Abstract
The Plant snoRNA database (http://www.scri.sari.ac.uk/plant_snoRNA/) provides information on small nucleolar RNAs from Arabidopsis and eighteen other plant species. Information includes sequences, expression data, methylation and pseudouridylation target modification sites, initial gene organization (polycistronic, single gene and intronic) and the number of gene variants. The Arabidopsis information is divided into box C/D and box H/ACA snoRNAs, and within each of these groups, by target sites in rRNA, snRNA or unknown. Alignments of orthologous genes and gene variants from different plant species are available for many snoRNA genes. Plant snoRNA genes have been given a standard nomenclature, designed wherever possible, to provide a consistent identity with yeast and human orthologues.
BACKGROUND
Small nucleolar RNAs (snoRNAs) are involved in cleavage of pre-cursor ribosomal RNA (pre-rRNA) and determine site-specific modification (2′-O-ribose methylation and pseudouridylation) in pre-rRNAs and snRNAs (1–4). In Archae, snoRNAs are responsible for modification of some tRNAs (5) and in human, brain-specific snoRNAs guide modification of mRNAs (6). SnoRNAs fall into two major groups, defined by the presence of conserved sequences: box C/D and box H/ACA snoRNAs (2–4). The box C/D snoRNAs have two phylogenetically conserved motifs: box C (RUGAUGA) and box D (CUGA), flanked by short inverted repeats at the 5′ and 3′ termini of the snoRNA, respectively. These structural elements are essential for snoRNA stability and nucleolar accumulation. Adjacent to the terminal box D, or to an internal box D′, there is a guide element of 10–21 bases that forms an snoRNA/rRNA duplex selecting the targeted nucleotide. In vivo, all box C/D snoRNAs are found within an snoRNP containing fibrillarin, the methylase and three other conserved core proteins (2–4). The box H/ACA snoRNAs have an ACA motif at the 3′ end of the snoRNA and a Hinge (H) box linking two stem structures. The nucleotide targeted for pseudouridylation is determined by an internal loop in the stem(s) with a pseudouridylation pocket formed by short snoRNA–rRNA duplexes of 4–10 bp flanking the target residue. All H/ACA snoRNA form an snoRNP with four core proteins including NAP57 in vertebrates (Cbf5p in yeast) which is the Ψ synthase (2–4). Thus, in eukaryotes, snoRNAs represent a family of small, stable RNAs with a variety of functions. SnoRNAs have been isolated from small RNA cDNA libraries or by computer algorithms applied to genomic DNA sequence (2–4;7–9). More recent isolation of small non-coding RNAs (ncRNAs) (10) have also identified snoRNAs, and in particular, box H/ACA snoRNAs. This is significant because their relatively short conserved sequences have made computer algorithms difficult to develop.
A major observation over the last five years, based largely on genomic analyses, has been the discovery of an unexpected high number of non-coding small RNAs in different organisms, playing pivotal roles at all levels of gene regulation from DNA replication to gene transcription and mRNA translation (11–13). Among the ncRNAs, snoRNAs represent a major family which have been well characterised both at the functional and expression level. Therefore, the information on snoRNAs is highly valuable in advancing the functional characterization and regulation of ncRNAs which remains largely unknown.
In plants, the availability of the Arabidopsis genome sequence led to three independent computer-assisted searches for box C/D snoRNA genes (14–16). Plant snoRNA genes differ from those of yeast and human by: (1) the presence of 2–4 gene variants in >50% of the genes, (2) polycistronic gene organization is the most common organisation, (3) entirely novel intronic polycistrons have been found in Arabidopsis and more frequently in rice and (4) processing of pre-snoRNA transcripts, both polycistronic and single gene, are splicing-independent (14–19). The Arabidopsis analyses identified 97 different box C/D and two different box H/ACA snoRNA genes with a total of 175 different gene variants (14–16). Of these, 133 were organized in 49 gene clusters distributed across the Arabidopsis genome. These genes were predicted to methylate ca. 120 rRNA target sites, agreeing well with earlier biochemical analyses (20,21), and demonstrating that plants have higher numbers of 2′-o-ribose methylated nucleotides than Archae, yeast and other higher eukaryotes.
