIL) PCR (Liu and Whittier, 1995) was used to obtain markers from the ends of eight selected BAC clones. These markers were mapped relative to Spr2 using recombination events in the TG590-TG118 interval. BAC end markers also were used to order the BAC clones relative to each other and to YAC149, resulting in construction of a contig spanning the Spr2 region (Figure 2). YAC149 and markers derived from it were described previously (Ling et al., 1996, 2002). The left end of BAC101L7, named 101L7L, mapped one recombination event away from Spr2. The right end of this BAC, 101L7R1, could not be mapped because of the presence of repetitive sequences. An additional round of TAIL-PCR using sequence information obtained from 101L7R1, however, yielded a second marker, designated 101L7R2, on the right end of BAC101L7. The sequences of the three 101L7R1-specific primers used for this TAIL-PCR procedure were as follows: A, 5′-GTGAGCGAACCAACCAATCTA-3′; B, 5′-GGAAGTCATCATCACAAGAAC-3′; and C, 5′-AGCTAGTCCATGCCAGAACTT-3′. Mapping experiments indicated that 101L7R2 is located one recombination event away from Spr2, on the opposite side of 101L7L. This information strongly indicated that Spr2 is located on BAC101L7.
DNA Sequence Analysis
DNA sequencing of tomato BAC101L7 was performed using a shotgun sequencing service provided by MWG-Biotech (High Point, NC). The assembled sequence consisted of seven contigs with a mean length of 16 kb and totaling 114 kb. The largest contig (43,170 bp) contained the Spr2/LeFad7 gene as determined by Basic Local Alignment Search Tool (BLAST) searches against GenBank and the tomato EST database. The sequence of this contig was deposited in GenBank. Oligonucleotide primers GPB and GPb (see below) were designed to amplify LeFad7 from wild-type and spr2 genomic DNA. The same primer set was used to obtain full-length LeFad7 cDNA clones using reverse transcription PCR of RNA isolated from wild-type and spr2 flowers. PCR-amplified fragments were cloned into the pGEM-T Easy plasmid vector. DNA sequencing of genomic and cDNA clones was performed at the Michigan State University Genomics Technology Support Facility. The sequence of the full-length LeFad7 cDNA obtained by reverse transcription PCR using RNA from wild-type plants was deposited in GenBank.
Agrobacterium tumefaciens–Mediated Transformation
A 2.05-kb genomic fragment containing the Spr2/LeFad7 gene was amplified from wild-type genomic DNA by PCR using the primer pair GPB (5′-ATCCCGGGTTCACCTAACCCAACATCTCATCT-3′) and GPb (5′-AAGAGCTCTGCCTTCCCCCATCTTCTA-3′). This fragment was cloned into the SmaI and SstI sites of the binary vector pBI121 (Clontech, Palo Alto, CA) under the control of the 35S promoter of Cauliflower mosaic virus. The resulting construct was transformed into Agrobacterium tumefaciens strain AGLO (Lazo et al., 1991). Agrobacterium-mediated transformation of spr2 cotyledon explants was performed as described previously (Li and Howe, 2001). Regenerated kanamycin-resistant transformants in which expression of the 35S:LeFad7 transgene was confirmed were potted into standard soil mix and grown in a growth chamber under standard conditions. Approximately 3 weeks after transfer to soil, these plants were used for assays of wound-induced PI-II accumulation and leaf fatty acid composition.
Quantification of JA
An approximately equal number of mature unopened flower buds (>6 mm long) and newly opened flowers were harvested from wild-type and spr2 plants grown side by side in a greenhouse. A quantity of flowers corresponding to 3 g fresh weight of tissue was used for each JA extraction, performed as described by Li et al. (2002b). Levels of JA in tissue extracts were quantified by gas chromatography–mass spectrometry using dihydro-JA as an internal standard (Li et al., 2002b). Extraction and quantification of JA was performed from three independent tissue samples from each plant genotype.