The Plant snoRNA Database brings together the information from the three Arabidopsis studies (14–16). Furthermore, it includes information from studies on ncRNAs in Arabidopsis (22,23, JPK and PG, unpublished results). These studies have identified a number of box C/D snoRNAs, helping to confirm the computer-assisted gene predictions. More importantly, they have identified 43 box H/ACA snoRNAs which guide pseudouridylation of rRNA. Prior to this, only two box H/ACA snoRNA genes had been identified (16). In addition, four box C/D and H/ACA snoRNAs which guide modification of snRNAs were discovered, in line with similar discoveries in yeast and human (24–26), and nine snoRNAs with no target site in rRNA or snRNA were also found (16,23). The database provides a unifying nomenclature for all of the above genes, which will be applied to newly discovered genes and genes from different plant species. To date, the Arabidopsis box C/D snoRNAs have been used to identify ∼250 genes from different non-Arabidopsis plant species (JWSB, unpublished results) and these sequences are included as annotated alignments in the database. The database will continue to expand, particularly with the release of the rice genome sequence, where snoRNA searches are already underway (19).
CONTENT OF THE DATABASE
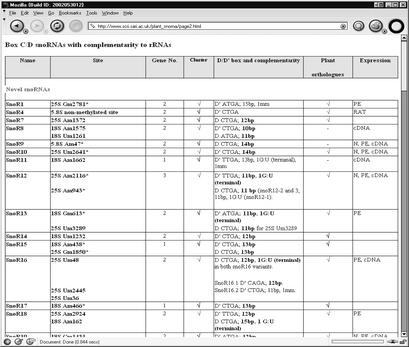
The database currently contains 475 snoRNA gene sequences from Arabidopsis and other plant species. The entry point to the database is through a number of topics on the Home Page. The first, ‘snoRNA genes in Arabidopsis’, presents summary Tables of Arabidopsis snoRNAs arranged in two groups: box C/D and box H/ACA snoRNAs, and by the target RNA for modification: rRNA, snRNA and unknown (Fig. 1 and Supplementary Material). For each snoRNA, information is provided on: (1) predicted/mapped modification sites, (2) the number of gene variants, (3) whether the gene is found as a single gene or in a polycistronic cluster, (4) whether the gene or cluster is intronic, (5) supporting expression data and (6) whether orthologues have been found in other plant species.
Figure 1.
Screen shot of Arabidopsis snoRNA gene information table ‘Box C/D snoRNAs with complementarity to rRNAs‘.
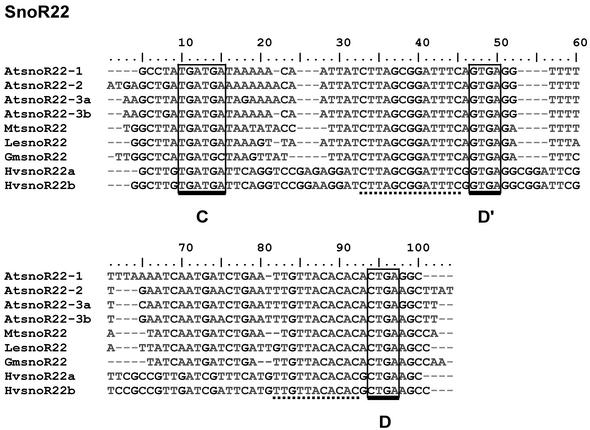
The second topic, ‘snoRNA genes in other plant species’, covers the snoRNA genes from the 18 plant species besides Arabidopsis, where snoRNA sequences have been found. These species include both dicotyledonous and monocotyledonous plants, a gymnosperm species, two moss species and an algal species. The number of snoRNA sequences so far identified for each species roughly reflects the total number of ESTs available, except for rice, where many sequences are of genomic origin. Under each species name, a list of snoRNA sequences identified and the number of variants found are provided, with links to annotated sequence alignments (e.g. Fig. 2). The alignments contain varying numbers of sequences, with some having up to 18 (e.g. snoR28). The alignments show conservation of boxes C, D′ and D, and complementary regions, while demonstrating substantial sequence variation among species and variants.