Fatty Acid and Nucleic Acid Analysis
Tomato leaf and root tissue was harvested from 16- to 18-day-old plants and frozen immediately in liquid nitrogen. Lipids were extracted as described by Dörmann et al. (1995). For quantitative analysis, individual lipids were separated by thin layer chromatography, scraped from the plates, and used to prepare fatty acid methyl esters (Xu et al., 2002). The relative proportions of methyl ester fatty acids were quantified by gas chromatography as described by Xu et al. (2002).
Total RNA was isolated from tomato tissues and analyzed by gel blot hybridization as described previously (Howe et al., 1996). Five micrograms of total RNA also was electrophoresed and stained with ethidium bromide to ensure equal loading of samples and intactness of the RNA. RNA gels were blotted to Hybond-N+ filters that were hybridized subsequently to a 32P-labeled cDNA for PI-II (Graham et al., 1985) and LeFad7. As an additional control for RNA loading, blots were hybridized to a cDNA probe (tomato EST clone cLED1D24) for translation initiation factor eIF4A. DNA gel blot analysis was performed as described by Li et al. (2002a).
Insect Feeding Trials
Tobacco hornworm (Manduca sexta) eggs and artificial diet for hornworm larvae were obtained from the Carolina Biological Supply Company (Burlington, NC). Eggs were hatched at 26°C as recommended by the supplier. Hatched larvae were reared on the artificial diet for 3 days before transfer to tomato plants. The average weight of larvae at the beginning of the feeding trial was ∼5 mg.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession number for the contig containing the Spr2/LeFad7 gene is AY248742. The accession number for the full-length LeFad7 cDNA is AY248741. Accession numbers for the sequences shown in Figure 4 are as follows: AtFAD3, AAL36322; AtFAD7, BAA03106; AtFAD8, AAA65621; StFAD7, T07685; OsFAD3, T03923; TaFAD3, T06238; TaFAD7, BAA07785; ZmFAD7, BAA22442; ZmFAD8, BAA22440; GmFAD3, BAB18135; GmFAD7, P48621; NtFAD3, P48623; NtFAD7, BAA11475; PfFAD7, AAB39387; SiFAD7, P48620; LeFAD7, AY248741; and putative FAD3, TC99736.
Acknowledgments
We gratefully acknowledge Mike Pollard and Xiaoming Bao for their helpful assistance with fatty acid analysis, Lei Li for assistance with RNA gel blot experiments, Liyan Liu for assistance with RFLP mapping, and Bonnie McCaig for help with phylogenetic analysis. We thank Steve Tanksley for providing the tomato RFLP markers used in this study and Clarence Ryan for providing systemin and OGA. The tomato BAC library and the tomato EST clone cLED1D24 were obtained from the Clemson University Genomics Institute. This research was supported by National Institutes of Health Grant GM57795 and U.S. Department of Energy Grant DE-FG02-91ER20021 to G.A.H.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012237.
References
- Allen, K.D. (2002). Assaying gene content in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 9568–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bate, N.J., and Rothstein, S.J. (1998). C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 16, 561–569. [DOI] [PubMed] [Google Scholar]
- Baydoun, E.A.H., and Fry, S.C. (1988). The immobility of pectic substances in injured tomato leaves and its bearing on the identity of the wound hormone. Planta 165, 269–276. [DOI] [PubMed] [Google Scholar]
- Bishop, P.D., Makus, D.J., Pearce, G., and Ryan, C.A. (1981). Proteinase inhibitor-inducing factor activity in tomato leaves resides in oligosaccharides enzymically released from cell walls. Proc. Natl. Acad. Sci. USA 78, 3536–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert, S., Bockelmann, C., Füsslein, M., Schrader, T.V., Stelmach, S.B., Niesel, U., and Weiler, E.W. (1999). Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207, 470–479. [Google Scholar]
- Blechert, S., Brodschelm, W., Holder, S., Kammerer, L., Kutchan, T.M., Mueller, M.J., Xia, Z.-Q., and Zenk, M.H. (1995). The octadecanoic pathway: Signal molecules for the regulation of secondary pathways. Proc. Natl. Acad. Sci. USA 92, 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse, J., McCourt, P., and Somerville, C.R. (1986). A mutant of Arabidopsis deficient in C18:3 and C16:3 leaf lipids. Plant Physiol. 81, 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse, J., and Somerville, C. (1991). Glycerolipid synthesis: Biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 467–506. [Google Scholar]
- Budiman, M.A., Mao, L., Wood, T.C., and Wing, R.A. (2000). A deep-coverage tomato BAC library and prospects towards development of an STC framework for genome sequencing. Genome Res. 10, 129–136. [PMC free article] [PubMed] [Google Scholar]