Figure 2.
Typical sequence alignment. SnoR22 orthologues from Arabidopsis thaliana (At), Medicago truncatula (Mt), Lycopersicon esculentum (Le), Glycine max (Gm) and Hordeum vulgare (Hv). Boxes C, D′ and D are boxed and regions of complementarity to rRNA are underlined with dashed lines.
The third topic highlights the conservation of modification sites among plant, yeast and human rRNAs. The Arabidopsis modification sites are organized by their position along rRNA. The information includes the position of corresponding modifications in yeast and human rRNA and the identity of the cognate snoRNAs (see Supplementary Material). Finally, a series of links to other snoRNA and ncRNA web pages, key references and a description of the nomenclature system are given.
NOMENCLATURE
Due to the number of Arabidopsis snoRNA genes with similarity to vertebrate and yeast genes, the Arabidopsis snoRNAs were named to identify relationships using the following criteria. When the complementary region(s) of an identified gene corresponded to that of a vertebrate or vertebrate/yeast snoRNA, the plant snoRNA was given the vertebrate name (e.g. U14, U34 etc.). If the complementary sequence corresponded only to that of a yeast snoRNA, then it is given the yeast number followed by ‘Y’ (e.g. snoR77Y). Novel plant genes were named following the nomenclature for the maize snoRNA genes, snoR1, snoR2 and snoR3 etc. (17). For a small number of genes, when a plant gene contained two guide sequences found separated in different single genes in human or yeast, the gene was considered novel. Similarly, if a plant gene contained a single guide sequence where the corresponding vertebrate gene contained two, the plant gene was given a plant snoRNA name (e.g. snoR21). When a plant gene contained two guide sequences, one of which corresponded to a vertebrate/yeast guide sequence, and the other was unique to plants, the plant gene was given the vertebrate/yeast name (e.g. U36a). When more than one gene variant was found at the same locus, the genes were given the suffix a, b, c etc. and when at different loci, they were given the suffix .1, .2 etc. or combinations thereof.
DATABASE ACCESS AND MANIPULATION
The database provides a structured interface to all of the published snoRNA gene sequences though a set of linked HTML tables and sequence files. Sequence information for individual genes is available in FASTA format and the entire set of sequences can be downloaded as a multiple sequence FASTA file.
LINKS
Links are provided to relevant snoRNA and ncRNA databases: yeast snoRNA databases (7,27), archael snoRNAs (9), non-coding RNAs (13) and non-coding RNAs in plants (22).
FUTURE OF THE DATABASE
In the near future, once analysis of particularly box H/ACA snoRNAs is complete, we will provide links from the Arabidopsis snoRNA tables to: (i) schematic diagrams of gene clusters from different plant species showing conservation and non-conservation of gene order, and intronic or non-intronic location of related clusters in different species, (ii) the position on Arabidopsis chromosomes of genes and gene clusters and (iii) schematic diagrams of complementary sequences base-paired with target RNAs for both box C/D and H/ACA snoRNAs. We are also developing a MySQL implementation of the database to allow more complex queries enabled through a Perl.DBI and Perl.CGI WWW interface and the entire sequence set will be made available as a Blastable database.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie J.P., Cavaillé,J. and Qu,L.-H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNA guides revisited. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, pp. 191–203.
- 3.Kiss T. (2002) Small nucleolar RNAs: An abundant group of non-coding RNAs with diverse cellular functions. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W. and Pogačić,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- 5.Clouet d'Orval B., Bortolin,M.-L., Gaspin,C. and Bachellerie,J.-P. (2001) Box C/D RNA guides for the ribose methylation of Archaeal tRNAs. The tRNATrp intron↔guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res., 29, 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavaillé J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.-P., Brosius,J. and Hüttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organisation. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 8.Gaspin C., Cavaillé,J., Erauso,G. and Bachellerie,J.-P. (2000) Archael homologues of eukaryotic methylation guide small nucleolar RNAs: lessons from Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 9.Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 10.Hüttenhofer A., Kiefmann,M., Meier-Ewart,S., O'Brien,J., Lehrach,H., Bachellerie,J.-P. and Brosius,J. (2001) Rnomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eddy S.R. (2001) Non-coding RNA genes and the modern RNA world. Nature Rev. Genet., 2, 919–929. [DOI] [PubMed] [Google Scholar]
- 12.Pasquinelli A.E. (2002) MicroRNAs: deviants no longer. Trends Genet., 18, 171–173. [DOI] [PubMed] [Google Scholar]
- 13.Erdmann V.A., Barciszewska,M.Z., Szymanski,M., Hochberg,A., de Groot,N. and Barciszewski,J. (2001) The non-coding RNAs as riboregulators. Nucleic Acids Res., 29, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu L.H., Meng,Q., Zhou,L. and Chen,Y.-Q. (2001) Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res., 29, 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barneche F., Gaspin,C., Guyot,R. and Echeverria,M. (2001) Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-o-methylation sites. J. Mol. Biol., 311, 57–73. [DOI] [PubMed] [Google Scholar]
- 16.Brown J.W.S., Clark,G.P., Simpson,C.G., Leader,D.J. and Lowe,T.M. (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA, 7, 5718–5732. [PMC free article] [PubMed] [Google Scholar]
- 17.Leader D.J., Clark,G.P., Watters,J.A., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1997) Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J., 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leader D.J., Clark,G.P., Watters,J.A., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1999) Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant Mol. Biol., 39, 1091–1100. [DOI] [PubMed] [Google Scholar]
- 19.Liang D., Zhou,H., Chen,Y.-P., Chen,X., Chen,C.-L. and Qu,L.-H. (2002) A novel gene organisation: intronic snoRNA gene clusters from Oryza sativa. Nucleic Acids Res., 30, 3262–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau R.Y., Kennedy,T.D. and Lane,B.G. (1974) Wheat embryo ribonucleates: III. Modified nucleotide constituents in each of the 5.8S, 18S and 26S ribonucleates. Can. J. Biochem., 52, 1110–1123. [DOI] [PubMed] [Google Scholar]
- 21.Cecchini J.P. and Miassod,R. (1979) Studies on the methylation of cytoplasmic ribosomal RNA from cultured higher plant cells. Eur. J. Biochem., 98, 203–214. [DOI] [PubMed] [Google Scholar]
- 22.MacIntosh G.C., Wilkerson,C. and Green,P.J. (2001) Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol., 127, 765–776. [PMC free article] [PubMed] [Google Scholar]
- 23.Marker C., Zemann,A., Terhörst,T., Kiefmann,M., Kastenmayer,J.P., Green,P.J., Bachellerie,J.-P., Brosius,J. and Hüttenhofer,A. (2002). Curr. Biol. (submitted). [DOI] [PubMed] [Google Scholar]
- 24.Jády B.E. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-o-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darzacq X., Jády,B.E., Verheggen,C., Kiss,A.M., Bertrand,E. and Kiss,T. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-o-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H., Chen,Y.-Q., Du,Y.-P. and Qu,L.-H. (2002) Schizosaccharomyces pombe mgU6-47 snoRNA is required for the methylation of U6 snRNA at 41. Nucleic Acids Res., 30, 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samarsky D.A. and Fournier,M.J. (1999) A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res., 27, 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.