- Conconi, C., Miquel, M., Browse, J.A., and Ryan, C.A. (1996). Intracellular levels of free linolenic and linoleic acids increase in tomato leaves in response to wounding. Plant Physiol. 111, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel, C.P., Yip, L., and Ryan, C.A. (1998). Prosystemin from potato, black nightshade, and bell pepper: Primary structure and biological activity of predicted systemin polypeptides. Plant Mol. Biol. 36, 55–62. [DOI] [PubMed] [Google Scholar]
- Doares, S.H., Syrovets, T., Weiler, E.W., and Ryan, C.A. (1995). Oligogalacturonides and chitosan activate plant defense genes through the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 92, 4095–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann, P., Hoffmann-Benning, S., Balbo, I., and Benning, C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7, 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., and Zamir, D. (1994). A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 79, 175–179. [Google Scholar]
- Farag, M.A., and Paré, P.W. (2002). C6-green leaf volatiles trigger local and systemic VOC emissions in tomato. Phytochemistry 61, 545–554. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E., and Ryan, C.A. (1992). Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G., and Boller, T. (1995). Systemin induces rapid ion fluxes and ethylene biosynthesis in Lycopersicon peruvianum cells. Plant J. 7, 381–389. [Google Scholar]
- Gatehouse, J.A. (2002). Plant resistance towards insect herbivores: A dynamic interaction. New Phytol. 156, 145–169. [DOI] [PubMed] [Google Scholar]
- Gibson, S., Arondel, V., Iba, K., and Somerville, C. (1994). Cloning of a temperature-regulated gene encoding a chloroplast ω-3 desaturase from Arabidopsis thaliana. Plant Physiol. 106, 1615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, J.S., Pearce, G., Merryweather, J., Titani, K., Ericsson, L.H., and Ryan, C.A. (1985). Wound-induced proteinase inhibitors from tomato leaves. II. The cDNA-deduced primary structure of pre-inhibitor II. J. Biol. Chem. 260, 6561–6564. [PubMed] [Google Scholar]
- Green, T.R., and Ryan, C.A. (1972). Wound inducible proteinase inhibitors in plant leaves: A possible defense mechanism against insects. Science 175, 776–777. [DOI] [PubMed] [Google Scholar]
- Hamada, T., Nishiuchi, T., Kodama, H., Nishimura, M., and Iba, K. (1996). cDNA cloning of a wounding-inducible gene encoding a plastid ω3 fatty acid desaturase from tobacco. Plant Cell Physiol. 37, 606–611. [DOI] [PubMed] [Google Scholar]
- Hause, B., Stenzel, I., Miersch, O., Maucher, H., Kramell, R., Ziegler, J., and Wasternack, C. (2000). Tissue-specific oxylipin signature of tomato flowers: Allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 24, 113–126. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., and Parker, R. (1999). Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33, 229–260. [DOI] [PubMed] [Google Scholar]
- Howe, G.A., Lee, G.I., Itoh, A., Li, L., and DeRocher, A. (2000). Cytochrome P450-dependent metabolism of oxylipins in tomato: Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 123, 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., Lightner, J., Browse, J., and Ryan, C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., and Ryan, C.A. (1999). Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153, 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugly, S., and Somerville, C.R. (1992). A role for membrane lipid polyunsaturation in chloroplast biogenesis at low temperature. Plant Physiol. 99, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba, K., Gibson, S., Nishiuchi, T., Fuse, T., Nishimura, M., Arondel, V., Hugly, S., and Somerville, C. (1993). A gene encoding a chloroplast ω-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana. J. Biol. Chem. 268, 24099–24105. [PubMed] [Google Scholar]
- Ishiguro, S., Kawai-Oda, A., Ueda, K., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo, K.R., Kleff, S., Amundsen, K.L., Zhang, X., Haake, V., Zhang, J.Z., Deits, T., and Thomashow, M.F. (2001). Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127, 910–917. [PMC free article] [PubMed] [Google Scholar]
- Jamieson, G.R., and Reid, E.H. (1971). The occurrence of hexadeca-7,10,13-trienoic acid in the leaf lipids of angiosperms. Phytochemistry 10, 1837–1843. [Google Scholar]
- Karban, R., and Baldwin, I.T. (1997). Induced Response to Herbivory. (Chicago, IL: University of Chicago Press).
- Keegstra, K., Olesen, L.J., and Theg, S.M. (1989). Chloroplastic precursors and their transport across the envelope membranes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 471–501. [Google Scholar]
- Kessler, A., and Baldwin, I.T. (2002). Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 53, 299–328. [DOI] [PubMed] [Google Scholar]
- Kubigsteltig, I., Laudert, D., and Weiler, E.W. (1999). Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta 208, 463–471. [DOI] [PubMed] [Google Scholar]
- Kwon, J.H., Lee, Y.M., and An, C.S. (2000). cDNA cloning of chloroplast omega-3 fatty acid desaturase from Capsicum annuum and its expression upon wounding. Mol. Cells 10, 493–497. [DOI] [PubMed] [Google Scholar]
- Lazo, G.R., Stein, P.A., and Ludwig, R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967. [DOI] [PubMed] [Google Scholar]
- LeBrasseur, N.D., MacIntosh, G.C., Perez-Amador, M.A., Saitoh, M., and Green, P.J. (2002). Local and systemic wound-induction of RNase and nuclease activities in Arabidopsis: RNS1 as a marker for a JA-independent systemic signaling pathway. Plant J. 29, 393–403. [DOI] [PubMed] [Google Scholar]
- Lee, G.I., and Howe, G.A. (2003). The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 33, 567–576. [DOI] [PubMed] [Google Scholar]
- León, J., Rojo, E., and Sánchez-Serrano, J.J. (2001). Wound signaling in plants. J. Exp. Bot. 52, 1–9. [DOI] [PubMed] [Google Scholar]
- Li, C., Williams, M.M., Loh, Y.-T., Lee, I.G., and Howe, G.A. (2002. a). Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 130, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and Howe, G.A. (2001). Alternative splicing of prosystemin pre-mRNA produces two isoforms that are active as signals in the wound response pathway. Plant Mol. Biol. 46, 409–419. [DOI] [PubMed] [Google Scholar]
- Li, L., Li, C., and Howe, G.A. (2001). Genetic analysis of wound signaling in tomato: Evidence for a dual role of jasmonic acid in defense and female fertility. Plant Physiol. 127, 1414–1417. [PMC free article] [PubMed] [Google Scholar]
- Li, L., Li, C., Lee, G.I., and Howe, G.A. (2002. b). Distinct roles for jasmonic acid synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 99, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti, R., and Farmer, E.E. (2002). The jasmonate pathway. Science 296, 1649–1650. [DOI] [PubMed] [Google Scholar]
- Ling, H.-Q., Bauer, P., Bereczky, Z., Keller, B., and Ganal, M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. USA 99, 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, H.Q., Pich, A., Scholz, G., and Ganal, M.W. (1996). Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Mol. Gen. Genet. 252, 87–92. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., and Whittier, R.F. (1995). Thermal asymmetric interlaced PCR-automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Malone, M. (1996). Rapid, long-distance signal transmission in higher plants. Adv. Bot. Res. 22, 163–228. [Google Scholar]
- Martín, M., León, J., Dammann, C., Albar, J.-P., Griffiths, G., and Serrano, J.J.S. (1999). Antisense-mediated depletion of potato leaf ω3 fatty acid desaturase lowers linolenic acid content and reduces gene activation in response to wounding. Eur. J. Biochem. 262, 283–290. [DOI] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., Hugly, S., Browse, J., and Somerville, C. (1994). A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast ω3 desaturase. Plant Physiol. 106, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch, S.R., Kochert, G., Yu, Z.H., Wang, Z.Y., Khush, G.S., Coffman, W.R., and Tanksley, S.D. (1988). Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76, 815–829. [DOI] [PubMed] [Google Scholar]
- McGurl, B., Orozco-Cardenas, M., Pearce, G., and Ryan, C.A. (1994). Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase inhibitor synthesis. Proc. Natl. Acad. Sci. USA 91, 9799–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurl, B., Pearce, G., Orozco-Cardenas, M., and Ryan, C.A. (1992). Structure, expression, and antisense inhibition of the systemin precursor gene. Science 255, 1570–1573. [DOI] [PubMed] [Google Scholar]
- Meindl, T., Boller, T., and Felix, G. (1998). The plant wound hormone systemin binds with the N-terminal part to its receptor but needs the C-terminal part to activate it. Plant Cell 10, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel, M., James, D.J., Dooner, H., and Browse, J. (1993). Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc. Natl. Acad. Sci. USA 90, 6208–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyen, C., Hammond-Kosack, K.E., Jones, J., Knight, M.R., and Johannes, E. (1998). Systemin triggers an increase of cytoplasmic calcium in tomato mesophyll cells: Ca2+ mobilization from intra- and extracellular compartments. Plant Cell Environ. 21, 1101–1111. [Google Scholar]
- Moyen, C., and Johannes, E. (1996). Systemin transiently depolarizes the tomato mesophyll cell membrane and antagonizes fusicoccin-induced extracellular acidification of mesophyll tissue. Plant Cell Environ. 19, 464–470. [Google Scholar]
- Narváez-Vásquez, J., Florin-Christensen, J., and Ryan, C.A. (1999). Positional specificity of a phospholipase A activity induced by wounding, systemin, and oligosaccharide elicitors in tomato leaves. Plant Cell 11, 2249–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi, T., Hamada, T., Kodoma, H., and Iba, K. (1997). Wounding changes the spatial expression pattern of the Arabidopsis plastid omega-3 fatty acid desaturase gene (FAD7) through different signal transduction pathways. Plant Cell 9, 1701–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitskaya, G.V., Suvorova, T.A., and Trunova, T.I. (2000). Lipid composition of tomato leaves as related to plant cold tolerance. Russian J. Plant Physiol. 47, 728–733. [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274, 1914–1917. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Truesdale, M.R., Calvert, C.M., Dorans, A., Roberts, M.R., and Bowles, D.J. (1998). A novel tomato gene that rapidly responds to wound- and pathogen-related signals. Plant J. 14, 137–142. [DOI] [PubMed] [Google Scholar]
- Orozco-Cárdenas, M., McGurl, B., and Ryan, C.A. (1993). Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larva. Proc. Natl. Acad. Sci. USA 90, 8273–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cárdenas, M.L., Narváez-Vásquez, J., and Ryan, C.A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13, 179–191. [PMC free article] [PubMed] [Google Scholar]
- Ouariti, O., Boussama, N., Zarrouk, M., Cherif, A., and Ghorbal, M.H. (1997). Cadmium- and copper-induced changes in tomato membrane lipids. Phytochemistry 45, 1343–1350. [DOI] [PubMed] [Google Scholar]
- Pearce, G., Moura, D.S., Stratmann, J., and Ryan, C.A. (2001). Production of multiple plant hormones from a single polyprotein precursor. Nature 411, 817–820. [DOI] [PubMed] [Google Scholar]
- Pearce, G., Strydom, D., Johnson, S., and Ryan, C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–898. [DOI] [PubMed] [Google Scholar]
- Rojo, E., León, J., and Sánchez-Serrano, J.J. (1999). Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 20, 135–142. [DOI] [PubMed] [Google Scholar]
- Roughan, P.G., Holland, R., and Slack, C.R. (1980). The role of chloroplasts and microsomal fractions in polar-lipid synthesis from [1-14C]acetate by cell-free preparations from spinach (Spinacia oleracea) chloroplasts. Biochem. J. 188, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul, J.M., Fischer, S.F., and Browse, J. (2000). Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol. 124, 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C.A. (1967). Quantitative determination of soluble cellular proteins by radial diffusion in agar gels containing antibodies. Anal. Biochem. 19, 434–440. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A. (2000). The systemin signaling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 1477, 112–121. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A., and Moura, D.S. (2002). Systemic wound signaling in plants: A new perception. Proc. Natl. Acad. Sci. USA 99, 6519–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C.A., Pearce, G., Scheer, J., and Moura, D.S. (2002). Polypeptide hormones. Plant Cell 14 (suppl.), S251.–S264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, A., Bergey, D.R., and Ryan, C.A. (1995). Induction of wound response genes in tomato leaves by bestatin, an inhibitor of aminopeptidases. Plant Cell 7, 1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, A., and Oecking, C. (1999). Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 11, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, J.M., and Ryan, C.A. (1999). A 160-kD systemin receptor on the surface of Lycopersicon peruvianum suspension-cultured cells. Plant Cell 11, 1525–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, J.M., and Ryan, C.A. (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99, 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C.R., and Browse, J. (1991). Plant lipids: Metabolism, mutants and membranes. Science 252, 82–87. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner, J., Schaller, F., Frick, U.B., Howe, G.A., Weiler, E.W., Amrhein, N., Macheroux, P., and Schaller, A. (2002). Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J. 32, 585–601. [DOI] [PubMed] [Google Scholar]
- Stratmann, J.W., and Ryan, C.A. (1997). Myelin basic protein kinase activity in tomato leaves is induced systemically by wounding and increases in response to systemin and oligosaccharide elicitors. Proc. Natl. Acad. Sci. USA 94, 11085–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M., Vos, P., Zabeau, M., Jones, D.A., Norcott, K.A., Chadwick, B.P., and Jones, J.D.G. (1995). Identification of amplified restriction fragment polymorphism (AFLP) markers tightly linked to the tomato Cf-9 gene for resistance to Cladosporium fulvum. Plant J. 8, 785–794. [DOI] [PubMed] [Google Scholar]
- Titarenko, E., Rojo, E., León, J., and Sánchez-Serrano, J.J. (1997). Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 11, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautman, R., Cowan, K.M., and Wagner, C.G. (1971). Data processing for radial immunodiffusion. Immunochemistry 8, 901–916. [DOI] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.), S153.–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancanneyt, G., Sanz, C., Farmaki, T., Paneque, M., and Ortego, F. (2001). Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc. Natl. Acad. Sci. USA 98, 8139–8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., and Zabeau, M. (1995). AFLP: A new technique for DNA finger printing. Nucleic Acids Res. 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling, L.L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19, 195–216. [DOI] [PubMed] [Google Scholar]
- Wallis, J.G., and Browse, J. (2002). Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 41, 254–278. [DOI] [PubMed] [Google Scholar]
- Wasternack, C., and Hause, B. (2002). Jasmonates and octadecanoids: Signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 72, 165–221. [DOI] [PubMed] [Google Scholar]
- Wasternack, C., Ortel, B., Miersch, O., Kramell, R., Beale, M., Greulich, F., Feussner, I., Hause, B., Krumm, T., Boland, W., and Parthier, B. (1998). Diversity in octadecanoid-induced gene expression in tomato. J. Plant Physiol. 152, 345–352. [Google Scholar]
- Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. [DOI] [PubMed] [Google Scholar]
- Weber, H., Vick, B.A., and Farmer, E.E. (1997). Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. USA 94, 10473–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker, B.D. (1986). Fatty-acid composition of polar lipids in fruit and leaf chloroplasts of “16:3” and “18:3” plant species. Planta 169, 313–319. [DOI] [PubMed] [Google Scholar]
- Xu, C., Härtel, H., Wada, H., Hagio, M., Yu, B., Eakin, C., and Benning, C. (2002). The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol. 129, 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.W., Lai, K.N., Tai, P.Y., and Li, W.H. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48, 597–604. [DOI] [PubMed] [Google Scholar]